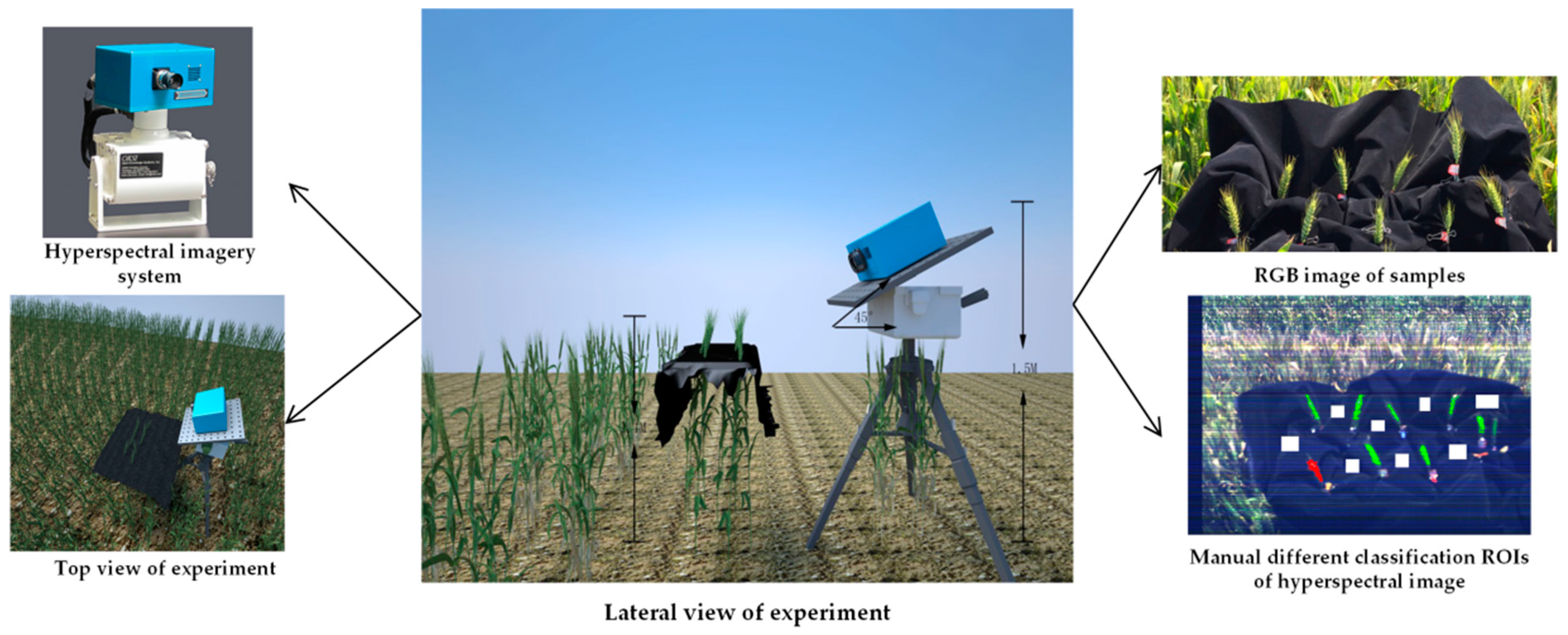
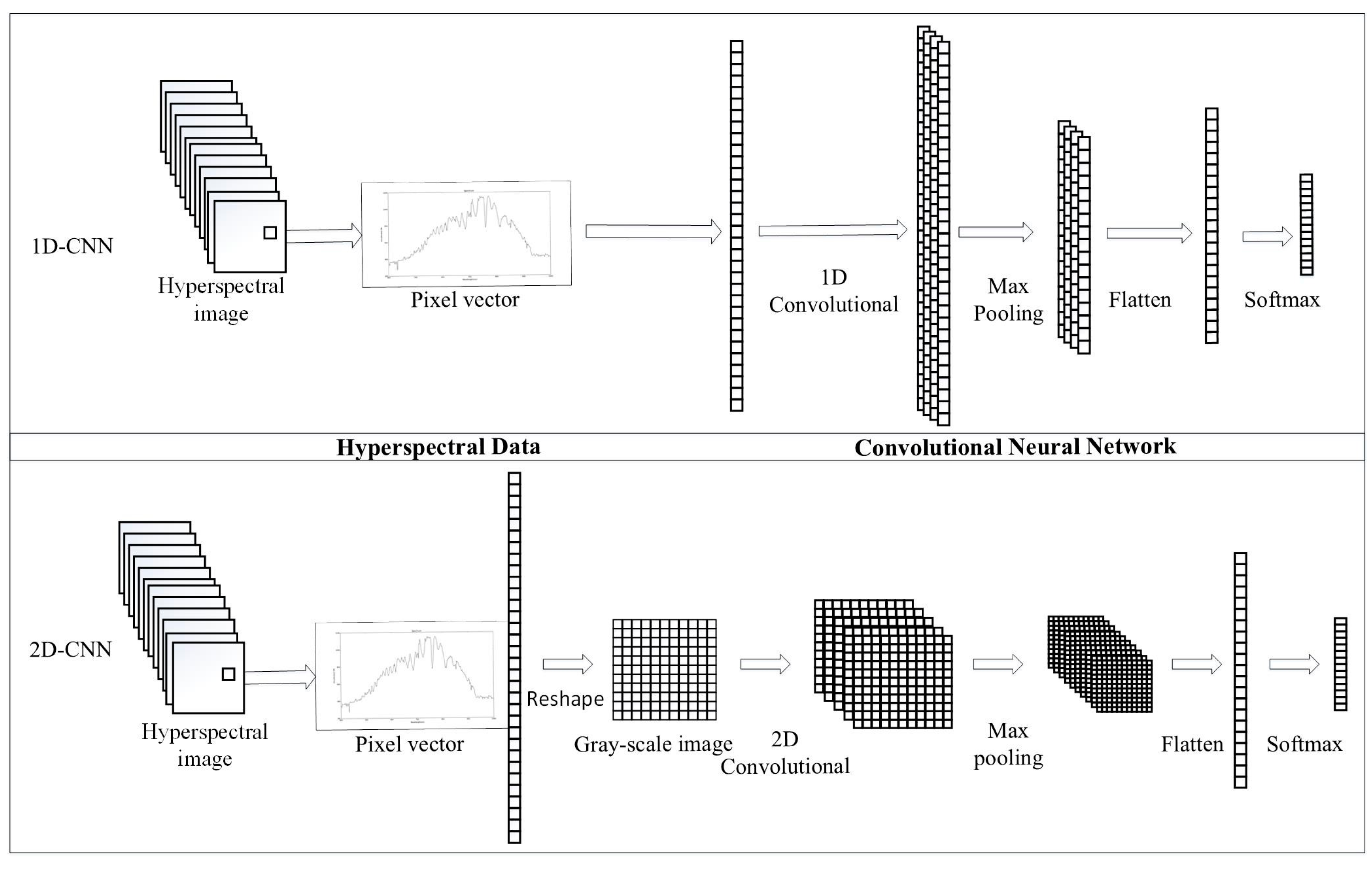
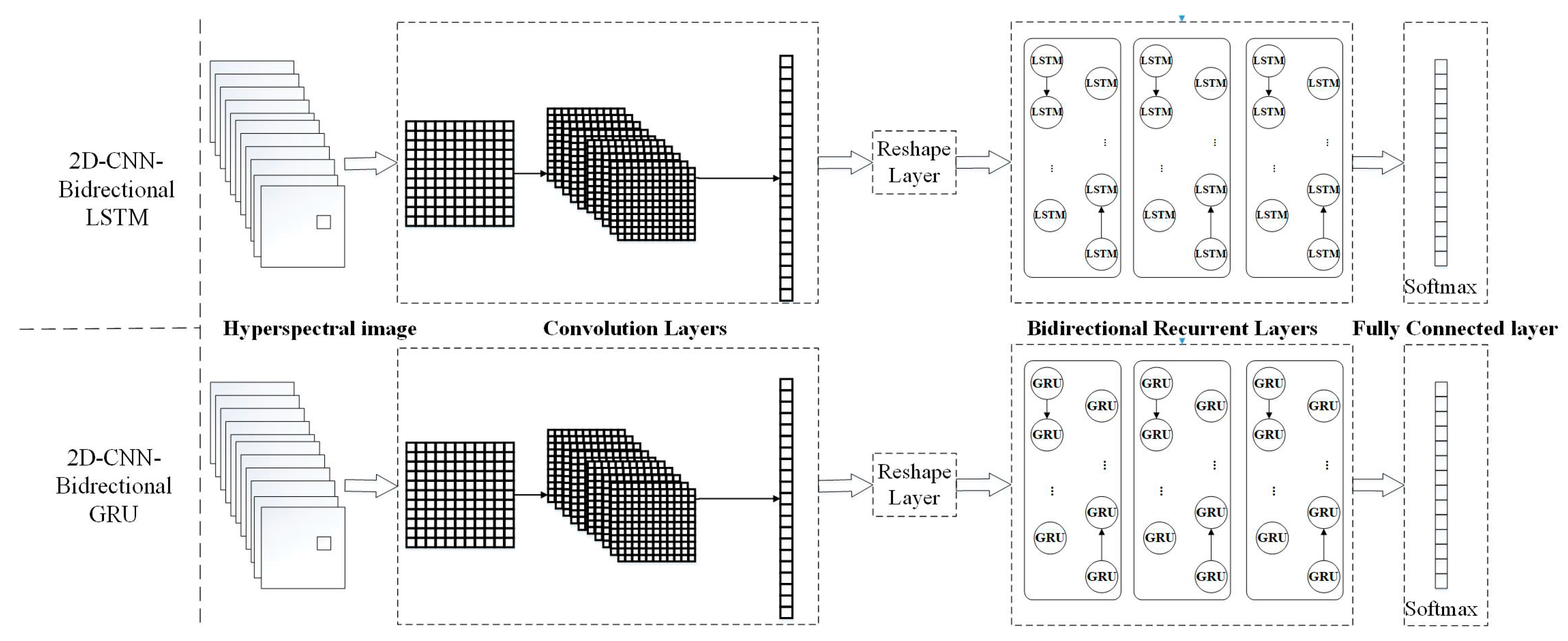
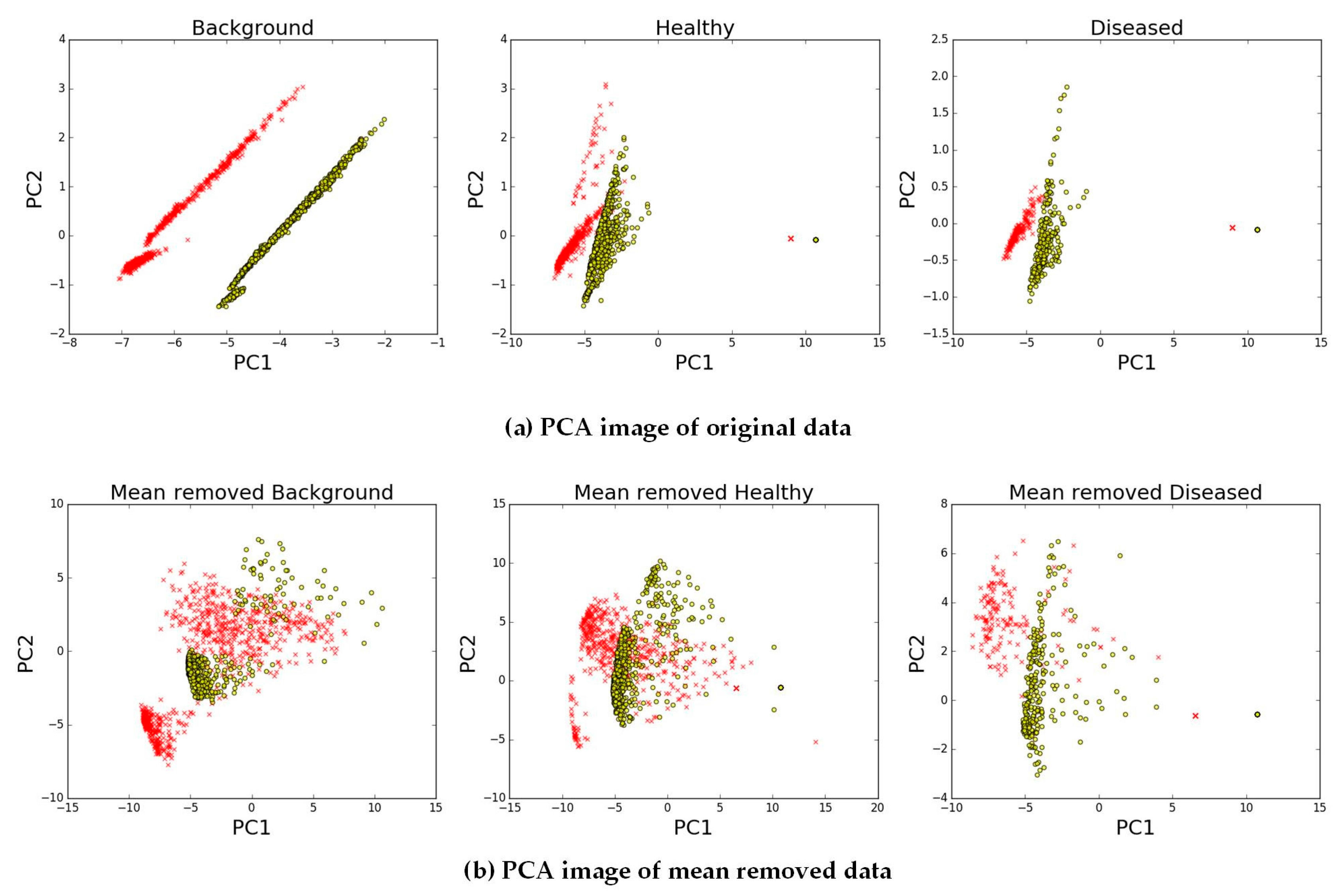
1. Introduction
Fusarium head blight disease is not evenly distributed across a wheat field, but occurs in patches with large areas of the field free of disease in the early stages of infestation, which will eventually result in significant losses. Fusarium head blight is an intrinsic infection by fungal organisms, after infestation, this disease can harm the normal physiological function of wheat, and cause changes in the form and internal physiological structure [
1,
2,
3]. After the wheat is infected, several fungal toxins will be produced. Deoxynivalenol (DON) is the most serious of these toxins. DON can lead to poisoning in people or animals and persists in the food chain for a long time [
4,
5]. If the disease in the field can be detected earlier and quicker, wheat containing the toxins can be isolated, which can reduce the loss caused by disease. Scabs appear on the wheat head due to chlorophyll content degradation and water losses after the wheat is infected [
6,
7]. Digital image and spectral analysis can be applied to detect the disease. The spectral reflectance of diseased plant tissue can be investigated as a function of the change in plant chlorophyll content, water, morphology, and structure during the development of the disease [
8,
9].
Hyperspectral imagery technology is a non-invasive method and consumes large amounts of memory and tranmission bandwidth, so that the very large data cubes can extract features and spectral identification, which represents a small subset. Hyperspectral imagery can include the spectral, spatial, textural, and contextual features of food and agricultural products [
10,
11,
12]. In addition, the application of hyperspectral imagery detection for wheat disease is relatively recent and offers interesting and potentially discerning opportunities [
13]. The occurrence
Fusarium head blight disease is a short time, and lasts approximately half of a month. Since the field environment has a strong influence on hyperspectral imagery, previous studies have mainly focused on laboratory conditions [
14]. Bauriegel detected the wavelength of the
Fusarium infection in wheat using hyperspectral imaging via Principal Component Analysis (PCA) under laboratory conditions. The degree of disease was correctly classified (87%) in the laboratory by utilizing the spectral angle mapper. Meanwhile, the medium milk stage was found to be the best time to detect the disease using spectral ranges of 665–675 nm and 550–560 nm [
14]. Karl-Heinz Dammer used the normalized differential vegetation index of the image pixels and the threshold segmentation to discriminate between infected and non-infected plant tissue. The grey threshold image of the disease ear shows a linear correlation between multispectral images and visually estimated disease levels [
15]. However, multispectral and RGB imagery are used to detect only infected ears with typical symptoms via sophisticated analysis of the image under uniform illumination conditions. Spectroscopy and imaging platforms for tractors, UAVs, aircraft and satellites are current innovative technologies for mapping disease in wild fields [
16].
The hyperspectral imagery classification model must be robust and generalizable to improve the disease detection accuracy in wild fields. The traditional commonly used algorithm is a Support Vector Machine (SVM), which has achieved remarkable results in statistical process control applications [
17]. Qiao uses the method of SVM to classify fungi-contaminated peanuts in hyperspectral image pixels, and the classification accuracy exceeded 90% [
18]. When using SVM to classify hyperspectral images, the spectral and spatial features should be extracted with reduced dimensionality. The overall accuracy increased from 83% without any feature reduction to 87% with feature reduction based on several principal components and morphological profiles [
19]. Feature reduction can be seen as a transformation from high dimensions to low dimensions to overcome the curse of dimensionality, which is a common phenomenon when conducting analyses in high-dimensional space. For a given number of available training samples, the curse of dimensionality decreases the classification accuracy as the dimensions of the input feature vectors increase [
20]. Therefore, the analysis of hyperspectral images that are large data cubes faces the major challenge of addressing redundant information.
The method of deep learning originates from an artificial neural network, the Multiple Layer Perceptron (MLP). Among the several applications of deep neural networks, Deep Convolutional Neural Nets (DCNN) have brought about breakthroughs in processing images, face detection, audio, and so on [
21,
22,
23]. In 2015, DCNN is firstly introduced into hyperspectral images classification And Wei Hu also proposed convolutional layers and max pooling layer to discriminate each spectral signature [
24,
25]. DCNN shows excellent performance for hyperspectral image classification [
26], including cancer classification [
27], and land-cover classification [
28]. A Deep Recurrent Neural Network (DRNN), another typical deep neural network, is a sequence network that contains the hidden layers and memory cells to remember the sequence state [
29,
30]. In 2017, Li was the first use the DRNN to treat the pixels of hyperspectral image as the sequence data. They developed a novel function, named PRetanh, for hyperspectral imagery [
31]. In addition, the deep convolutional recurrent neural network for hyperspectral image was firstly for hyperspectral images that were used by Hao Wu in 2017. They constructed a few convolutional layers and followed recurrent layers to extract the contextual spectra information using the features of the convolutional layers. By combining convolutional and recurrent layers, the DCRNN model achieves results that are superior to those of other methods [
32].
Up to now, research on wheat
Fusarium infection was primarily focused on approaches to classify the fungal disease of wheat kernels by grey threshold segmentation, or extract head blight symptoms based on PCA [
33,
34,
35]. In the early development stage of
Fusarium head blight disease, infected and healthy grains are easier to separate, but the disease symptoms are difficult to diagnose. [
14,
15]. Moreover, in a wild field, the complicated environmental conditions and irregular disease patterns limit the classification accuracy of hyperspectral imagery experiments.
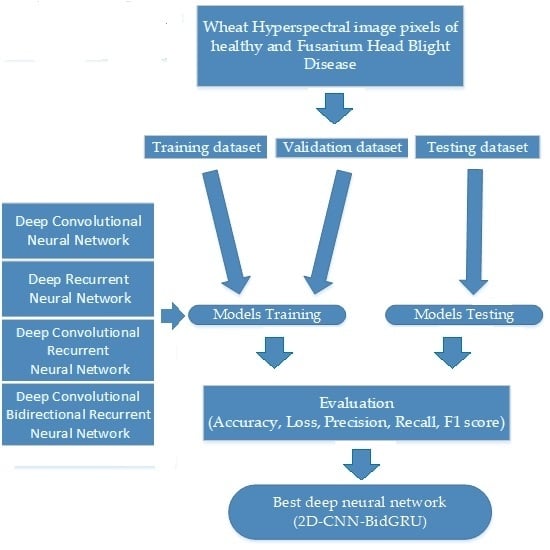
Therefore, the current study’s main aim is to develop a robust and generalizable classification model for hyperspectral image pixels to detect early-stage Fusarium head blight disease in a wild field. Specifically, the main work are listed, as follows.
Design and complete a hyperspectral image classification experiment for healthy head and Fusarium head blight disease in the wild field. The hyperspectral images are divided by pixels of different classes into a training dataset, a validation dataset, and a testing dataset to training the model.
Compare and improve the different deep neural networks for hyperspectral image classification. These neural networks include DCNN with two input data structures, DRNN with Long Short Term (LSTM), and the Gated Recurrent Unit (GRU), and an improved hybrid CRNN.
Take advantage of these assessment methods to determine the best model for classifying hyperspectral image pixels. Different SVM and deep neural network models are assessed and analysed on training dataset, validation dataset, and the testing dataset.
The remainder of this paper is organized as follows. An introduction to the experiment for obtaining hyperspectral imagery of
Fusarium head blight disease is briefly given in
Section 2. The details of the classification algorithms, including modeling and evaluating, are described in
Section 3. The experimental results and a comparison with different approaches are provided in
Section 4.
Section 5 discusses the effectiveness of the early diagnosis of
Fusarium head blight disease by different hyperspectral classification models; finally,
Section 6 concludes this paper.
5. Discussion
The main purpose of this work was to study classification algorithms of hyperspectral image pixels and analyse the performance of deep models for diagnosing
Fusarium head blight disease in wheat. Previous studies attempted to classify the disease symptoms based on spatial and spectral features via machine learning algorithms [
14,
15]; for example, PCA [
33,
34,
35], random forest, and SVM [
17,
18,
19]. Recently, the study of deep algorithms for hyperspectral imagery has becomes increasingly intensive, such as DCNN [
24,
25,
26,
27], DRNN [
31], and hybrid neural networks [
32]. The complicated hybrid structure [
50,
51] of a bidirectional recurrent layer [
53,
54] will help to improve the generalization of the classification model for prediction in testing datasets. This study, therefore, indicates that the benefits that are gained from deep characteristics of the disease spectra and identifies the best hybrid model for the diagnosis of
Fusarium head blight disease in the field.
5.1. Extracting and Representing Deep Characteristics for Disease Symptoms
For high-resolution spectral instrumentation, the number of bands obtained by hyperspectral images is greater than that by multispectral images [
12]. Therefore, the valid features of hyperspectral image pixels for disease detection in wild fields are difficult to determine. Many classifiers of hyperspectral images are based on specific wavelet features or vegetation indices that are related to pathological characteristics [
7,
10]. Deep feature extraction for disease detection by deep neural networks in the spectral domain is presented to acquire deeper information from hyperspectral images [
26].
Among the abundance of deep learning methods, DCNN is commonly used to classify two-dimensional data-type images in visual tasks [
42]. Our work on training and testing 1D-CNN and 2D-CNN shows that two-dimensional data can capture the intrinsic features of hyperspectral images better than one-dimensional data. Another significant branch of the deep learning family is the DRNN, which is designed to address sequential data [
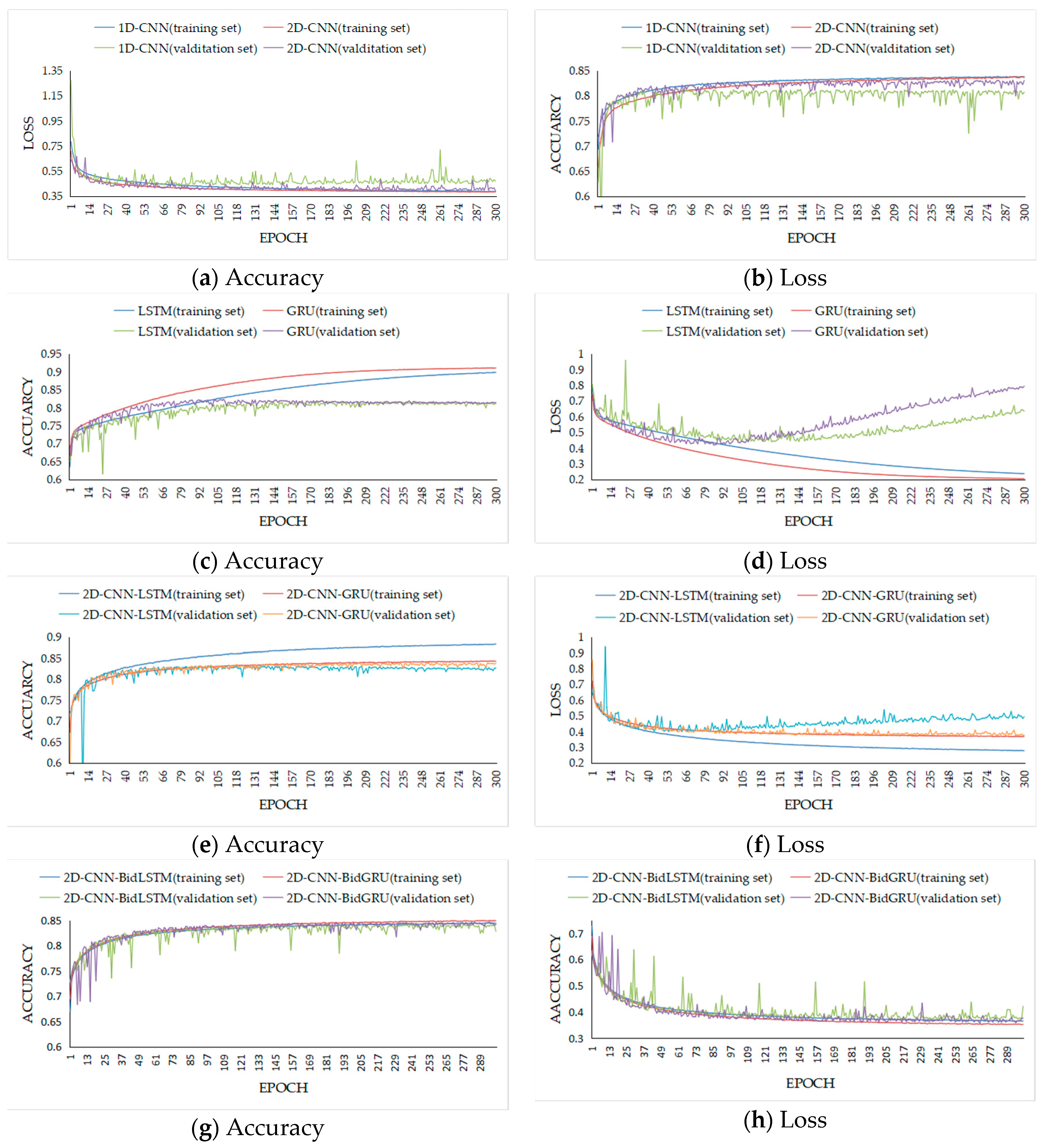
31]. By contrast to the CNN, the RNN maintains all of the spectral information in a recurrent procedure with a sequence-based data structure to characterize spectral correlation and band-to-band variability. As a result, to integrate the advantages of these deep neural networks, novel models that combine recurrent neural networks are proposed. However, in this work, the accuracy of second hybrid neural network with bidirectional recurrent layer for validation data is the better than first one with recurrent layer. The bidirectional RNNs that can incorporate contextual information from both past and future inputs [
54]. The assessment results of all the deep models on the validation data show that 2D-CNN-BidGRU, with an accuracy of 0.846, is the most effective method for disease feature extraction from hyperspectral image pixels.
5.2. Assessing the Performance of Different Models for Hyperspectral Image Pixel Classification
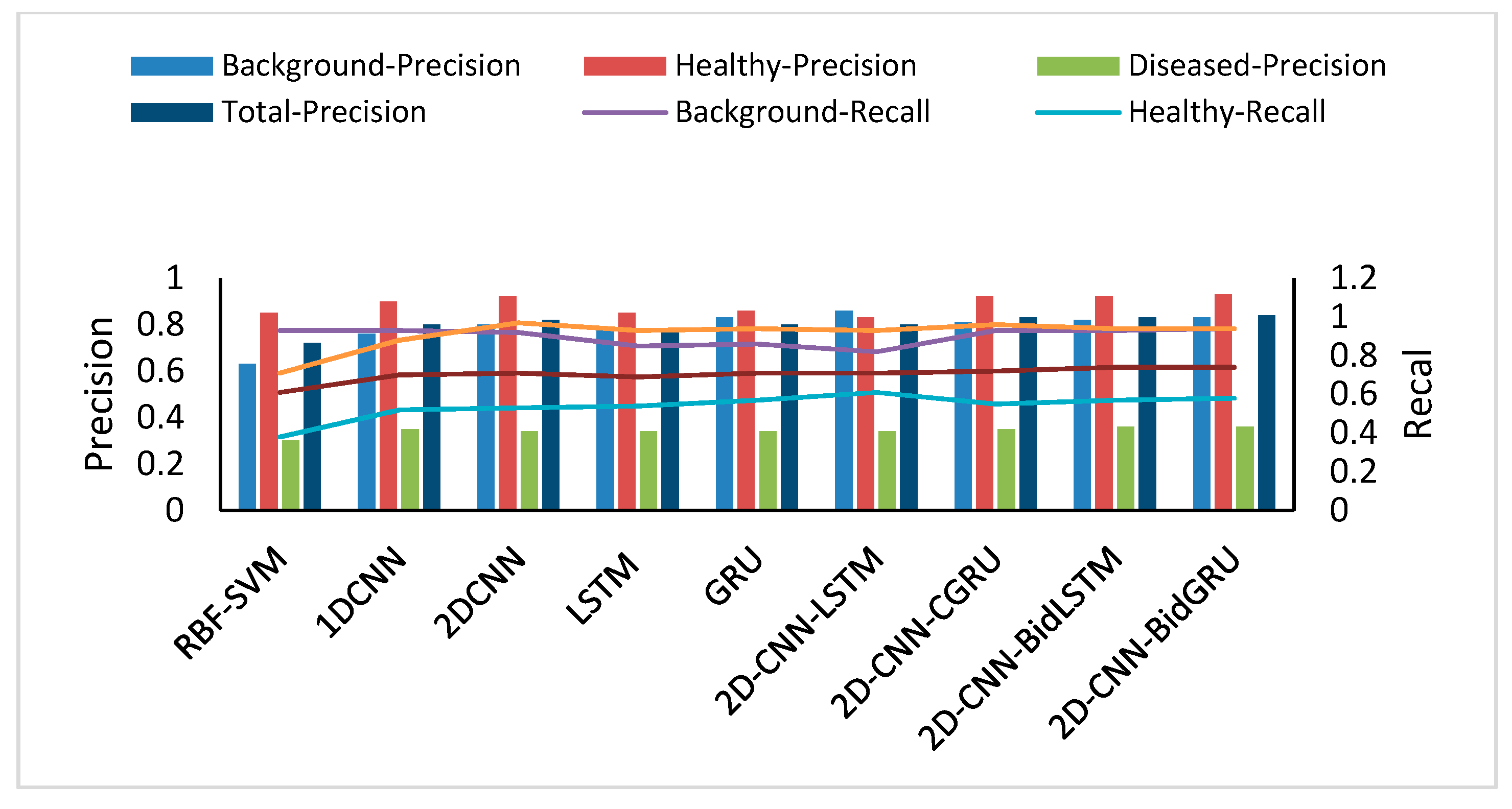
Robustness and generalizability are very important for large-scale disease detection in hyperspectral images. Therefore, the superior performance of a model must be assessed for a large number of testing datasets. The assessment metrics of these classification models include not accuracy, but the confusion matrix, F1 score, precision and recall [
57]. The precision, recall, and F1 score for the testing dataset indicate that both the background and diseased classes are efficient for hyperspectral image pixel classification, but part of the healthy dataset is misclassified as diseased by all of the models.
The assessment results show that the deep models are better than SVM for the performance. With these deep models, despite applying regularization [
71] and dropout [
72] for the deep neural networks, the LSTM model, which has the worst performance, has an over-fitting problem. When a deep model learns a concept when there is noise in the training data, the problem of over-fitting will occur to such an extent that it negatively impacts the performance of the model on the testing data [
73]. The performance of the hybrid neural networks are better than that of other deep neural networks. For comparing the assessment result of the two novel hybrid models on training and testing dataset, the first hybrid neural network with LSTM has the same over-fitting problem, but the second hybrid model with bidirectional LSTM and GRU can restrain the problem. The F1 score (0.75) and accuracy (0.743) on the testing dataset provide compelling evidence that the hybrid structure of 2D-CNN-BidGRU is the best for improving the model with respect to robustness and generalizability.
5.3. Next Steps
Notably, the application of deep neural networks to hyperspectral image pixel classification for Fusarium head blight disease is a new idea and has a great potential for disease diagnosis by remote sensing. Despite some insufficiencies, these results show that a deep neural network with a convolutional layer and bidirectional recurrent layer can classify the hyperspectral image pixels to diagnose the Fusarium head blight disease and improve the generalization and robustness of the classification model. In a wild wheat field, many types of objects have strong impacts on the generalization of the classification model. Therefore, future studies of deep models should apply more in-deep neural networks and customized loss functions to optimize these algorithms for a large number of testing datasets and more types of objects. Future study of application will focus on airborne hyperspectral remote sensing, which could be used to develop large-scale Fusarium head blight disease monitoring and mapping.