Leaf and Canopy Level Detection of Fusarium Virguliforme (Sudden Death Syndrome) in Soybean
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area and Experimental Design
2.2. Data Collection and Processing
2.2.1. Canopy Spectral Data Collection and Processing
2.2.2. Leaf Spectral Data Collection and Processing
2.2.3. Root Samples for Quantitative Real-Time Polymerase Chain Reaction (qPCR)
2.3. Data Analyses
2.3.1. Normalized Difference Spectral Indices
2.3.2. Partial Least Squares
3. Results and Discussion
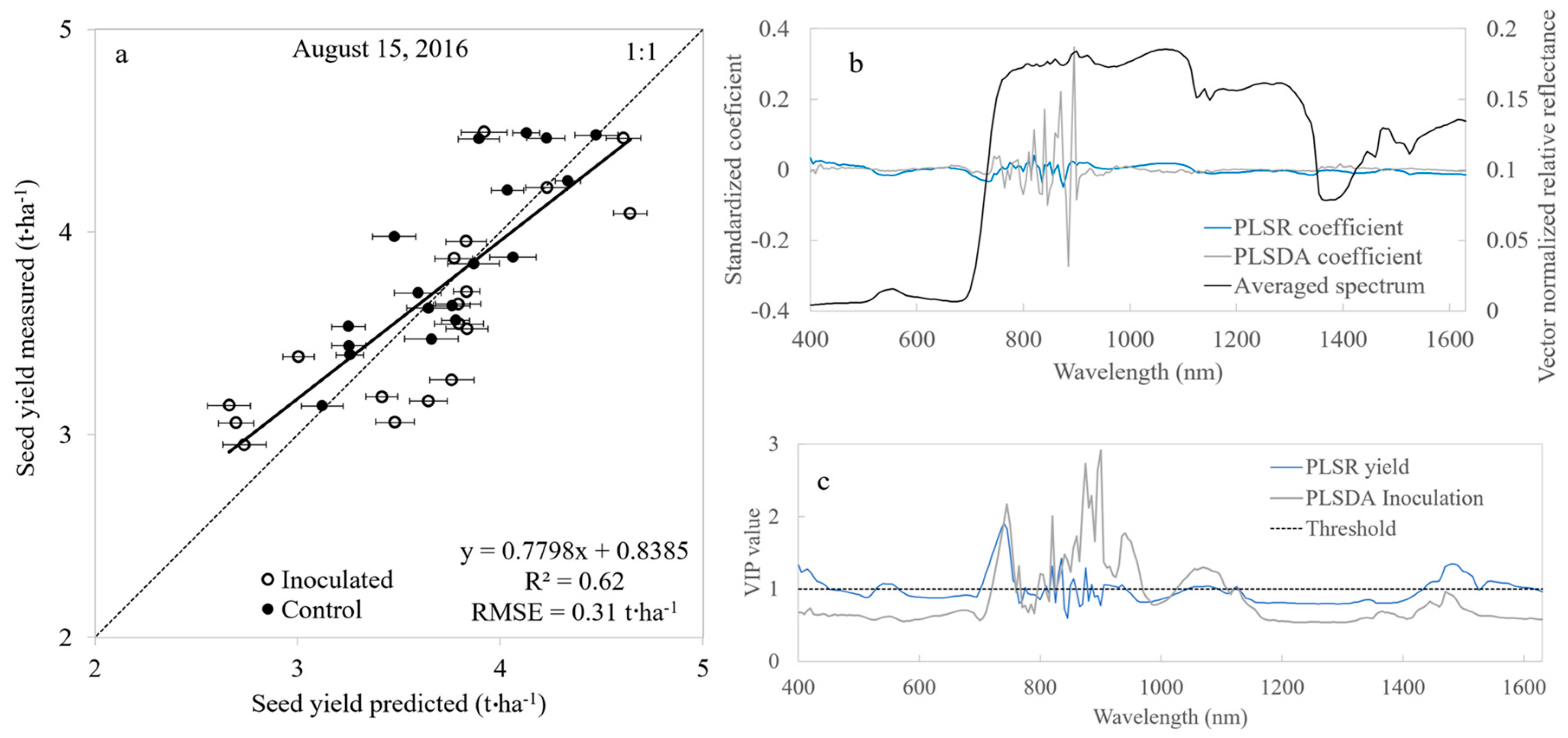
3.1. Spectral Identification of Inoculation—Canopy Level
3.2. Leaf Level Spectral Analyses
3.2.1. Spectral Identification of Inoculation—Leaf Level
3.2.2. Spectral Assessment of Fv Abundance in Root by Leaf Measurement
3.3. Piccolo Performance
4. Conclusions
- Fv inoculation that accrues in the roots can be spectrally detected in the soybean foliage at the canopy and leaf scales as demonstrated by distinguishing between inoculated and control plots and plants, respectively.
- Early reproductive stage is the recommended timing for canopy level measurements to distinguish between inoculated and control plots.
- Late vegetative and early reproductive stages are the recommended timings to distinguish between inoculated and control plants at the leaf level.
- Fv abundance in soybean roots can be spectrally assessed by leaf hyperspectral data.
- The dual field-of-view system produced canopy spectral data resulting in our ability to spectrally distinguish between Fv inoculation treatments and assess seed yield, thus, showing feasibility of operation for precision agriculture research and potential commercial applications.
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Mulla, D.J. Twenty five years of remote sensing in precision agriculture: Key advances and remaining knowledge gaps. Biosyst. Eng. 2013, 114, 358–371. [Google Scholar] [CrossRef]
- Weber, V.S.; Araus, J.L.; Cairns, J.E.; Sanchez, C.; Melchinger, A.E.; Orsini, E. Prediction of grain yield using reflectance spectra of canopy and leaves in maize plants grown under different water regimes. Field Crop. Res. 2012, 128, 82–90. [Google Scholar] [CrossRef]
- Bonfil, D.J.; Karnieli, A.; Raz, M.; Mufradi, I.; Asido, S.; Egozi, H.; Hoffman, A.; Schmilovitch, Z. Rapid assessing of water and nitrogen status in wheat flag leaves. J. Food Agric. Environ. 2005, 3, 148–153. [Google Scholar]
- Christensen, L.K.; Bennedsen, B.S.; Jorgensen, R.N.; Nielsen, H. Modelling nitrogen and phosphorus content at early growth stages in spring barley using hyperspectral line scanning. Biosyst. Eng. 2004, 88, 19–24. [Google Scholar] [CrossRef]
- Herrmann, I.; Karnieli, A.; Bonfil, D.J.; Cohen, Y.; Alchanatis, V. SWIR-based spectral indices for assessing nitrogen content in potato fields. Int. J. Remote Sens. 2010, 31, 5127–5143. [Google Scholar] [CrossRef]
- Pimstein, A.; Karnieli, A.; Bansal, S.K.; Bonfil, D.J. Exploring remotely sensed technologies for monitoring wheat potassium and phosphorus using field spectroscopy. Field Crop. Res. 2011, 121, 125–135. [Google Scholar] [CrossRef]
- Asrar, G.; Fuchs, M.; Kanemasu, E.T.; Hatfield, J.L. Estimating absorbed photosynthetic radiation and leaf-area index from spectral reflectance in wheat. Agron. J. 1984, 76, 300–306. [Google Scholar] [CrossRef]
- Gitelson, A.A. Wide dynamic range vegetation index for remote quantification of biophysical characteristics of vegetation. J. Plant Physiol. 2004, 161, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Nguy-Robertson, A.L.; Peng, Y.; Gitelson, A.A.; Arkebauer, T.J.; Pimstein, A.; Herrmann, I.; Karnieli, A.; Rundquist, D.C.; Bonfil, D.J. Estimating green LAI in four crops: Potential of determining optimal spectral bands for a universal algorithm. Agric. For. Meteorol. 2014, 192, 140–148. [Google Scholar] [CrossRef]
- Herrmann, I.; Pimstein, A.; Karnieli, A.; Cohen, Y.; Alchanatis, V.; Bonfil, D.J. LAI assessment of wheat and potato crops by VENμS and Sentinel-2 bands. Remote Sens. Environ. 2011, 115, 2141–2151. [Google Scholar] [CrossRef]
- Mahlein, A.K.; Rumpf, T.; Welke, P.; Dehne, H.W.; Plumer, L.; Steiner, U.; Oerke, E.C. Development of spectral indices for detecting and identifying plant diseases. Remote Sens. Environ. 2013, 128, 21–30. [Google Scholar] [CrossRef]
- Mahlein, A.-K.; Oerke, E.-C.; Steiner, U.; Dehne, H.-W. Recent advances in sensing plant diseases for precision crop protection. Eur. J. Plant Pathol. 2012, 133, 197–209. [Google Scholar] [CrossRef]
- Herrmann, I.; Shapira, U.; Kinast, S.; Karnieli, A.; Bonfil, D.J. Ground-level hyperspectral imagery for detecting weeds in wheat fields. Precis. Agric. 2013, 14, 637–659. [Google Scholar] [CrossRef]
- Lan, Y.; Zhang, H.; Hoffmann, W.C.; Lopez, J.J.D. Spectral response of spider mite infested cotton: Mite density and miticide rate study. Int. J. Agric. Biol. Eng. 2013, 6, 48–52. [Google Scholar]
- Nansen, C. The potential and prospects of proximal remote sensing of arthropod pests. Pest Manag. Sci. 2016, 72, 653–659. [Google Scholar] [CrossRef] [PubMed]
- Shapira, U.; Herrmann, I.; Karnieli, A.; Bonfil, D.J. Field spectroscopy for weed detection in wheat and chickpea fields. Int. J. Remote Sens. 2013, 34, 6094–6108. [Google Scholar] [CrossRef]
- King, L.; Adusei, B.; Stehman, S.V.; Potapov, P.V.; Song, X.P.; Krylov, A.; Di Bella, C.; Loveland, T.R.; Johnson, D.M.; Hansen, M.C. A multi-resolution approach to national-scale cultivated area estimation of soybean. Remote Sens. Environ. 2017, 195, 13–29. [Google Scholar] [CrossRef]
- Wrather, A.; Koenning, S. Effects of diseases on soybean yields in the United States 1996 to 2007. Plant Health Prog. 2009. [Google Scholar] [CrossRef]
- Ji, J.; Scott, M.P.; Bhattacharyya, M.K. Light is essential for degradation of ribulose-1.5-bisphosphate carboxylase-oxygenase large subunit during sudden death syndrome development in soybean. Plant Biol. 2006, 8, 597–605. [Google Scholar] [CrossRef] [PubMed]
- Marburger, D.A.; Venkateshwaran, M.; Conley, S.P.; Esker, P.D.; Lauer, J.G.; Ane, J.M. Crop Rotation and Management Effect on Fusarium spp. Populations. Crop Sci. 2015, 55, 365–376. [Google Scholar] [CrossRef]
- Aoki, T.; O’Donnell, K.; Homma, Y.; Lattanzi, A.R. Sudden-death syndrome of soybean is caused by two morphologically and phylogenetically distinct species within the Fusarium solani species complex-F-virguliforme in North America and F-tucumaniae in South America. Mycologia 2003, 95, 660–684. [Google Scholar] [PubMed]
- Chilvers, M.I.; Brown-Rytlewski, D.E. First Report and Confirmed Distribution of Soybean Sudden Death Syndrome Caused by Fusarium virguliforme in Southern Michigan. Plant Dis. 2010, 94, 1164. [Google Scholar] [CrossRef]
- Vosberg, S.K.; Marburger, D.A.; Smith, D.L.; Conley, S.P. Planting Date and Fluopyram Seed Treatment Effect on Soybean Sudden Death Syndrome and Seed Yield. Agron. J. 2017, 109, 2570–2578. [Google Scholar] [CrossRef]
- Hartman, G.L.; Chang, H.X.; Leandro, L.F. Research advances and management of soybean sudden death syndrome. Crop Prot. 2015, 73, 60–66. [Google Scholar] [CrossRef]
- Roy, K.W.; Lawrence, G.W.; Hodges, H.H.; McLean, K.S.; Killebrew, J.F. Sudden-death syndrome of soybean-Fusarium solani as intant and relation of Heterodera glycines to disease sevirity. Phytopathology 1989, 79, 191–197. [Google Scholar] [CrossRef]
- Chang, H.X.; Domier, L.L.; Radwan, O.; Yendrek, C.R.; Hudson, M.E.; Hartman, G.L. Identification of Multiple Phytotoxins Produced by Fusarium virguliforme Including a Phytotoxic Effector (FvNIS1) Associated With Sudden Death Syndrome Foliar Symptoms. Mol. Plant-Microbe Interact. 2016, 29, 96–108. [Google Scholar] [CrossRef] [PubMed]
- Brar, H.K.; Swaminathan, S.; Bhattacharyya, M.K. The Fusarium virguliforme Toxin FvTox1 Causes Foliar Sudden Death Syndrome-Like Symptoms in Soybean. Mol. Plant-Microbe Interact. 2011, 24, 1179–1188. [Google Scholar] [CrossRef] [PubMed]
- Scherm, H.; Yang, X.B. Development of sudden death syndrome of soybean in relation to soil temperature and soil water matric potential. Phytopathology 1996, 86, 642–649. [Google Scholar] [CrossRef]
- Abdelsamad, N.A.; Baumbach, J.; Bhattacharyya, M.K.; Leandro, L.F. Soybean Sudden Death Syndrome Caused by Fusarium virguliforme is Impaired by Prolonged Flooding and Anaerobic Conditions. Plant Dis. 2017, 101, 712–719. [Google Scholar] [CrossRef]
- Mueller, D.; Robertson, A.; Sisson, A.; Tylka, G. Soybean Diseases—Soybean Research & Information Initiative; Iowa State University of Science and Technology: Ames, IA, USA, 2010; p. 40. [Google Scholar]
- Wang, J.; Jacobs, J.L.; Byrne, J.M.; Chilvers, M.I. Improved Diagnoses and Quantification of Fusarium virguliforme, Causal Agent of Soybean Sudden Death Syndrome. Phytopathology 2015, 105, 378–387. [Google Scholar] [CrossRef] [PubMed]
- Kandel, Y.R.; Wise, K.A.; Bradley, C.A.; Tenuta, A.U.; Mueller, D.S. Effect of Planting Date, Seed Treatment, and Cultivar on Plant Population, Sudden Death Syndrome, and Yield of Soybean. Plant Dis. 2016, 100, 1735–1743. [Google Scholar] [CrossRef]
- Yang, S.; Li, X.; Chen, C.; Kyveryga, P.; Yang, X.B. Assessing Field-Specific Risk of Soybean Sudden Death Syndrome Using Satellite Imagery in Iowa. Phytopathology 2016, 106, 842–853. [Google Scholar] [CrossRef] [PubMed]
- Duggin, M.J. The field measurement of reflectance factors. Photogramm. Eng. Remote Sens. 1980, 46, 643–647. [Google Scholar]
- Milton, E.J.; Schaepman, M.E.; Anderson, K.; Kneubuhler, M.; Fox, N. Progress in field spectroscopy. Remote Sens. Environ. 2009, 113, S92–S109. [Google Scholar] [CrossRef]
- Anderson, K.; Milton, E.J.; Rollin, E.M. Calibration of dual-beam spectroradiometric data. Int. J. Remote Sens. 2006, 27, 975–986. [Google Scholar] [CrossRef]
- Huber, S.; Tagesson, T.; Fensholt, R. An automated field spectrometer system for studying VIS, NIR and SWIR anisotropy for semi-arid savanna. Remote Sens. Environ. 2014, 152, 547–556. [Google Scholar] [CrossRef]
- Pimstein, A.; Notesco, G.; Ben-Dor, E. Performance of Three Identical Spectrometers in Retrieving Soil Reflectance under Laboratory Conditions. Soil Sci. Soc. Am. J. 2011, 75, 746–759. [Google Scholar] [CrossRef]
- MacLellan, C.J.; Malthus, T.J. High performance dual field of view spectrometer with novel input optics for, autonomous reflectance measurements over an extended spectral range. IEEE Int. Geosci. Remote Sens. Symp. 2009, 3, 2163–2166. [Google Scholar]
- Meroni, M.; Barducci, A.; Cogliati, S.; Castagnoli, F.; Rossini, M.; Busetto, L.; Migliavacca, M.; Cremonese, E.; Galvagno, M.; Colombo, R.; et al. The hyperspectral irradiometer, a new instrument for long-term and unattended field spectroscopy measurements. Rev. Sci. Instrum. 2011, 82, 043106. [Google Scholar] [CrossRef] [PubMed]
- MacArthur, A.; Robinson, I.; Rossini, M.; Davis, N.; Mac Donald, K. A dual-field-of-view spectrometer system for reflectance and fluorescence measurements (Piccolo Doppio) and correction of etaloning. Proceedings of 5th International Workshop on Remote Sensing of Vegetation Fluorescence, Paris, France, 22–24 April 2014. [Google Scholar]
- Hilker, T.; Nesic, Z.; Coops, N.C.; Lessard, D. A new automated, multiangular radiometer instrument for tower-based observations of canopy reflectance (AMSPEC II). Instrum. Sci. Technol. 2010, 38, 319–340. [Google Scholar] [CrossRef]
- Pacheco-Labrador, J.; Martin, M.P. Characterization of a Field Spectroradiometer for Unattended Vegetation Monitoring. Key Sensor Models and Impacts on Reflectance. Sensors 2015, 15, 4154–4175. [Google Scholar] [CrossRef] [PubMed]
- Savitzky, A.; Golay, M. Smoothing and differentiation of data by simplified least squares procedures. Anal. Chem. 1964, 36, 1627–1639. [Google Scholar] [CrossRef]
- Gautam, R.; Vanga, S.; Ariese, F.; Umapathy, S. Review of multidimensional data processing approaches for Raman and infrared spectroscopy. EPJ Tech. Instrum. 2015, 2. [Google Scholar] [CrossRef]
- Inoue, Y.; Penuelas, J.; Miyata, A.; Mano, M. Normalized difference spectral indices for estimating photosynthetic efficiency and capacity at a canopy scale derived from hyperspectral and CO2 flux measurements in rice. Remote Sens. Environ. 2008, 112, 156–172. [Google Scholar] [CrossRef]
- Atzberger, C.; Guerif, M.; Baret, F.; Werner, W. Comparative analysis of three chemometric techniques for the spectroradiometric assessment of canopy chlorophyll content in winter wheat. Comput. Electron. Agric. 2010, 73, 165–173. [Google Scholar] [CrossRef]
- Wold, S.; Johansson, E.; Cocchi, M. PLS-partial least squars projections to latent structures. In 3D QSAR in Drug Design: Theory, Methods, and Applications; Kubinyi, H., Ed.; ESCOM: Leiden, The Netherlands, 1993; pp. 523–550. [Google Scholar]
- Wold, S.; Sjostrom, M.; Eriksson, L. PLS-regression: A basic tool of chemometrics. Chemom. Intell. Lab. Syst. 2001, 58, 109–130. [Google Scholar] [CrossRef]
- Musumarra, G.; Barresi, V.; Condorelli, D.F.; Fortuna, C.G.; Scire, S. Potentialities of multivariate approaches in genome-based cancer research: Identification of candidate genes for new diagnostics by PLS discriminant analysis. J. Chemom. 2004, 18, 125–132. [Google Scholar] [CrossRef]
- Mevik, B.H.; Wehrens, R.; Liland, K.H. Pls: Partial Least Squares and Principal Component Regression, R Package Version 2.6-0, 2016. Available online: https://CRAN.R-project.org/package=pls (accessed on 25 November 2017).
- Ortiz-Ribbing, L.M.; Eastburn, D.M. Soybean root systems and sudden death syndrome severity: Taproot and lateral root infection. Plant Dis. 2004, 88, 1011–1016. [Google Scholar] [CrossRef]
- Gausman, H. Plant Leaf Optical Properties in Visible and Near Infrared light; Texas Tech Press: Lubbock, TX, USA, 1985. [Google Scholar]
- Fan, J.L.; McConkey, B.; Wang, H.; Janzen, H. Root distribution by depth for temperate agricultural crops. Field Crop. Res. 2016, 189, 68–74. [Google Scholar] [CrossRef]
- Baret, F.; Jacquemoud, S.; Guyot, G.; Leprieur, C. Modeled analysis of the biophysical nature of spectral shifts and comparison with information-content of broad bands. Remote Sens. Environ. 1992, 41, 133–142. [Google Scholar] [CrossRef]
- Clevers, J.G.P.W.; de Jong, S.M.; Ephama, G.F.; Van der Meer, F.; Bakker, W.H.; Skidmore, A.; Addink, E.A. MERIS and the red-edge position. Int. J. Appl. Earth Obs. Geoinf. 2001, 3, 313–320. [Google Scholar] [CrossRef]
- Delegido, J.; Fernandez, G.; Gandia, S.; Moreno, J. Retrieval of chlorophyll content and LAI of crops using hyperspectral techniques: Application to PROBA/CHRIS data. Int. J. Remote Sens. 2008, 29, 7107–7127. [Google Scholar] [CrossRef]
- Townsend, P.A.; Green, R.O.; Campbell, P.K.; Cavender-Bares, J.; Clark, M.L.; Couture, J.J.; Desai, A.R.; Gamon, J.A.; Gaunter, L.; Kruger, E.L.; et al. Global terrestrial ecosystem functioning and biogeochemical processes. 2016. Available online: https://hyspiri.jpl.nasa.gov/downloads/RFI2_HyspIRI_related_160517/RFI2_final_Ecosystem_TownsendPhilipA.pdf (accessed on 25 November 2017).
- Gitelson, A.; Merzlyak, M.N. Quantitative estimation of chlorophyll-a using reflectance spectra: Experiments with autumn chestnut and maple leaves. J. Photochem. Photobiol. B Biol. 1994, 22, 247–252. [Google Scholar] [CrossRef]
- Roth, M.G.; Noel, Z.A.; Wang, J.; Byrne, A.M.; TerAvest, D.; Kramer, D.M.; Chilvers, M.I. Assessment and utilization of risk factors in predicting soybean yield and sudden death syndrome development. Phytopathology 2018, in press. [Google Scholar]
- Pauli, D.; Chapman, S.C.; Bart, R.; Topp, C.N.; Lawrence-Dill, C.J.; Poland, J.; Gore, M.A. The Quest for Understanding Phenotypic Variation via Integrated Approaches in the Field Environment. Plant Physiol. 2016, 172, 622–634. [Google Scholar] [CrossRef] [PubMed]
- Peyraud, R.; Dubiella, U.; Barbacci, A.; Genin, S.; Raffaele, S.; Roby, D. Advances on plant-pathogen interactions from molecular toward systems biology perspectives. Plant J. 2017, 90, 720–737. [Google Scholar] [CrossRef] [PubMed]
- Curran, P.J. Remote Sensing of Foliar Chemistry. Remote Sens. Environ. 1989, 30, 271–278. [Google Scholar] [CrossRef]
- Kuhlgert, S.; Austic, G.; Zegarac, R.; Osei-Bonsu, I.; Hoh, D.; Chilvers, M.I.; Roth, M.G.; Bi, K.; TerAvest, D.; Weebadde, P.; et al. MultispeQ Beta: A tool for large-scale plant phenotyping connected to the open PhotosynQ network. R. Soc. Open Sci. 2016, 3, 160592. [Google Scholar] [CrossRef] [PubMed]
- Christenson, B.S.; Schapaugh, W.T.; An, N.; Price, K.P.; Prasad, V.; Fritz, A.K. Predicting Soybean Relative Maturity and Seed Yield Using Canopy Reflectance. Crop Sci. 2016, 56, 625–643. [Google Scholar] [CrossRef]
- Ma, B.L.; Dwyer, L.M.; Costa, C.; Cober, E.R.; Morrison, M.J. Early prediction of soybean yield from canopy reflectance measurements. Agron. J. 2001, 93, 1227–1234. [Google Scholar] [CrossRef]
- Yu, N.; Li, L.J.; Schmitz, N.; Tiaz, L.F.; Greenberg, J.A.; Diers, B.W. Development of methods to improve soybean yield estimation and predict plant maturity with an unmanned aerial vehicle based platform. Remote Sens. Environ. 2016, 187, 91–101. [Google Scholar] [CrossRef]
- Atzberger, C.; Eilers, P.H.C. Evaluating the effectiveness of smoothing algorithms in the absence of ground reference measurements. Int. J. Remote Sens. 2011, 32, 3689–3709. [Google Scholar] [CrossRef]
- Eilers, P.H.C. A perfect smoother. Anal. Chem. 2003, 75, 3631–3636. [Google Scholar] [CrossRef] [PubMed]
- Kirchgessner, N.; Liebisch, F.; Yu, K.; Pfeifer, J.; Friedli, M.; Hund, A.; Walter, A. The ETH field phenotyping platform FIP: A cable-suspended multi-sensor system. Funct. Plant Biol. 2016, 44, 154–168. [Google Scholar] [CrossRef]
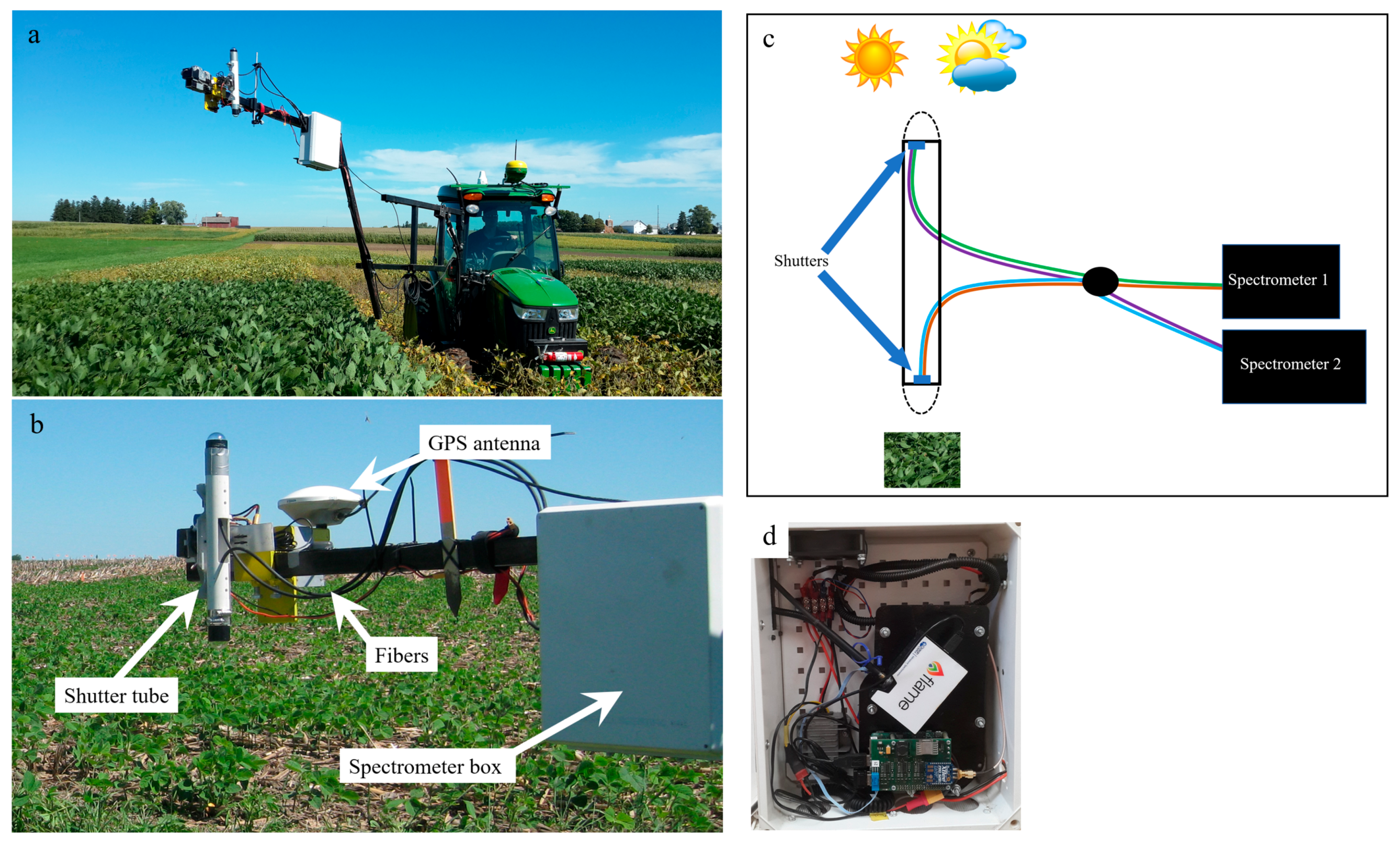





| Date | Site | Development Stage * | Activity | Number of Samples ** | Comments |
|---|---|---|---|---|---|
| Canopy experiment | |||||
| 30 April 2016 | Arlington | - | Planting I | - | |
| 18 May 2016 | Arlington | - | Planting II | - | |
| 8 June 2016 | Arlington | Vc V1 | Spectra | 96 | Planting dates I and II |
| 10 June 2016 | Arlington | - | Planting III | - | |
| 28 June 2016 | Arlington | V1 V3 V4 | Spectra | 144 | |
| 18 July 2016 | Arlington | V5 R3 R4 | Spectra | 144 | |
| 26 July 2016 | Arlington | R2 R4 R5 | Spectra | 144 | |
| 15 August 2016 | Arlington | R5 R6 | Spectra | 144 | |
| 22 August 2016 | Arlington | R6 | Spectra | 144 | |
| 2 September 2016 | Arlington | R6 | Spectra | 144 | Early senescence visual for planting dates I and II |
| 12 September 2016 | Arlington | R6 | Spectra | 48 | Planting date III |
| 18 October 2016 | Arlington | R8 | Harvest | - | All planting dates |
| Leaf experiment | |||||
| 2 May 2016 | Hancock | - | Planting | - | |
| 3 May 2016 | Arlington | - | Planting | - | |
| 9 May 2016 | Arlington fill | - | Planting | - | |
| 13 Jun 2016 | Arlington | V3 | Spectra | 245 | |
| 14 and 15 Jun 2016 | Hancock | V3 | Spectra | 238 | Weather did not allow to obtain all data in one day |
| 20 June 2016 | Hancock | V4 | Spectra | 242 | |
| 21 June 2016 | Arlington | V4 | Spectra | 250 | |
| 27 June 2016 | Hancock | V5 | Spectra | 249 | |
| 28 June 2016 | Arlington | V5 | Spectra | 248 | |
| 1 July 2016 | Hancock | V6 | Leaf tagging | - | |
| 5 July 2016 | Arlington | V6 | Spectra | 250 | |
| 6 July 2016 | Arlington | V6 | Leaf tagging | - | |
| 6 July 2016 | Hancock | R1 | Spectra | 250 | |
| 11 July 2016 | Arlington | R1 | Spectra | 247 | |
| 12 July 2016 | Hancock | R2 | Spectra | 248 | |
| 18 Jul 2016 | Hancock | R3 | Spectra | 250 | |
| 19 July 2016 | Arlington | R3 | Spectra | 249 | |
| 26 July 2016 | Arlington | R4 | Spectra | 248 | |
| 29 July 2016 | Hancock | R4 | Spectra | 250 | |
| 1 August 2016 | Arlington | R5 | Spectra | 189 | Weather did not allow to obtain all data |
| 8 August 2016 | Hancock | R5 | Spectra | 244 | |
| 18 August 2016 | Arlington fill | R6 | Spectra | 197 | |
| 22 August 2016 | Arlington fill | R6 | Root sampling | 195 (6) | |
| 25 August 2016 | Hancock | R6 | Spectra | 225 | |
| 26 August 2016 | Hancock | R6 | Root sampling | 248 (37) | |
| 30 August 2016 | Arlington | R6 | Spectra | 231 | |
| 1 September 2016 | Arlington | R6 | Root sampling | 250 (36) | |
| PLSDA Model | % Total Accuracy (Number of Samples; Kappa) | ||
|---|---|---|---|
| Calibration | Cross-Validation | Validation | |
| All dates | 62 (527; 0.25) | 58 (228; 0.17) | 63 (252; 0.26) |
| All vegetative stages | 76 (200; 0.52) | 70 (88; 0.40) | 77 (96; 0.55) |
| All reproductive stages | 65 (326; 0.30) | 59 (141; 0.17) | 61 (156;0.21) |
| 18 July 2016 | 88 (74; 0.77) | 79 (34; 0.57) | 82 (36; 0.66) |
| 26 July 2016 | 86 (74; 0.72) | 71 (34; 0.43) | 82 (36; 0.65) |
| 18 and 26 July 2016 | 78 (150; 0.56) | 67 (66; 0.35) | 75 (72; 0.50) |
| PLSDA Model | Development Stage * | % Total Accuracy (Number of Samples; Kappa) | ||
|---|---|---|---|---|
| Calibration | Cross-Validation | Validation | ||
| Arlington and Hancock | ||||
| All dates | V and R | 67 (2531; 0.33) | 63 (1086; 0.25) | 61 (1227; 0.21) |
| Vegetative stages | V | 58 (903; 0.16) | 56 (389; 0.12) | 55 (430; 0.10) |
| Reproductive stages | R | 76 (1626; 0.48) | 70 (699; 0.40) | 66 (797; 0.33) |
| Arlington | ||||
| All dates | V and R | 76 (1255; 0.52) | 71 (539; 0.43) | 72 (589; 0.43) |
| Vegetative stages | V | 74 (525; 0.48) | 70 (226; 0.39) | 67 (242; 0.34) |
| Reproductive stages | R | 83 (729; 0.66) | 77 (314; 0.53) | 77 (347; 0.55) |
| Late vegetative stages | V5 V6 | 91 (263; 0.82) | 87 (115; 0.74) | 92 (120; 0.83) |
| Early reproductive stages | R1 R3 | 89 (254; 0.79) | 81 (111; 0.63) | 81 (131; 0.61) |
| Hancock | ||||
| All dates | V and R | 78 (1275; 0.55) | 74 (548; 0.46) | 68 (616; 0.36) |
| Vegetative stages | V | 71 (378; 0.42) | 64 (163; 0.28) | 63 (188; 0.26) |
| Reproductive stages | R | 78 (897; 0.55) | 75 (385; 0.48) | 67 (428; 0.35) |
| Late vegetative stages | V5 V6 | 98 (265; 0.97) | 90 (111; 0.81) | 91 (132; 0.82) |
| Early reproductive stages | R2 R3 | 92 (263; 0.84) | 82 (114; 0.64) | 75 (121; 0.49) |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Herrmann, I.; Vosberg, S.K.; Ravindran, P.; Singh, A.; Chang, H.-X.; Chilvers, M.I.; Conley, S.P.; Townsend, P.A. Leaf and Canopy Level Detection of Fusarium Virguliforme (Sudden Death Syndrome) in Soybean. Remote Sens. 2018, 10, 426. https://doi.org/10.3390/rs10030426
Herrmann I, Vosberg SK, Ravindran P, Singh A, Chang H-X, Chilvers MI, Conley SP, Townsend PA. Leaf and Canopy Level Detection of Fusarium Virguliforme (Sudden Death Syndrome) in Soybean. Remote Sensing. 2018; 10(3):426. https://doi.org/10.3390/rs10030426
Chicago/Turabian StyleHerrmann, Ittai, Steven K. Vosberg, Prabu Ravindran, Aditya Singh, Hao-Xun Chang, Martin I. Chilvers, Shawn P. Conley, and Philip A. Townsend. 2018. "Leaf and Canopy Level Detection of Fusarium Virguliforme (Sudden Death Syndrome) in Soybean" Remote Sensing 10, no. 3: 426. https://doi.org/10.3390/rs10030426






