Differences in Rate and Direction of Shifts between Phytoplankton Size Structure and Sea Surface Temperature
Abstract
:1. Introduction
2. Materials and Methods
2.1. In Situ Data
2.1.1. Size Fractionated Chla Concentration
2.1.2. Absorption Coefficient of Phytoplankton
2.1.3. Retrieval of IOPs from In Situ Spectral Radiation Data
2.2. Satellite Remote Sensing Data
2.3. Model Description
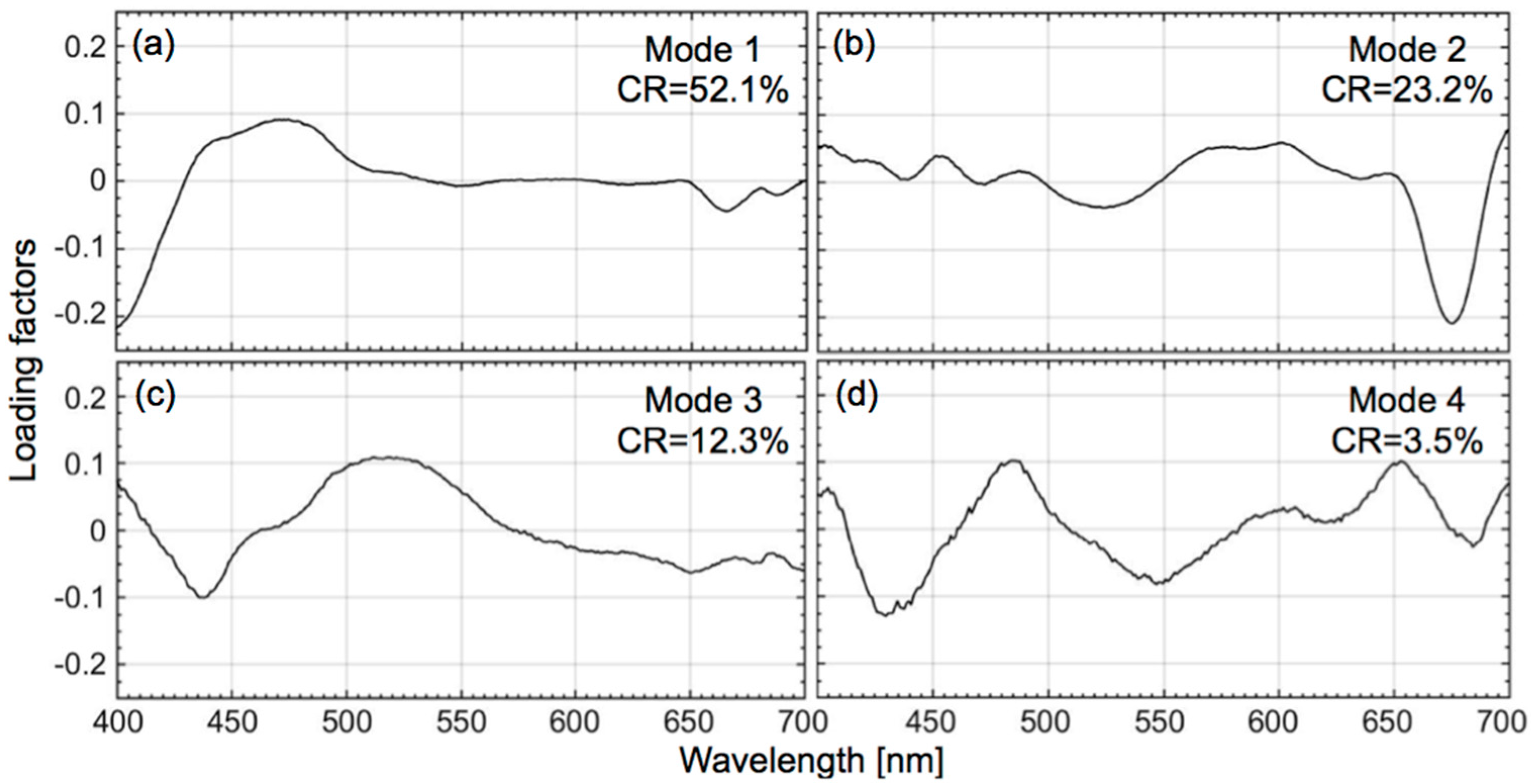
2.4. Quantification of CSD Slope Using Phytoplankton Absorption Spectra
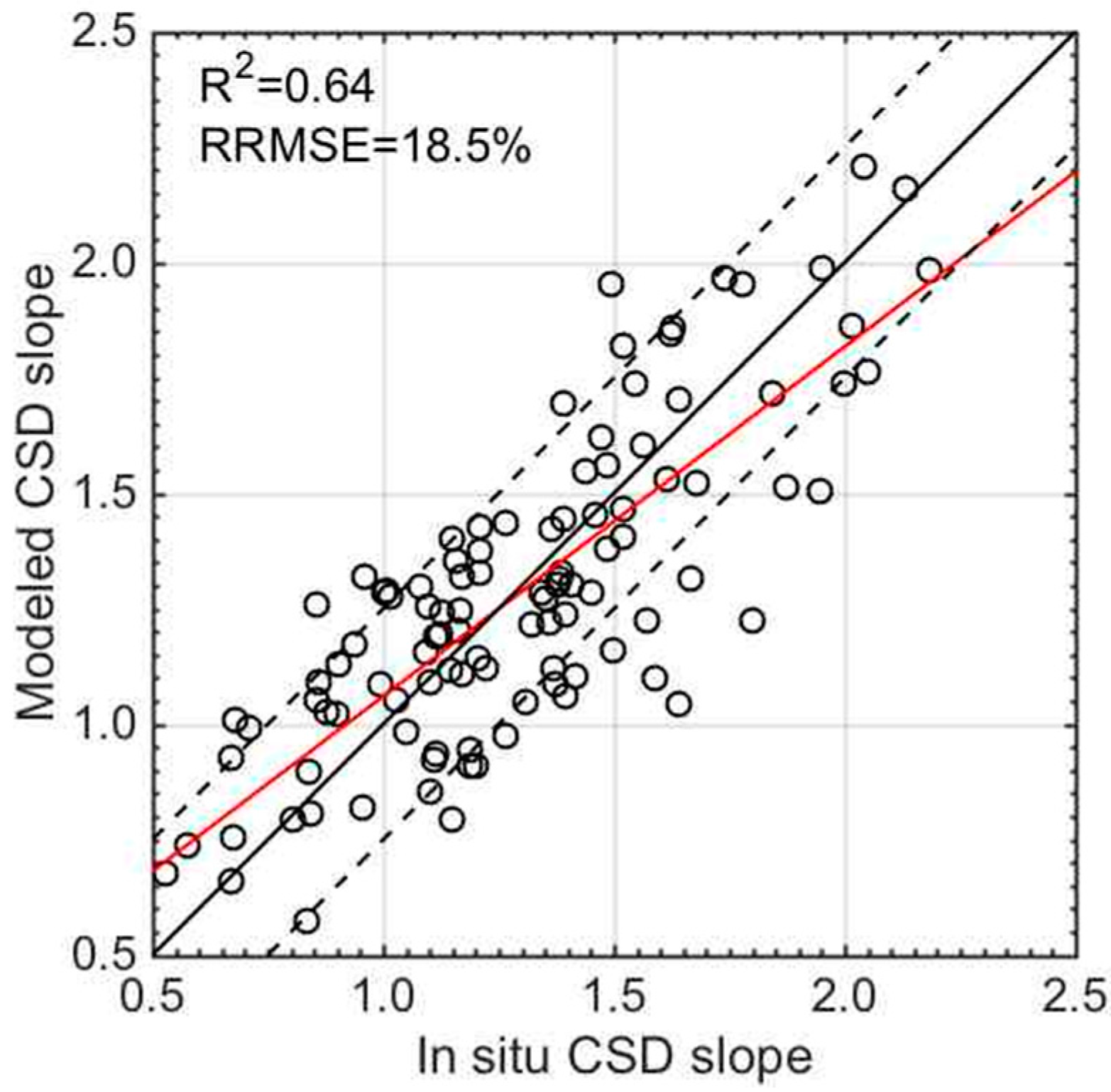
2.5. Evaluation of Estimate Accuracy
2.6. Velocities of the Shift in Oceanographic Parameters
3. Results
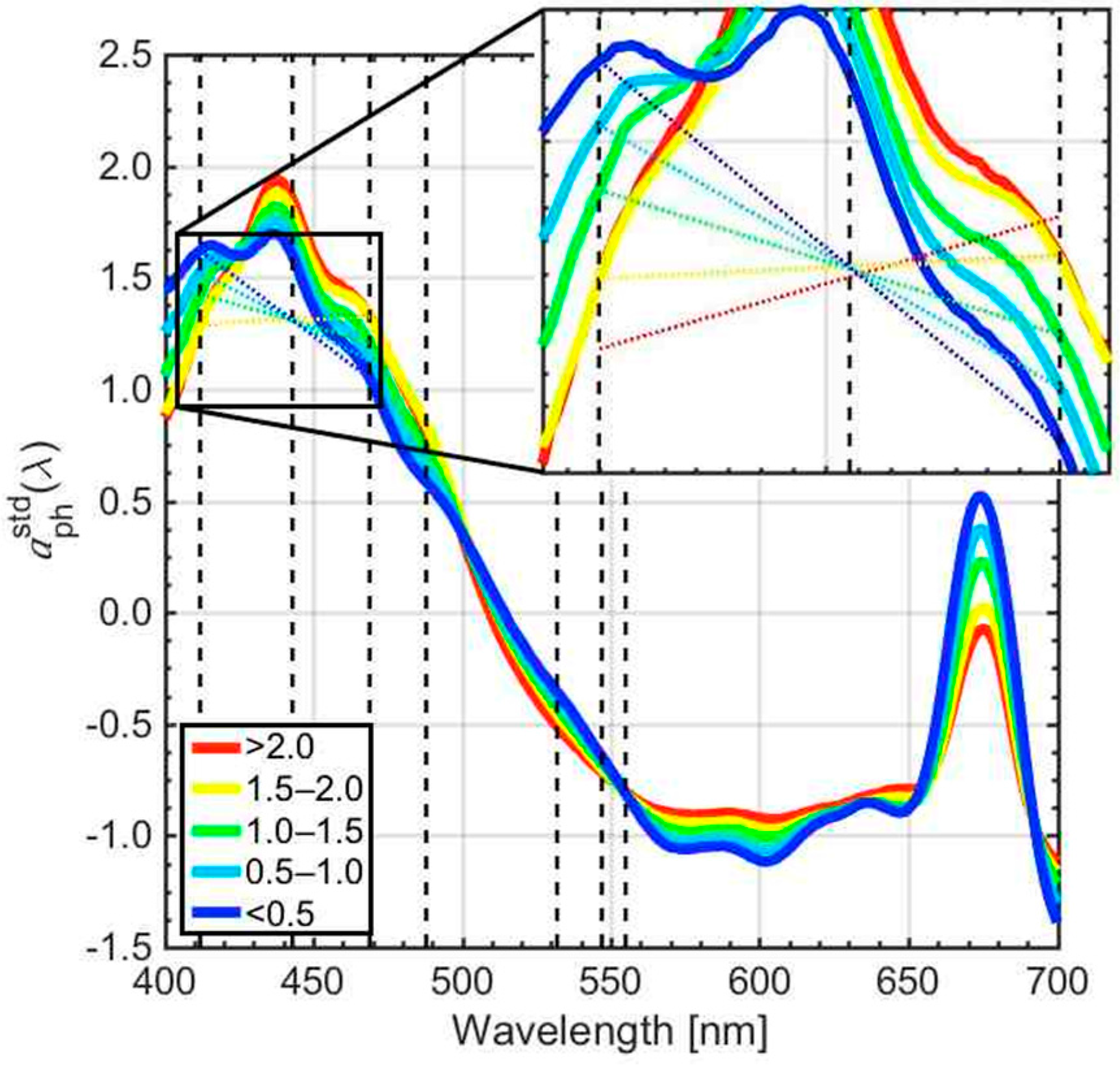
3.1. Retrieval of the CSD Slope from In Situ Chlasize
3.2. Empirical Development of the CSD Model
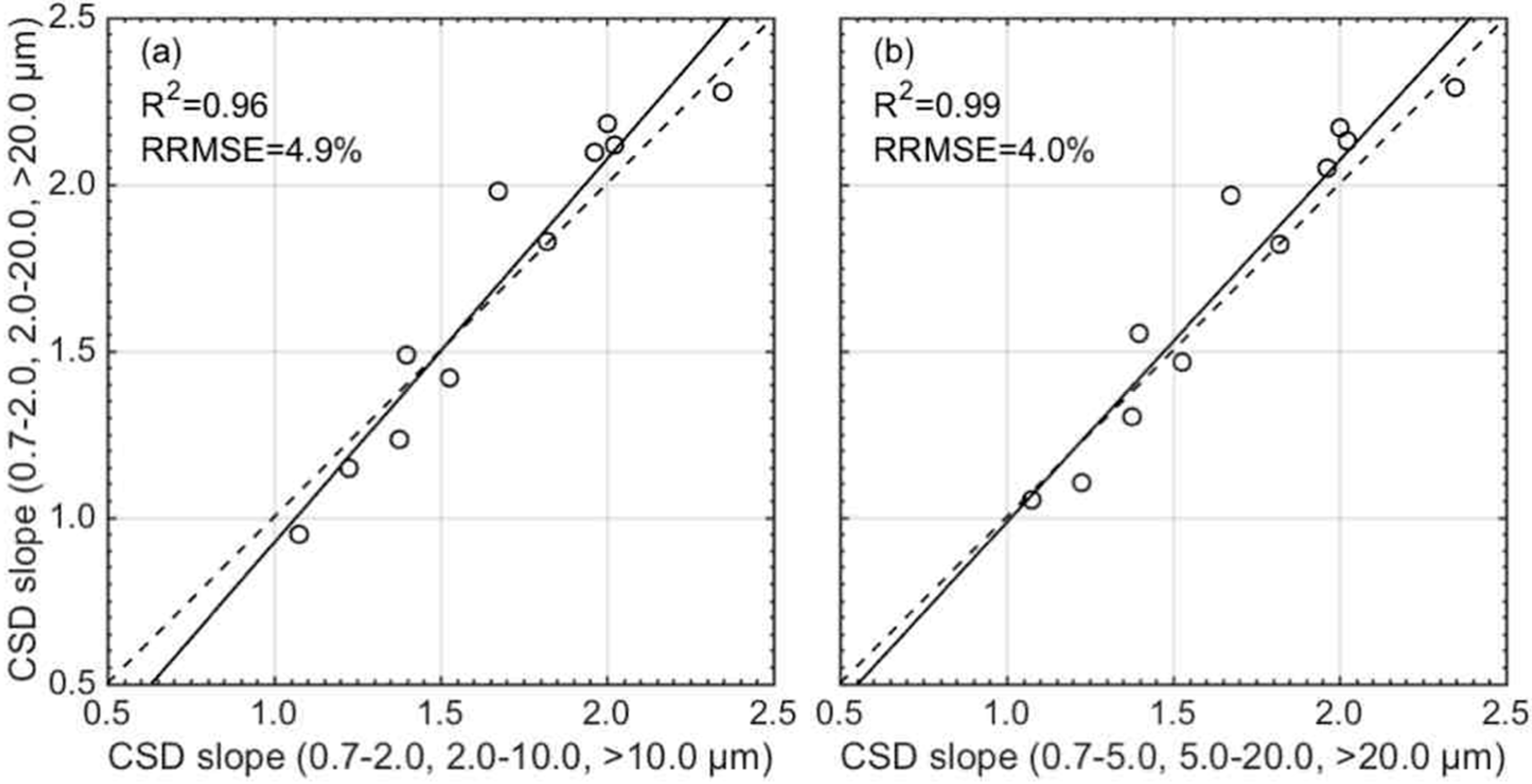
3.3. Validation of the CSD Model
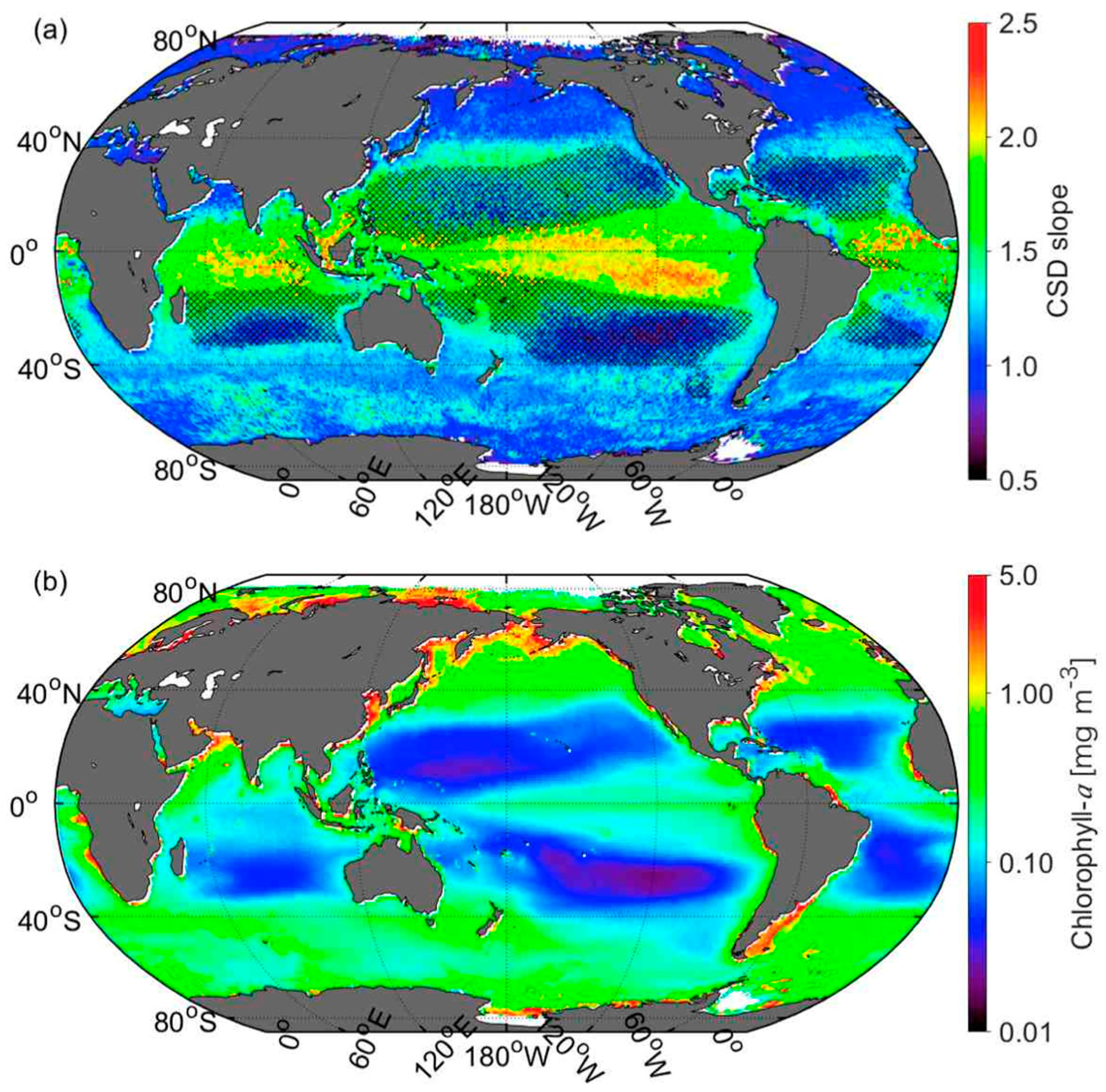
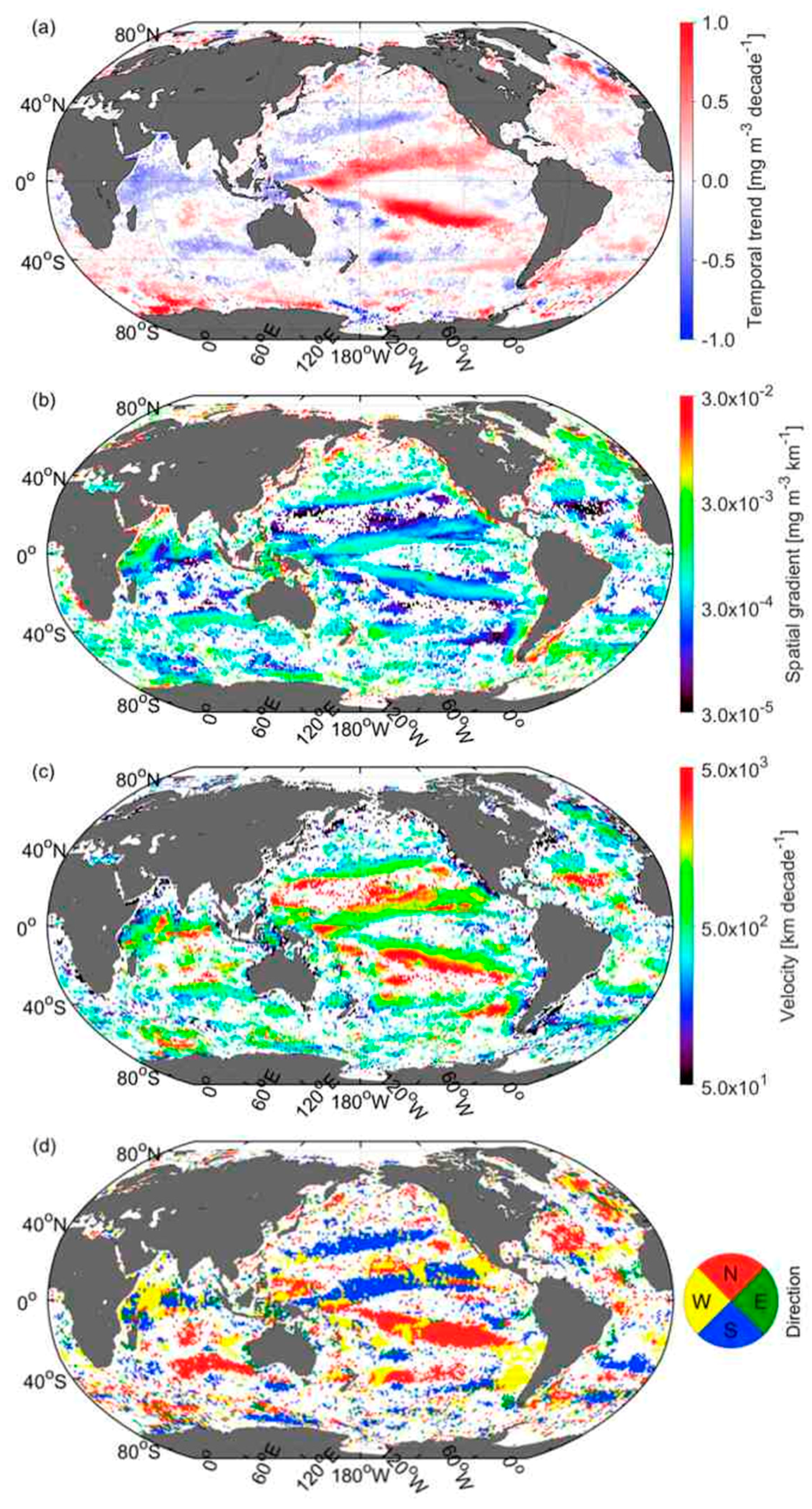
3.4. Global Distribution of CSD Slope and Chla
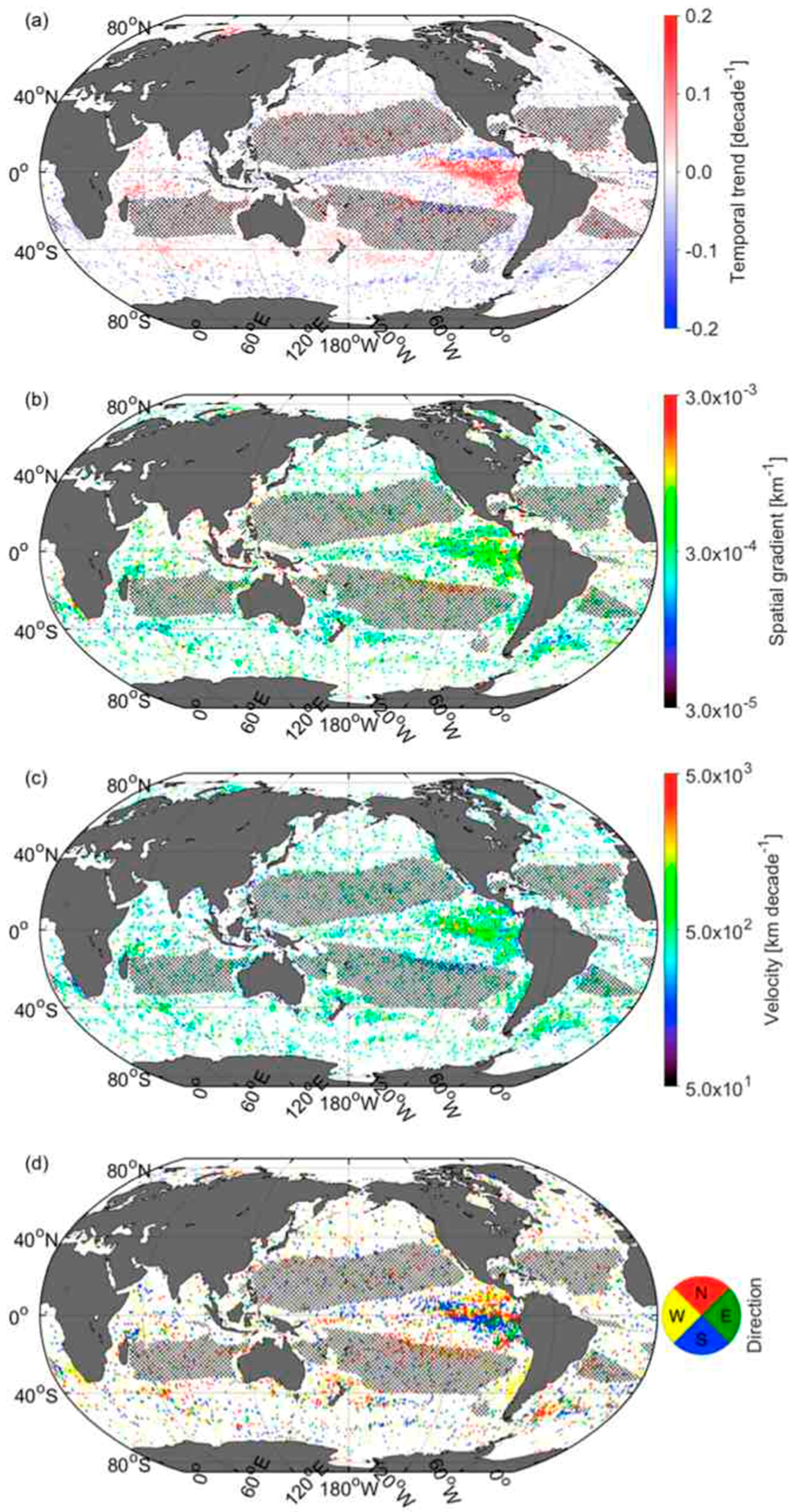
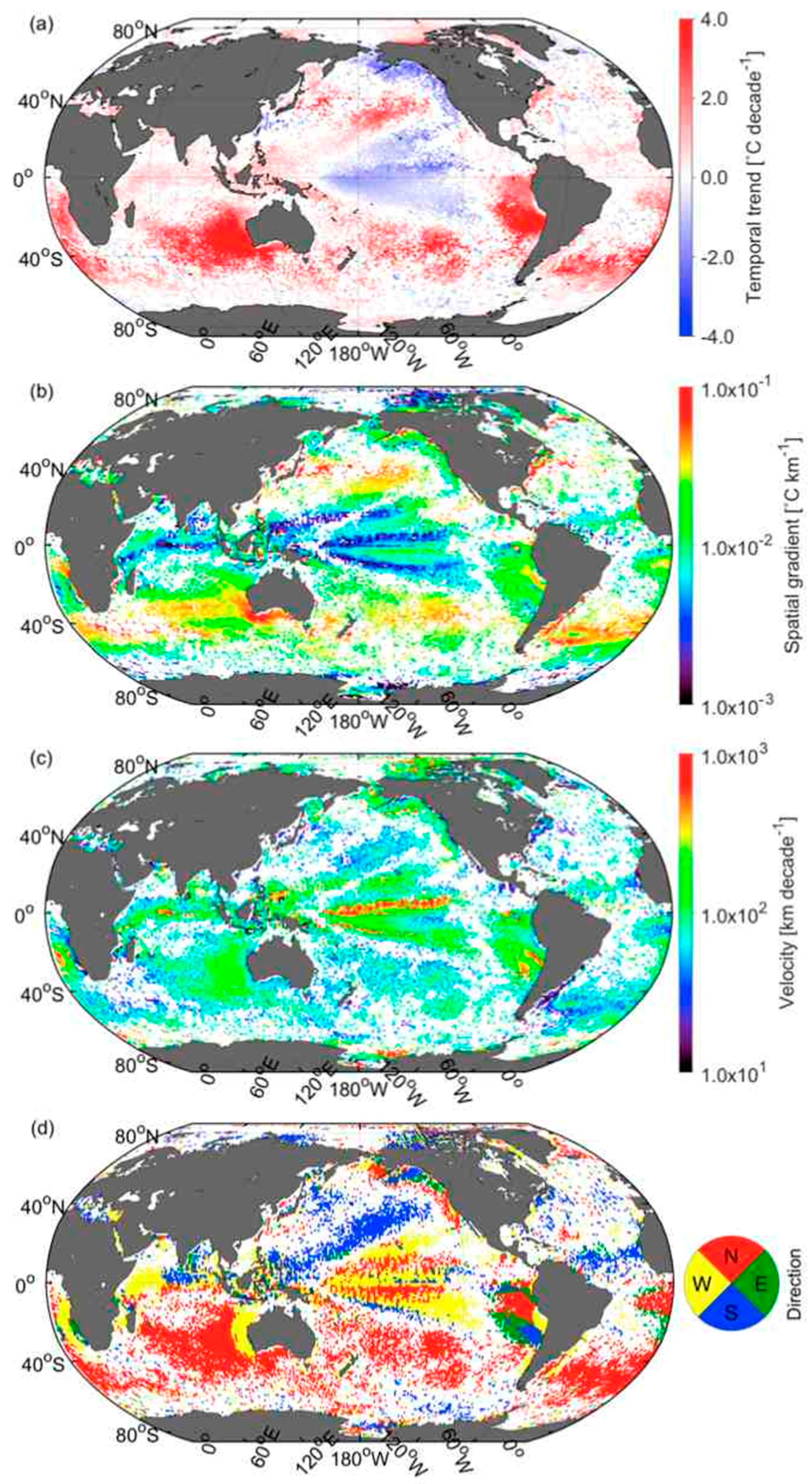
3.5. Velocities of Shift in CSD Slope and Chla
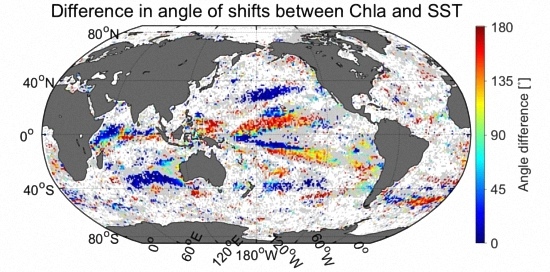
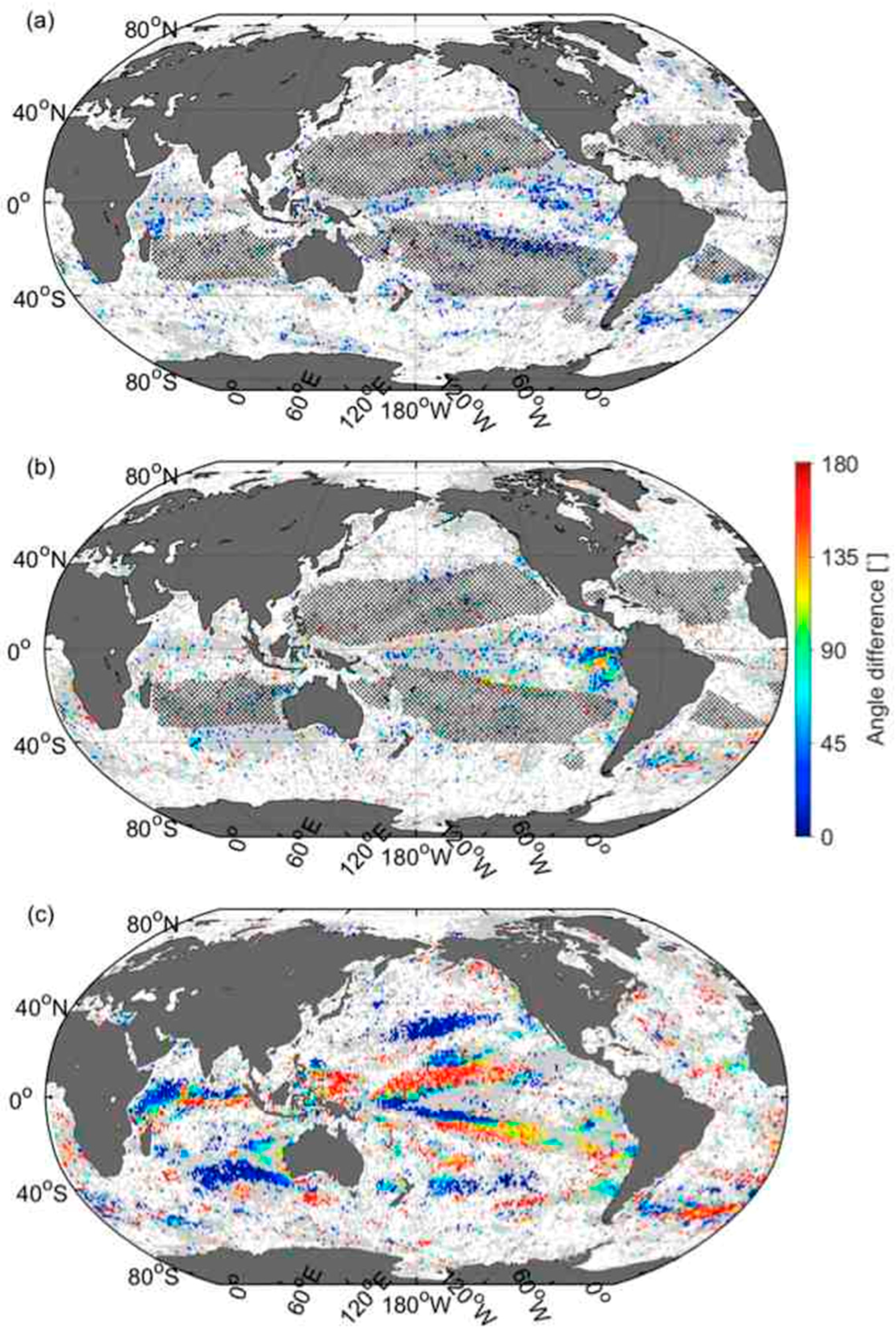
3.6. Comparison of Velocities Derived from CSD Slope and Chla with That of SST
4. Discussion
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Levitus, S.; Antonov, J.I.; Boyer, T.P.; Locarnini, R.A.; Garcia, H.E.; Mishonov, A.V. Global ocean heat content 1955–2008 in light of recently revealed instrumentation problems. Geophys. Res. Lett. 2009, 36, 1–5. [Google Scholar] [CrossRef]
- Wassmann, P.; Kosobokova, K.N.; Slagstad, D.; Drinkwater, K.F.; Hopcroft, R.R.; Moore, S.E.; Ellingsen, I.; Nelson, R.J.; Carmack, E.; Popova, E.; et al. The contiguous domains of Arctic Ocean advection: Trails of life and death. Prog. Oceanogr. 2015, 139, 42–65. [Google Scholar] [CrossRef]
- Grebmeier, J.M.; Bluhm, B.A.; Cooper, L.W.; Danielson, S.L.; Arrigo, K.R.; Blanchard, A.L.; Clarke, J.T.; Day, R.H.; Frey, K.E.; et al. Ecosystem characteristics and processes facilitating persistent macrobenthic biomass hotspots and associated benthivory in the Pacific Arctic. Prog. Oceanogr. 2015, 136, 92–114. [Google Scholar] [CrossRef]
- Mueter, F.J.; Litzow, M.A. Sea ice retreat alters the biogeography of the Bering Sea continental shelf. Ecol. Appl. 2008, 18, 309–320. [Google Scholar] [CrossRef] [PubMed]
- Spencer, P.D. Density-independent and density-dependent factors affecting temporal changes in spatial distributions of eastern Bering Sea flatfish. Fish. Oceanogr. 2008, 17, 396–410. [Google Scholar] [CrossRef]
- Thackeray, S.J.; Sparks, T.H.; Frederiksen, M. Trophic level asynchrony in rates of phenological change for marine, freshwater and terrestrial environments. Glob. Chang. Biol. 2010, 16, 3304–3313. [Google Scholar] [CrossRef]
- Dawson, T.P.; Jackson, S.T.; House, J.I.; Prentice, I.C.; Mace, G.M. Beyond Predictions: Biodiversity Conservation in a Changing Climate. Science 2011, 332, 53–58. [Google Scholar] [CrossRef] [PubMed]
- Loarie, S.R.; Duffy, P.B.; Hamilton, H.; Asner, G.P.; Field, C.B.; Ackerly, D.D. The velocity of climate change. Nature 2009, 462, 1052–1055. [Google Scholar] [CrossRef] [PubMed]
- Burrows, M.T.; Schoeman, D.S.; Buckley, L.B.; Moore, P.; Poloczanska, E.S.; Brander, K.M.; Brown, C.; Bruno, J.F.; Duarte, C.M.; Halpern, B.S.; et al. The Pace of Shifting Climate in Marine and Terrestrial Ecosystems. Science 2011, 334, 652–655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- VanDerWal, J.; Murphy, H.T.; Kutt, A.S.; Perkins, G.C.; Bateman, B.L.; Perry, J.L.; Reside, A.E. Focus on poleward shifts in species’ distribution underestimates the fingerprint of climate change. Nat. Clim. Chang. 2012, 3, 239–243. [Google Scholar] [CrossRef]
- Philippart, C.J.M.; Anadon, R.; Danovaro, R.; Dippner, J.W.; Drinkwater, K.F.; Hawkins, S.J.; Oguz, T.; O’Sullivan, G.; Reid, P.C. Impacts of climate change on European marine ecosystems: Observations, expectations and indicators. J. Exp. Mar. Biol. Ecol. 2011, 400, 52–69. [Google Scholar] [CrossRef]
- Poloczanska, E.S.; Brown, C.J.; Sydeman, W.J.; Kiessling, W.; Schoeman, D.S.; Moore, P.J.; Brander, K.; Bruno, J.F.; Buckley, L.B.; Burrows, M.T.; et al. Global imprint of climate change on marine life. Nat. Clim. Chang. 2013, 3, 919–925. [Google Scholar] [CrossRef]
- Pinsky, M.L.; Worm, B.; Fogarty, M.J.; Sarmiento, J.L.; Levin, S.A. Marine Taxa Track Local Climate Velocities. Science 2013, 341, 1239–1242. [Google Scholar] [CrossRef] [PubMed]
- McClain, C.R. A decade of satellite ocean color observations. Annu. Rev. Mar. Sci. 2009, 1, 19–42. [Google Scholar] [CrossRef] [PubMed]
- Lalli, C.M.; Parsons, T.R. Biological Oceanography: An Introduction, 2nd ed.; Elsevier: New York, NY, USA, 1997; p. 314. [Google Scholar]
- Kędra, M.; Moritz, C.; Choy, E.S.; David, C.; Degen, R.; Duerksen, S.; Ellingsen, I.; Górska, B.; Grebmeier, J.M.; Kirievskaya, D.; et al. Status and trends in the structure of Arctic benthic food webs. Polar Res. 2015, 34, 1–23. [Google Scholar] [CrossRef] [Green Version]
- Kostadinov, T.S.; Siegel, D.A. Retrieval of the particle size distribution from satellite ocean color observations. J. Geophys. Res. 2009, 114, 1–22. [Google Scholar] [CrossRef]
- Fujiwara, A.; Hirawake, T.; Suzuki, K.; Saitoh, S.I. Remote sensing of size structure of phytoplankton communities using optical properties of the Chukchi and Bering Sea shelf region. Biogeosciences 2011, 8, 3567–3580. [Google Scholar] [CrossRef]
- Roy, S.; Sathyendranath, S.; Bouman, H.; Platt, T. The global distribution of phytoplankton size spectrum and size classes from their light-absorption spectra derived from satellite data. Remote Sens. Environ. 2013, 139, 185–197. [Google Scholar] [CrossRef]
- Bricaud, A.; Morel, A. Light attenuation and scattering by phytoplanktonic cells: A theoretical modeling. Appl. Opt. 1986, 25, 571–580. [Google Scholar] [CrossRef] [PubMed]
- Stramski, D.; Boss, E.; Bogucki, D.; Voss, K.J. The role of seawater constituents in light backscattering in the ocean. Prog. Oceanogr. 2004, 61, 27–56. [Google Scholar] [CrossRef]
- Lee, Z.P.; Carder, K.L.; Arnone, R.A. Deriving inherent optical properties from water color: A multiband quasi-analytical algorithm for optically deep waters. Appl. Opt. 2002, 41, 5755–5772. [Google Scholar] [CrossRef] [PubMed]
- Craig, S.E.; Jones, C.T.; Li, W.K.W.; Lazin, G.; Horne, E.; Caverhill, C.; Cullen, J.J. Deriving optical metrics of coastal phytoplankton biomass from ocean colour. Remote Sens. Environ. 2012, 119, 72–83. [Google Scholar] [CrossRef]
- Bracher, A.; Taylor, M.; Taylor, B.; Dinter, T.; Roettgers, R. Using empirical orthogonal functions derived from remote sensing reflectance for the prediction of phytoplankton pigments concentrations. Ocean Sci. 2015, 11, 139–158. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Ishizaka, J.; Hirawake, T.; Watanabe, Y.; Zhu, Y.; Hayashi, M.; Yoo, S. Remote estimation of phytoplankton size fractions using the spectral shape of light absorption. Opt. Express 2015, 23, 10301–10318. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, R.; Ishimaru, T. An improved method for the determination of phytoplankton chlorophyll using N, N-dimethylformamide. J. Oceanogr. Soc. Jpn. 1990, 46, 190–194. [Google Scholar] [CrossRef]
- Welshmeyer, N.A. Fluorometric analysis of chlorophyll budgets: Zooplankton grazing and phytoplankton growth in a temperate fjord and the central Pacific gyres. Limnol. Oceangr. 1994, 39, 1985–1992. [Google Scholar]
- Holm-Hansen, O.; Lorenzen, C.J.; Holmes, R.W.; Strickland, J.D.H. Fluorometric Determination of Chlorophyll. J. Du Cons. 1965, 30, 3–15. [Google Scholar] [CrossRef]
- Mitchell, B.G. Algorithms for Determining the Absorption Coefficient for Aquatic Particulates Using the Quantitative Filter Technique (QFT). Proc. SPIE 1990, 1302, 137–148. [Google Scholar]
- Kishino, M.; Takahashi, M.; Okami, N.; Ichimura, S. Estimation of the spectral absorption coefficients of phytoplankton in the sea. Bull. Mar. Sci. 1985, 37, 634–642. [Google Scholar]
- Tassan, S.; Ferrari, G.M. An alternative approach to absorption measurements of aquatic particles retained on filters. Limnol. Oceangr. 1995, 40, 1358–1368. [Google Scholar] [CrossRef]
- Darecki, M.; Stramski, D. An evaluation of MODIS and SeaWiFS bio-optical algorithms in the Baltic Sea. Remote Sens. Environ. 2004, 89, 326–350. [Google Scholar] [CrossRef]
- Lee, Z.; Hu, C.; Shang, S.; Du, K.; Lewis, M.; Arnone, R.; Brewin, R. Penetration of UV-visible solar radiation in the global oceans: Insights from ocean color remote sensing. J. Geophys. Res. Oceans 2013, 118, 4241–4255. [Google Scholar] [CrossRef]
- Casey, K.S.; Brandon, T.B.; Evans, R. The Past, Present, and Future of the AVHRR Pathfinder SST Program. In Oceanography from Space; Springer: Dordrecht, The Netherlands, 2010; pp. 273–287. [Google Scholar]
- Junge, C.E. Air Chemistry and Radioactivity; Academic Press Inc.: New York, NY, USA; London, UK, 1963; p. 382. [Google Scholar]
- Bader, H. The hyperbolic distribution of particle sizes. J. Geophys. Res. 1970, 75, 2822–2830. [Google Scholar] [CrossRef]
- Dussart, B.H. Les différentes catégories de plancton. Hydrobiologia 1965, 26, 72–74. [Google Scholar] [CrossRef]
- Ciotti, A.M.; Bricaud, A. Retrievals of a size parameter for phytoplankton and spectral light absorption by colored detrital matter from water-leaving radiances at SeaWiFS channels in a continental shelf region off Brazil. Limnol. Oceanogr. Methods 2006, 4, 237–253. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, M.; van Dijk, M.A.; Zhu, G.; Gong, Z.; Li, Y.; Qin, B. Measured and numerically partitioned phytoplankton spectral absorption coefficients in inland waters. J. Plankton Res. 2009, 31, 311–323. [Google Scholar] [CrossRef]
- Brewin, R.J.W.; Sathyendranath, S.; Hirata, T.; Lavender, S.J.; Barciela, R.M.; Hardman-Mountford, N.J. A three-component model of phytoplankton size class for the Atlantic Ocean. Ecol. Model. 2010, 221, 1472–1483. [Google Scholar] [CrossRef]
- Devred, E.; Sathyendranath, S.; Stuart, V.; Platt, T. A three component classification of phytoplankton absorption spectra: Application to ocean-color data. Remote Sens. Environ. 2011, 115, 2255–2266. [Google Scholar] [CrossRef]
- Ciotti, A.M.; Lewis, M.R.; Cullen, J.J. Assessment of the relationships between dominant cell size in natural phytoplankton communities and the spectral shape of the absorption coefficient. Limnol. Oceangr. 2002, 47, 404–417. [Google Scholar] [CrossRef]
- Wang, J.; Cota, G.F.; Ruble, D.A. Absorption and backscattering in the Beaufort and Chukchi Seas. J. Geophys. Res. 2005, 110, 1–12. [Google Scholar] [CrossRef]
- Matsuoka, A.; Hill, V.; Huot, Y.; Babin, M.; Bricaud, A. Seasonal variability in the light absorption properties of western Arctic waters: Parameterization of the individual components of absorption for ocean color applications. J. Geophys. Res. 2011, 116, 1–15. [Google Scholar] [CrossRef]
- Maritorena, S.; Siegel, D.A.; Peterson, A.R. Optimization of a semianalytical ocean color model for global-scale applications. Appl. Opt. 2002, 41, 2705–2714. [Google Scholar] [CrossRef] [PubMed]
- Boss, E.; Roesler, C. Over Constrained Linear Matrix Inversion with Statistical Selection. In Remote Sensing of Inherent Optical Properties: Fundamentals, Tests of Algorithms, and Applications; Lee, Z.P., Ed.; International Ocean-Colour Coordinating Group (IOCCG): Dartmouth, NS, Canada, 2006; Volume 5, pp. 57–61. [Google Scholar]
- Li, Z.; Li, L.; Song, K.; Cassar, N. Estimation of phytoplankton size fractions based on spectral features of remote sensing ocean color data. J. Geophys. Res. Oceans 2013, 118, 1445–1458. [Google Scholar] [CrossRef]
- Alvain, S.; Moulin, C.; Dandonneau, Y.; Bréon, F.-M. Remote sensing of phytoplankton groups in case 1 waters from global SeaWiFS imagery. Deep-Sea Res. Part I Oceanogr. Res. Pap. 2005, 52, 1989–2004. [Google Scholar] [CrossRef] [Green Version]
- Hirata, T.; Hardman-Mountford, N.J.; Brewin, R.J.W.; Aiken, J.; Barlow, R.; Suzuki, K.; Isada, T.; Howell, E.; Hashioka, T.; Noguchi-Aita, M.; et al. Synoptic relationships between surface Chlorophyll-a and diagnostic pigments specific to phytoplankton functional types. Biogeosciences 2011, 8, 311–327. [Google Scholar] [CrossRef] [Green Version]
- Brewin, R.J.; Sathyendranath, S.; Lange, P.K.; Tilstone, G. Comparison of two methods to derive the size-structure of natural populations of phytoplankton. Deep-Sea Res. I 2014, 85, 72–79. [Google Scholar] [CrossRef]
- Gregg, W.W.; Rousseaux, C.S. Decadal trends in global pelagic ocean chlorophyll: A new assessment integrating multiple satellites, in situ data, and models. J. Geophys. Res. Oceans 2014, 119, 5921–5933. [Google Scholar] [CrossRef] [PubMed]
- Gregg, W.W. Recent trends in global ocean chlorophyll. Geophys. Res. Lett. 2005, 32, 1–5. [Google Scholar] [CrossRef]
- Przeslawski, R.; Falkner, I.; Ashcroft, M.B.; Hutchings, P. Using rigorous selection criteria to investigate marine range shifts. Estuar. Coast. Shelf Sci. 2012, 113, 205–212. [Google Scholar] [CrossRef]
- Parmesan, C. Influences of species, latitudes and methodologies on estimates of phenological response to global warming. Glob. Chang. Biol. 2007, 13, 1860–1872. [Google Scholar] [CrossRef]
- Chen, I.C.; Hill, J.K.; Ohlemuller, R.; Roy, D.B.; Thomas, C.D. Rapid Range Shifts of Species Associated with High Levels of Climate Warming. Science 2011, 333, 1024–1026. [Google Scholar] [CrossRef] [PubMed]
- Lenoir, J.; Gégout, J.C.; Guisan, A.; Vittoz, P.; Wohlgemuth, T.; Zimmermann, N.E.; Dullinger, S.; Pauli, H.; Willner, W.; Svenning, J.C. Going against the flow: Potential mechanisms for unexpected downslope range shifts in a warming climate. Ecography 2010, 33, 295–303. [Google Scholar] [CrossRef]
- Bates, A.E.; Pecl, G.T.; Frusher, S.; Hobday, A.J.; Wernberg, T.; Smale, D.A.; Sunday, J.M.; Hill, N.A.; Dulvy, N.K.; Colwell, R.K.; et al. Defining and observing stages of climate-mediated range shifts in marine systems. Glob. Environ. Chang. 2014, 26, 27–38. [Google Scholar] [CrossRef]
- Sunday, J.M.; Pecl, G.T.; Frusher, S.; Hobday, A.J.; Hill, N.; Holbrook, N.J.; Edgar, G.J.; Stuart-Smith, R.; Barrett, N.; Wernberg, T.; et al. Species traits and climate velocity explain geographic range shifts in an ocean-warming hotspot. Ecol. Lett. 2015, 18, 944–953. [Google Scholar] [CrossRef] [PubMed]
- Parmesan, C.; Yohe, G. A globally coherent fingerprint of climate change impacts across natural systems. Nature 2003, 421, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Sunday, J.M.; Bates, A.E.; Dulvy, N.K. Thermal tolerance and the global redistribution of animals. Nat. Clim. Chang. 2012, 54, 14–19. [Google Scholar] [CrossRef]
- Van der Putten, W.H.; Macel, M.; Visser, M.E. Predicting species distribution and abundance responses to climate change: Why it is essential to include biotic interactions across trophic levels. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2010, 365, 2025–2034. [Google Scholar] [CrossRef] [PubMed]
- Bates, A.E.; McKelvie, C.M.; Sorte, C.J.B.; Morley, S.A.; Jones, N.A.R.; Mondon, J.A.; Bird, T.J.; Quinn, G. Geographical range, heat tolerance and invasion success in aquatic species. Proc. R. Soc. B 2013, 280, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Lenoir, J.; Svenning, J.C. Climate-related range shifts—A global multidimensional synthesis and new research directions. Ecography 2015, 38, 15–28. [Google Scholar] [CrossRef]
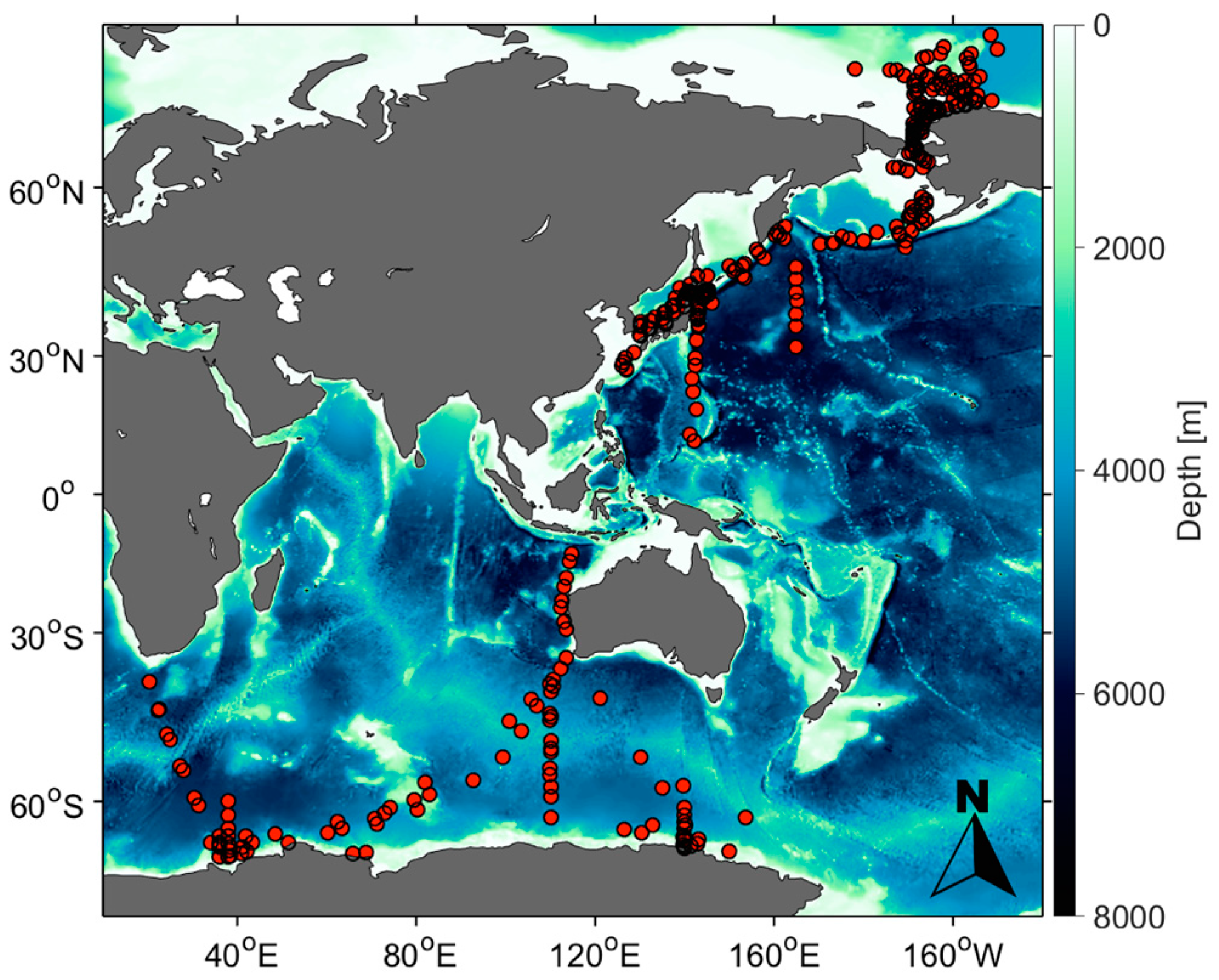

| Cruise Period | Cruise | Vessel Names | Number of Data | |
|---|---|---|---|---|
| Water Sample | Spectral Radiation | |||
| 22 November–12 December 2000 | JARE-42 | JMSDF Shirase | 19 | 6 |
| 16–25 February 2002 | JARE-43 | JMSDF Shirase | 10 | 9 |
| 25 January–9 February 2003 | UM0203 | T/S Umitaka-maru | 15 | 14 |
| 22 February–6 March 2003 | JARE-44 | R/V Tangaroa | 9 | 6 |
| 1–21 January 2005 | UM0405 | T/S Umitaka-maru | 16 | 11 |
| 5–22 January 2006 | UM0506 | T/S Umitaka-maru | 10 | 7 |
| 25 July–14 August 2007 | OS180 | T/S Oshoro-maru | 20 | 20 |
| 27 December 2007–12 February 2008 | UM0708 | T/S Umitaka-maru | 21 | 15 |
| 13–25 January 2009 | UM0809 | T/S Umitaka-maru | 9 | 7 |
| 18 August–18 September 2009 | KH-09-4 | R/V Hakuho-maru | 16 | 16 |
| 11 September–10 October 2009 | MR09-03 | R/V Mirai | 12 | 12 |
| 3 June–7 July 2010 | OS216 | T/S Oshoro-maru | 21 | 17 |
| 4 September–13 October 2010 | MR10-05 | R/V Mirai | 28 | 28 |
| 9–11 May 2011 | KT-11-07 | R/V Tansei-maru | 8 | 7 |
| 8 June–7 July 2011 | OS229 | T/S Oshoro-maru | 25 | 24 |
| 17–18 June 2012 | US260 | T/S Ushio-maru | 8 | 7 |
| 6–8 August 2012 | US263 | T/S Ushio-maru | 10 | 8 |
| 13 September–2 October 2012 | MR12-E03 | R/V Mirai | 12 | 12 |
| 23 June–17 July 2013 | OS255 | T/S Oshoro-maru | 26 | 24 |
| 31 August–4 October 2013 | MR13-06 | R/V Mirai | 32 | 32 |
| 10–12 October 2013 | ARAON-EJS | Icebreaker ARAON | 9 | 7 |
| 8–30 June 2014 | Mu14 | R/V Multanovski | 16 | 16 |
| 8–23 March 2015 | KH-15-1 | R/V Hakuho-maru | 12 | 12 |
| Cruise | 20 μm | 10 μm | 5 μm | 2 μm | GF/F |
|---|---|---|---|---|---|
| JARE-42 | × | × | × | × | |
| JARE-43 | × | × | × | × | |
| UM0203 | × | × | × | × | |
| JARE-44 | × | × | × | × | |
| UM0405 | × | × | × | ||
| UM0506 | × | × | × | ||
| OS180 | × | × | × | ||
| UM0708 | × | × | × | ||
| UM0809 | × | × | × | ||
| KH-09-4 | × | × | × | ||
| MR09-03 | × | × | × | × | |
| OS216 | × | × | × | × | |
| MR10-05 | × | × | × | × | |
| KT-11-07 | × | × | × | × | |
| OS229 | × | × | × | × | |
| US260 | × | × | × | ||
| US263 | × | × | × | ||
| MR12-E03 | × | × | × | × | × |
| OS255 | × | × | × | ||
| MR13-06 | × | × | × | × | |
| ARAON-EJS | × | × | × | ||
| Mu14 | × | × | × | ||
| KH-15-1 | × | × | × |
| Wavelength (nm) | ||||||||||
| 412 | 443 | 469 | 488 | 531 | 547 | 555 | 645 | 667 | 678 | |
| Number of minus value | 31 | 31 | 36 | 28 | 78 | 77 | 86 | 14 | 44 | 83 |
| R2 | 0.68 | 0.71 | 0.59 | 0.61 | 0.57 | 0.62 | 0.57 | 0.00 | 0.01 | 0.01 |
| RRMSE (%) | 20.3 | 19.0 | 19.7 | 21.2 | 20.5 | 15.8 | 15.4 | 67.2 | 77.9 | 58.1 |
| Model Parameters | ||||||||
| β0 | C1 | C2 | C3 | C4 | C5 | C6 | C7 | |
| aph(412) ≥ aph(469) | −0.221 | 0.324 | 0.021 | −0.780 | 0.243 | 1.714 | −0.190 | −1.308 |
| aph(412) < aph(469) | −0.314 | 0.245 | 0.026 | −0.693 | 0.209 | 1.813 | −0.191 | −1.311 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Waga, H.; Hirawake, T.; Fujiwara, A.; Kikuchi, T.; Nishino, S.; Suzuki, K.; Takao, S.; Saitoh, S.-I. Differences in Rate and Direction of Shifts between Phytoplankton Size Structure and Sea Surface Temperature. Remote Sens. 2017, 9, 222. https://doi.org/10.3390/rs9030222
Waga H, Hirawake T, Fujiwara A, Kikuchi T, Nishino S, Suzuki K, Takao S, Saitoh S-I. Differences in Rate and Direction of Shifts between Phytoplankton Size Structure and Sea Surface Temperature. Remote Sensing. 2017; 9(3):222. https://doi.org/10.3390/rs9030222
Chicago/Turabian StyleWaga, Hisatomo, Toru Hirawake, Amane Fujiwara, Takashi Kikuchi, Shigeto Nishino, Koji Suzuki, Shintaro Takao, and Sei-Ichi Saitoh. 2017. "Differences in Rate and Direction of Shifts between Phytoplankton Size Structure and Sea Surface Temperature" Remote Sensing 9, no. 3: 222. https://doi.org/10.3390/rs9030222