Towards an Integrative Understanding of tRNA Aminoacylation–Diet–Host–Gut Microbiome Interactions in Neurodegeneration
Abstract
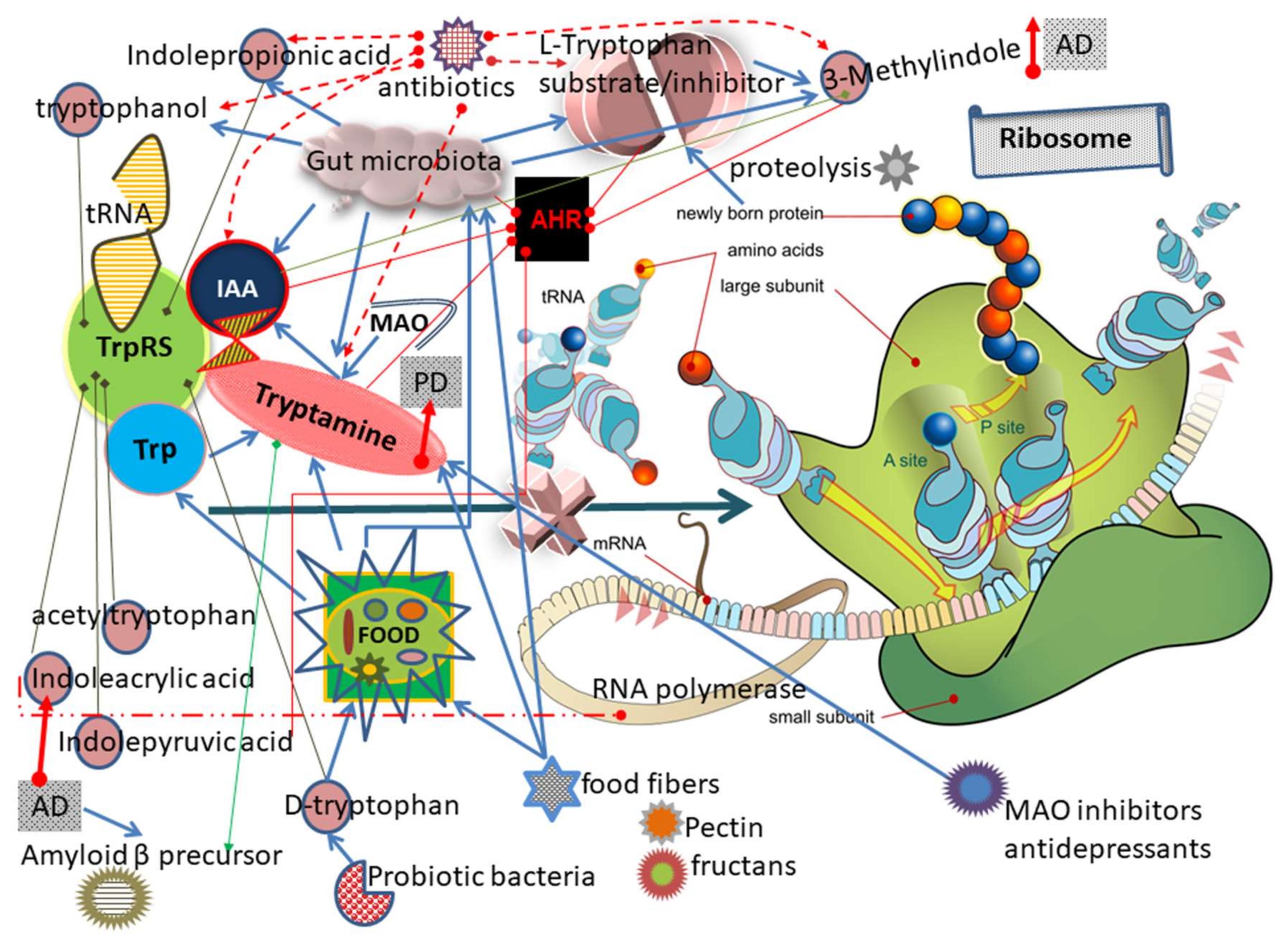
:1. Introduction
2. Materials and Methods
2.1. BLAST for Short Peptides Altered in Mild Cognitive Impairment and Alzheimer’s disease
2.2. Search for Tryptophan-Free Proteins
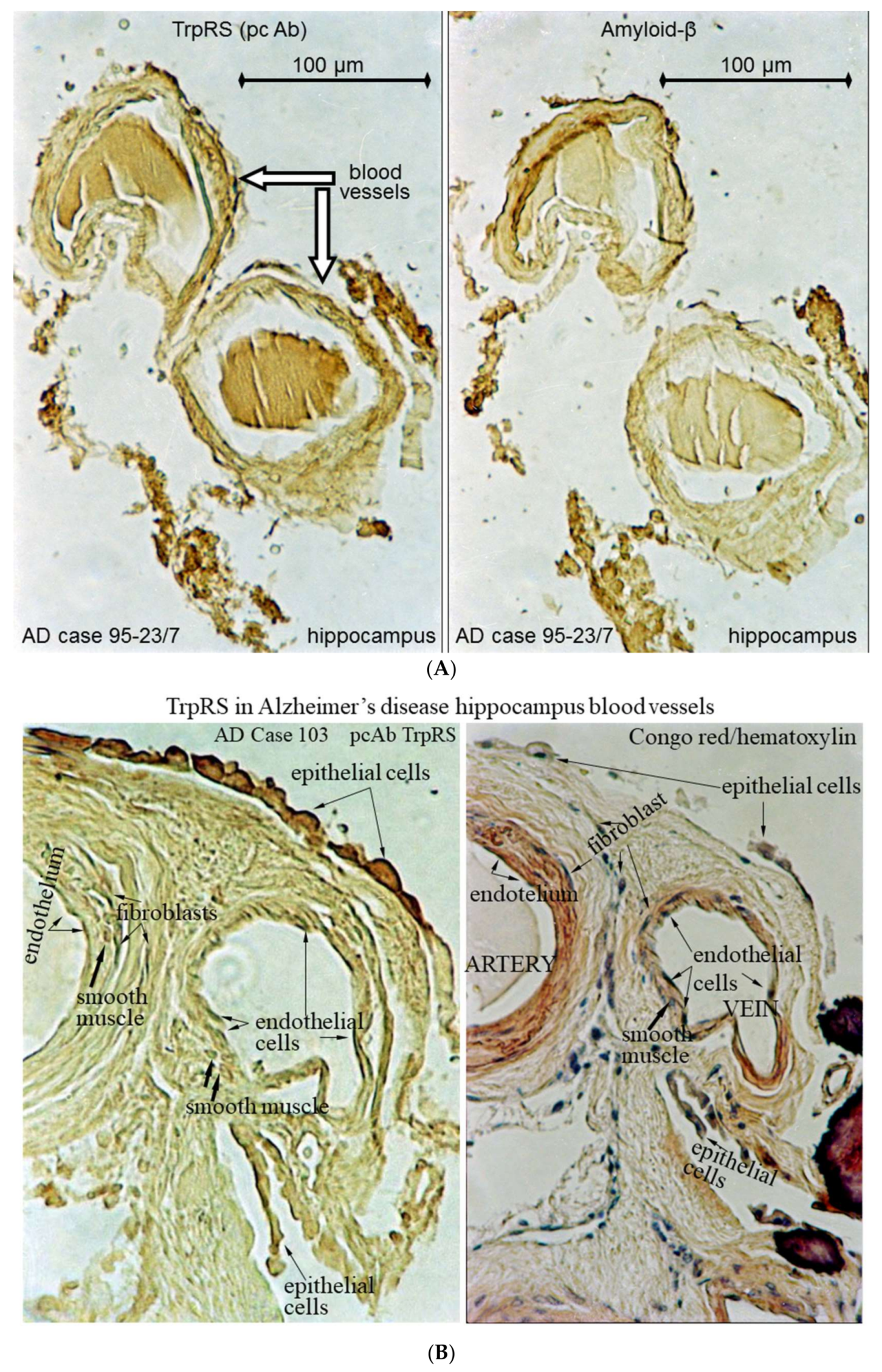
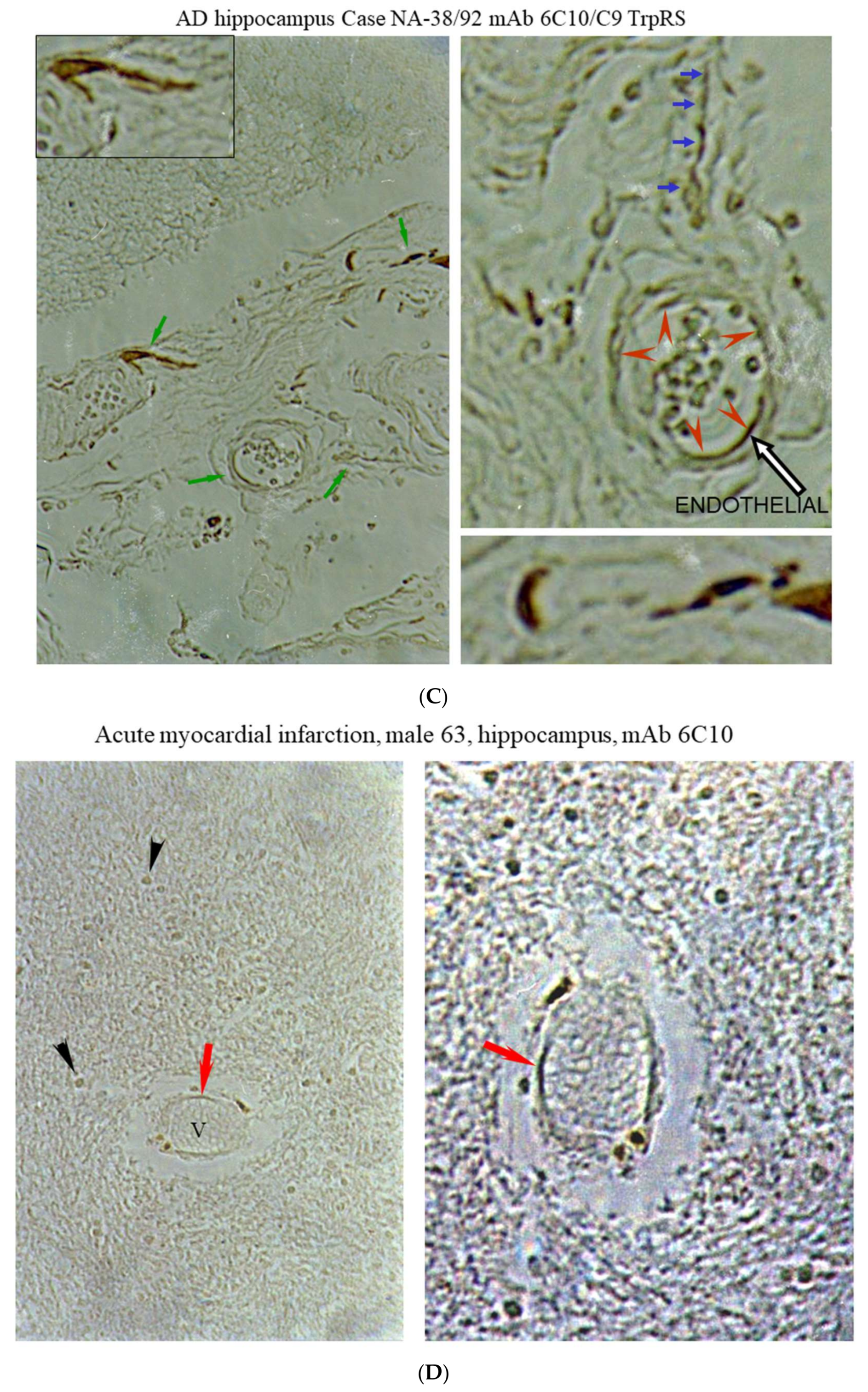
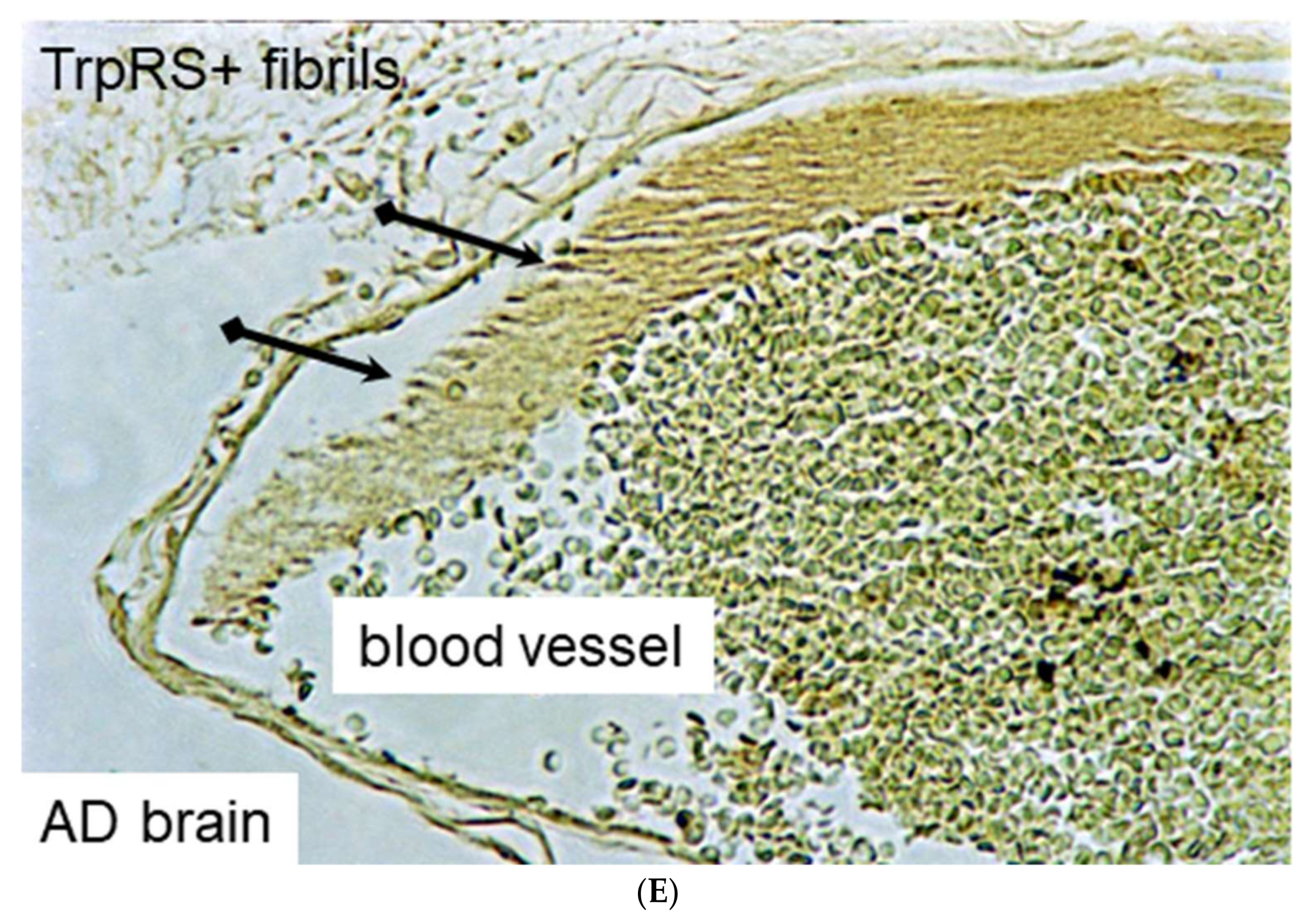
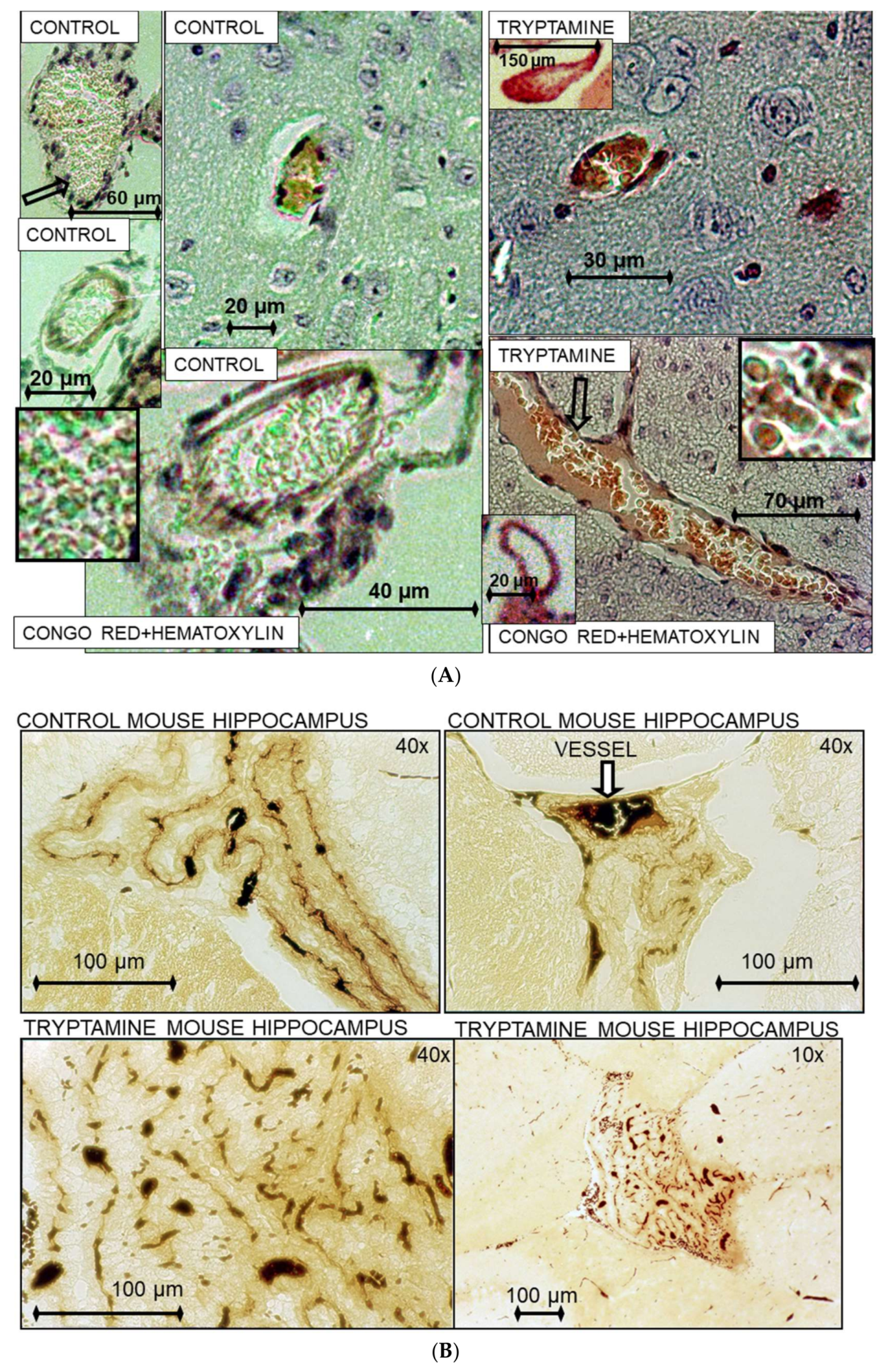
2.3. Histochemical Analysis of Vasculopathies
3. Results and Discussion
3.1. TrpRS Inhibition and Aggregation Can Occur Physiologically
3.2. Mechanism of TrpRS Inhibition by Tryptamine and Tryptophanol
3.3. Tryptamine and Tryptophanol Cytotoxicity and Bioavailability
3.4. Tryptamine and Tryptophanol Increases Following Antibiotic Treatment
3.5. Dietary Exposure of Human Population to Tryptamine
3.6. Concentration-Dependent Tryptamine Effects in Animal Experiments: Seizures, Death
3.7. Tryptamine in Healthy Human Population and in Diseases
3.8. Tryptamine Upregulates Transcription of Genes Including Gene Encoding Aβ Precursor
3.9. The Diet Additives Increase Tryptamine Content in Animals
3.10. Other Trp and Tryptamine Metabolites Can Inhibit TrpRS
3.10.1. IAA
3.10.2. Indolepyruvic Acid
3.10.3. IPA
3.10.4. d-Tryptophan
3.10.5. Indoleacrylic Acid (IAcrA)
3.10.6. 3-Methylindole (3MI)
3.11. Acetyltryptophan in Macular Degeneration
3.12. Di- and Tripeptides in AD, Mild Cognitive Impairment (MCI) and Cognitively Normal (CN)
3.13. Link of Trp Frequency in Proteins to AD and Related Disorders
3.14. Ribosomal Frameshifting and Bypassing
3.15. Trp Levels Decreased in the Human Bodily Fluids of Diseased Individuals
3.16. TrpRS Inhibition Leads to Both Translation and Transcription Impairments
3.17. Mutations in Genes Encoding Cytoplasmic (WARS) and Mitochondrial (WARS2) TrpRS in Humans
3.18. Bacterial Toxic Analog of Amino Acid Alanine Causes Neurodegeneration in Monkey
3.19. TrpRS, Trp and Tryptamine in Vascular Dysfunctions and Pathological Changes
4. Conclusions
Lethal Tryptamine Drug Combinations.
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Appendix A
References
- Abbott, A.; Dolgin, E. Failed alzheimer’s trial does not kill leading theory of disease. Nature 2016, 540, 15–16. [Google Scholar] [CrossRef] [PubMed]
- Trushina, E.; Dutta, T.; Persson, X.M.; Mielke, M.M.; Petersen, R.C. Identification of altered metabolic pathways in plasma and CSF in mild cognitive impairment and alzheimer’s disease using metabolomics. PLoS ONE 2013, 8, e63644. [Google Scholar]
- Paley, E.L.; Denisova, G.; Sokolova, O.; Posternak, N.; Wang, X.; Brownell, A.L. Tryptamine induces tryptophanyl-tRNA synthetase-mediated neurodegeneration with neurofibrillary tangles in human cell and mouse models. Neuromolecular Med. 2007, 9, 55–82. [Google Scholar] [CrossRef]
- Paley, E.L. Tryptamine-induced tryptophanyl-tRNAtrp deficiency in neurodifferentiation and neurodegeneration interplay: Progenitor activation with neurite growth terminated in alzheimer’s disease neuronal vesicularization and fragmentation. J. Alzheimers Dis. 2011, 26, 263–298. [Google Scholar] [PubMed]
- Paley, E.L.; Perry, G.; Sokolova, O. Tryptamine induces axonopathy and mitochondriopathy mimicking neurodegenerative diseases via tryptophanyl-tRNA deficiency. Curr. Alzheimer Res. 2013, 10, 987–1004. [Google Scholar] [CrossRef] [PubMed]
- Lowe, G.; Tansley, G. An investigation of the mechanism of activation of tryptophan by tryptophanyl-tRNA synthetase from beef pancreas. Eur. J. Biochem. 1984, 138, 597–602. [Google Scholar] [CrossRef] [PubMed]
- Favorova, O.O.; Khochkina, L.L.; Shaigo, M.; Parin, A.V.; Khil’ko, S.H.; Prasolov, V.S.; Kiselev, L.L. Truptophanyl tRNA synthetase. Isolation and characterization of the 2 enzyme forms. Mol. Biol. 1974, 8, 729–740. [Google Scholar] [PubMed]
- Swanson, K.S.; Grieshop, C.M.; Flickinger, E.A.; Bauer, L.L.; Wolf, B.W.; Chow, J.; Garleb, K.A.; Williams, J.A.; Fahey, G.C., Jr. Fructooligosaccharides and Lactobacillus acidophilus modify bowel function and protein catabolites excreted by healthy humans. J. Nutr. 2002, 132, 3042–3050. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Xie, G.; Zhao, A.; Zhao, L.; Yao, C.; Chiu, N.H.; Zhou, Z.; Bao, Y.; Jia, W.; Nicholson, J.K.; et al. The footprints of gut microbial-mammalian co-metabolism. J. Proteome Res. 2011, 10, 5512–5522. [Google Scholar] [CrossRef] [PubMed]
- Luqman, A.; Nega, M.; Nguyen, M.T.; Ebner, P.; Gotz, F. Sada-expressing staphylococci in the human gut show increased cell adherence and internalization. Cell Rep. 2018, 22, 535–545. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Jin, U.H.; Allred, C.D.; Jayaraman, A.; Chapkin, R.S.; Safe, S. Aryl hydrocarbon receptor activity of tryptophan metabolites in young adult mouse colonocytes. Drug Metab. Dispos. 2015, 43, 1536–1543. [Google Scholar] [CrossRef] [PubMed]
- Martin, F.P.; Wang, Y.; Sprenger, N.; Yap, I.K.; Lundstedt, T.; Lek, P.; Rezzi, S.; Ramadan, Z.; van Bladeren, P.; Fay, L.B.; et al. Probiotic modulation of symbiotic gut microbial-host metabolic interactions in a humanized microbiome mouse model. Mol. Syst. Biol. 2008, 4, 157. [Google Scholar] [CrossRef] [PubMed]
- Atanassova, M.R.; Fernandez-Otero, C.; Rodriguez-Alonso, P.; Fernandez-No, I.C.; Garabal, J.I.; Centeno, J.A. Characterization of yeasts isolated from artisanal short-ripened cows’ cheeses produced in Galicia (NW Spain). Food Microbiol. 2016, 53, 172–181. [Google Scholar] [CrossRef] [PubMed]
- Powell, J.J.; Carere, J.; Fitzgerald, T.L.; Stiller, J.; Covarelli, L.; Xu, Q.; Gubler, F.; Colgrave, M.L.; Gardiner, D.M.; Manners, J.M.; et al. The Fusarium crown rot pathogen Fusarium pseudograminearum triggers a suite of transcriptional and metabolic changes in bread wheat (Triticum aestivum L.). Ann. Bot. 2017, 119, 853–867. [Google Scholar] [PubMed]
- Kulkarni, G.B.; Sanjeevkumar, S.; Kirankumar, B.; Santoshkumar, M.; Karegoudar, T.B. Indole-3-acetic acid biosynthesis in fusarium delphinoides strain GPK, a causal agent of wilt in chickpea. Appl. Biochem. Biotechnol. 2013, 169, 1292–1305. [Google Scholar] [CrossRef] [PubMed]
- Suhr, M.J.; Banjara, N.; Hallen-Adams, H.E. Sequence-based methods for detecting and evaluating the human gut mycobiome. Lett. Appl. Microbiol. 2016, 62, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Gummer, J.P.; Trengove, R.D.; Oliver, R.P.; Solomon, P.S. Dissecting the role of G-protein signalling in primary metabolism in the wheat pathogen stagonospora nodorum. Microbiology 2013, 159, 1972–1985. [Google Scholar] [CrossRef] [PubMed]
- Paley, E.L.; Merkulova-Rainon, T.; Faynboym, A.; Shestopalov, V.I.; Aksenoff, I. Geographical distribution and diversity of gut microbial NADH: Ubiquinone oxidoreductase sequence associated with alzheimer’s disease. J. Alzheimers Dis. 2018, 61, 1531–1540. [Google Scholar] [CrossRef] [PubMed]
- Vogt, N.M.; Kerby, R.L.; Dill-McFarland, K.A.; Harding, S.J.; Merluzzi, A.P.; Johnson, S.C.; Carlsson, C.M.; Asthana, S.; Zetterberg, H.; Blennow, K.; et al. Gut microbiome alterations in alzheimer’s disease. Sci. Rep. 2017, 7, 13537. [Google Scholar] [CrossRef] [PubMed]
- Lagerkvist, U.; Akesson, B.; Branden, R. Aminoacyl adenylate, a normal intermediate or a dead end in aminoacylation of transfer ribonucleic acid. J. Biol. Chem. 1977, 252, 1002–1006. [Google Scholar] [PubMed]
- Shalaby, A.R. Changes in biogenic amines in mature and germinating legume seeds and their behavior during cooking. Die Nahr. 2000, 44, 23–27. [Google Scholar] [CrossRef]
- Ly, D.; Kang, K.; Choi, J.Y.; Ishihara, A.; Back, K.; Lee, S.G. HPLC analysis of serotonin, tryptamine, tyramine, and the hydroxycinnamic acid amides of serotonin and tyramine in food vegetables. J. Med. Food 2008, 11, 385–389. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Ding, X.; Qin, Y.; Zeng, Y. Safety assessment of the biogenic amines in fermented soya beans and fermented bean curd. J. Agric. Food. Chem. 2014, 62, 7947–7954. [Google Scholar] [CrossRef] [PubMed]
- Wust, N.; Rauscher-Gabernig, E.; Steinwider, J.; Bauer, F.; Paulsen, P. Risk assessment of dietary exposure to tryptamine for the Austrian population. Food Addit. Contam. Part A Chem. Anal. Control Expo. Risk Assess. 2017, 34, 404–420. [Google Scholar] [CrossRef] [PubMed]
- Renes, E.; Diezhandino, I.; Fernandez, D.; Ferrazza, R.E.; Tornadijo, M.E.; Fresno, J.M. Effect of autochthonous starter cultures on the biogenic amine content of ewe’s milk cheese throughout ripening. Food Microbiol. 2014, 44, 271–277. [Google Scholar] [CrossRef] [PubMed]
- Barry, K.A.; Wojcicki, B.J.; Middelbos, I.S.; Vester, B.M.; Swanson, K.S.; Fahey, G.C., Jr. Dietary cellulose, fructooligosaccharides, and pectin modify fecal protein catabolites and microbial populations in adult cats. J. Anim. Sci. 2010, 88, 2978–2987. [Google Scholar] [CrossRef] [PubMed]
- Maxwell, D.R.; Gray, W.R.; Taylor, E.M. Relative activity of some inhibitors of mono-amine oxidase in potentiating the action of tryptamine in vitro and in vivo. Br. J. Pharmacol. Chemother. 1961, 17, 310–320. [Google Scholar] [CrossRef] [PubMed]
- Yu, M.; Zhang, C.; Yang, Y.; Mu, C.; Su, Y.; Yu, K.; Zhu, W. Long-term effects of early antibiotic intervention on blood parameters, apparent nutrient digestibility, and fecal microbial fermentation profile in pigs with different dietary protein levels. J. Anim. Sci. Biotechnol. 2017, 8, 60. [Google Scholar] [CrossRef] [PubMed]
- Yao, K.; De Luca, V.; Brisson, N. Creation of a metabolic sink for tryptophan alters the phenylpropanoid pathway and the susceptibility of potato to phytophthora infestans. Plant Cell 1995, 7, 1787–1799. [Google Scholar] [CrossRef] [PubMed]
- Rubin, B.Y.; Anderson, S.L.; Xing, L.; Powell, R.J.; Tate, W.P. Interferon induces tryptophanyl-tRNA synthetase expression in human fibroblasts. J. Biol. Chem. 1991, 266, 24245–24248. [Google Scholar] [PubMed]
- Bianchi, L.; Puglia, M.; Landi, C.; Matteoni, S.; Perini, D.; Armini, A.; Verani, M.; Trombetta, C.; Soldani, P.; Roncada, P.; et al. Solubilization methods and reference 2-de map of cow milk fat globules. J. Proteomics 2009, 72, 853–864. [Google Scholar] [CrossRef] [PubMed]
- Nissen, A.; Bendixen, E.; Ingvartsen, K.L.; Rontved, C.M. Expanding the bovine milk proteome through extensive fractionation. J. Dairy Sci. 2013, 96, 7854–7866. [Google Scholar] [CrossRef] [PubMed]
- D’Alessandro, A.; Zolla, L.; Scaloni, A. The bovine milk proteome: Cherishing, nourishing and fostering molecular complexity. An interactomics and functional overview. Mol. Biosyst. 2011, 7, 579–597. [Google Scholar] [CrossRef] [PubMed]
- Paley, E.L.; Alexandrova, N.; Smelansky, L. Tryptophanyl-tRNA synthetase as a human autoantigen. Immunol. Lett. 1995, 48, 201–207. [Google Scholar] [CrossRef]
- Paley, E.L.; Paley, D.E.; Merkulova-Rainon, T.; Subbarayan, P.R. Hypoxia signature of splice forms of tryptophanyl-tRNA synthetase marks pancreatic cancer cells with distinct metastatic abilities. Pancreas 2011, 40, 1043–1056. [Google Scholar] [CrossRef] [PubMed]
- Paley, E.L.; Smelyanski, L.; Malinovskii, V.; Subbarayan, P.R.; Berdichevsky, Y.; Posternak, N.; Gershoni, J.M.; Sokolova, O.; Denisova, G. Mapping and molecular characterization of novel monoclonal antibodies to conformational epitopes on NH2 and COOH termini of mammalian tryptophanyl-tRNA synthetase reveal link of the epitopes to aggregation and Alzheimer’s disease. Mol. Immunol. 2007, 44, 541–557. [Google Scholar] [CrossRef] [PubMed]
- Nevinsky, G.A.; Favorova, O.O.; Lavrik, O.I.; Petrova, T.D.; Kochkina, L.L.; Savchenko, T.I. Fluorinated tryptophans as substrates and inhibitors of the ATP-[32P] PPi exchange reaction catalysed by tryptophanyl tRNA synthetase. FEBS Lett. 1974, 43, 135–138. [Google Scholar] [CrossRef]
- Efimov, I.; Basran, J.; Sun, X.; Chauhan, N.; Chapman, S.K.; Mowat, C.G.; Raven, E.L. The mechanism of substrate inhibition in human indoleamine 2,3-dioxygenase. J. Am. Chem. Soc. 2012, 134, 3034–3041. [Google Scholar] [CrossRef] [PubMed]
- Nienhaus, K.; Nienhaus, G.U. Different mechanisms of catalytic complex formation in two l-tryptophan processing dioxygenases. Front. Mol. Biosci. 2017, 4, 94. [Google Scholar] [CrossRef] [PubMed]
- Tidemand, K.D.; Peters, G.H.; Harris, P.; Stensgaard, E.; Christensen, H.E.M. Isoform-specific substrate inhibition mechanism of human tryptophan hydroxylase. Biochemistry 2017, 56, 6155–6164. [Google Scholar] [CrossRef] [PubMed]
- Ito, J.; Cox, E.C.; Yanofsky, C. Anthranilate synthetase, an enzyme specified by the tryptophan operon of escherichia coli: Purification and characterization of component I. J. Bacteriol. 1969, 97, 725–733. [Google Scholar] [PubMed]
- Singh, M.; Widholm, J.M. Study of a corn (Zea Mays L.) mutant (blue fluorescent-1) which accumulates anthranilic acid and its beta-glucoside. Biochem. Genet. 1975, 13, 357–367. [Google Scholar] [CrossRef] [PubMed]
- Malygin, E.G.; Zinoviev, V.V.; Fasiolo, F.; Kisselev, L.L.; Kochkina, L.L.; Achverdyan, V.Z. Interaction of aminoacyl-tRNA synthetases and tRNA: Positive and negative cooperativity of their active centres. Mol. Biol. Rep 1976, 2, 445–454. [Google Scholar] [CrossRef] [PubMed]
- Paley, E.L.; Baranov, V.N.; Alexandrova, N.M.; Kisselev, L.L. Tryptophanyl-tRNA synthetase in cell lines resistant to tryptophan analogs. Exp. Cell Res. 1991, 195, 66–78. [Google Scholar] [CrossRef]
- Paley, E.L. Chaperon-like activation of serum-inducible tryptophanyl-tRNA synthetase phosphorylation through refolding as a tool for analysis of clinical samples. Transl. Oncol. 2011, 4, 377–389. [Google Scholar] [CrossRef] [PubMed]
- Kisselev, L.L.; Favorova, O.O.; Kovaleva, G.K. Tryptophanyl-tRNA synthetase from beef pancreas. Methods Enzymol. 1979, 59, 234–257. [Google Scholar] [PubMed]
- Tuzikov, F.V.; Tuzikova, N.A.; Vavilin, V.I.; Zinov’ev, V.V.; Malygin, E.G.; Favorova, O.O.; Zargarova, T.A.; Sudomoina, M.A.; Kiselev, L.L. Aggregation of tryptophanyl-tRNA synthetase depending on temperature. Study by a low-angle scatter X-ray method. Mol. Biol. 1991, 25, 740–751. [Google Scholar]
- Iborra, F.; Dorizzi, M.; Labouesse, J. Tryptophanyl-transfer ribonucleic-acid synthetase from beef pancreas. Ligand binding and dissociation equilibrium between the active dimeric and inactive monomeric structures. Eur. J. Biochem. 1973, 39, 275–282. [Google Scholar] [CrossRef] [PubMed]
- Ahn, Y.H.; Park, S.; Choi, J.J.; Park, B.K.; Rhee, K.H.; Kang, E.; Ahn, S.; Lee, C.H.; Lee, J.S.; Inn, K.S.; et al. Secreted tryptophanyl-tRNA synthetase as a primary defence system against infection. Nat. Microbiol. 2016, 2, 16191. [Google Scholar] [CrossRef] [PubMed]
- Sharon, N.; Lipmann, F. Reactivity of analogs with pancreatic tryptophan-activating enzyme. Arch. Biochem. Biophys. 1957, 69, 219–227. [Google Scholar] [CrossRef]
- Dorizzi, M.; Labouesse, B.; Labouesse, J. Isolation and stoichiometry of beef pancreas tryptophanyl-tRNA synthetase complexes with tryptophan and tryptophanyladenylate. Eur. J. Biochem. 1971, 19, 563–572. [Google Scholar] [CrossRef] [PubMed]
- Herrera, F.; Martin, V.; Carrera, P.; Garcia-Santos, G.; Rodriguez-Blanco, J.; Rodriguez, C.; Antolin, I. Tryptamine induces cell death with ultrastructural features of autophagy in neurons and glia: Possible relevance for neurodegenerative disorders. Anat. Rec. A Discov. Mol. Cell. Evol. Biol. 2006, 288, 1026–1030. [Google Scholar] [CrossRef] [PubMed]
- Nelson, T.M.; Borgogna, J.C.; Michalek, R.D.; Roberts, D.W.; Rath, J.M.; Glover, E.D.; Ravel, J.; Shardell, M.D.; Yeoman, C.J.; Brotman, R.M. Cigarette smoking is associated with an altered vaginal tract metabolomic profile. Sci. Rep. 2018, 8, 852. [Google Scholar] [CrossRef] [PubMed]
- Cataldo, J.K.; Prochaska, J.J.; Glantz, S.A. Cigarette smoking is a risk factor for Alzheimer’s disease: An analysis controlling for tobacco industry affiliation. J. Alzheimers Dis. 2010, 19, 465–480. [Google Scholar] [CrossRef] [PubMed]
- Okuyama, S.; Chaki, S.; Yoshikawa, R.; Suzuki, Y.; Ogawa, S.; Imagawa, Y.; Kawashima, N.; Ikeda, Y.; Kumagai, T.; Nakazato, A.; et al. In vitro and in vivo characterization of the dopamine D4 receptor, serotonin 5-HT2A receptor and alpha-1 adrenoceptor antagonist (R)-(+)-2-Amino-4-(4-Fluorophenyl)-5-[1-[4-(4-Fluorophenyl)-4-Oxobutyl]Pyrrolidin-3-yl]Thiazole (NRA0045). J. Pharmacol. Exp. Ther. 1997, 282, 56–63. [Google Scholar] [PubMed]
- Mousseau, D.D.; McManus, D.J.; Baker, G.B.; Juorio, A.V.; Dewhurst, W.G.; Greenshaw, A.J. Effects of age and of chronic antidepressant treatment on [3H]tryptamine and [3H]dihydroalprenolol binding to rat cortical membranes. Cell. Mol. Neurobiol. 1993, 13, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Mousseau, D.D. Tryptamine: A metabolite of tryptophan implicated in various neuropsychiatric disorders. Metab. Brain Dis. 1993, 8, 1–44. [Google Scholar] [CrossRef] [PubMed]
- Samonina-Kosicka, J.; Kanska, M. Mechanistic studies of reactions catalysed by diamine oxidase using isotope effects. Isotopes Environ. Health Stud. 2013, 49, 357–364. [Google Scholar] [CrossRef] [PubMed]
- Paley, E.L. Tryptamine-mediated stabilization of tryptophanyl-tRNA synthetase in human cervical carcinoma cell line. Cancer Lett. 1999, 137, 1–7. [Google Scholar] [CrossRef]
- Juorio, A.V.; Durden, D.A. The distribution and turnover of tryptamine in the brain and spinal cord. Neurochem. Res. 1984, 9, 1283–1293. [Google Scholar] [CrossRef] [PubMed]
- Fontanilla, D.; Johannessen, M.; Hajipour, A.R.; Cozzi, N.V.; Jackson, M.B.; Ruoho, A.E. The hallucinogen N,N-dimethyltryptamine (DMT) is an endogenous sigma-1 receptor regulator. Science 2009, 323, 934–937. [Google Scholar] [CrossRef] [PubMed]
- Brown, T.; Shao, W.; Ayub, S.; Chong, D.; Cornelius, C. A physician’s attempt to self-medicate bipolar depression with N,N-dimethyltryptamine (DMT). J. Psychoact. Drugs 2017, 49, 294–296. [Google Scholar] [CrossRef] [PubMed]
- Sankaranarayanan, K.; Ozga, A.T.; Warinner, C.; Tito, R.Y.; Obregon-Tito, A.J.; Xu, J.; Gaffney, P.M.; Jervis, L.L.; Cox, D.; Stephens, L.; et al. Gut microbiome diversity among cheyenne and arapaho individuals from Western Oklahoma. Curr. Biol. 2015, 25, 3161–3169. [Google Scholar] [CrossRef] [PubMed]
- Demling, J.; Langer, K.; Mehr, M.Q. Age dependence of large neutral amino acid levels in plasma. Focus on tryptophan. Adv. Exp. Med. Biol. 1996, 398, 579–582. [Google Scholar] [PubMed]
- Luan, H.; Liu, L.F.; Meng, N.; Tang, Z.; Chua, K.K.; Chen, L.L.; Song, J.X.; Mok, V.C.; Xie, L.X.; Li, M.; et al. LC-MS-based urinary metabolite signatures in idiopathic Parkinson’s disease. J. Proteome Res. 2015, 14, 467–478. [Google Scholar] [CrossRef] [PubMed]
- Candia, O.A.; Alvarez, L.J.; Lanzetta, P.A.; Cook, P. Tryptamine in the vertebrate lens. Biochim. Biophys. Acta 1983, 762, 232–240. [Google Scholar] [CrossRef]
- Lai, S.W.; Lin, C.L.; Liao, K.F. Cataract may be a non-memory feature of Alzheimer’s disease in older people. Eur. J. Epidemiol. 2014, 29, 405–409. [Google Scholar] [CrossRef] [PubMed]
- Lai, S.W.; Lin, C.L.; Liao, K.F.; Chang-Ou, K.C. Increased risk of parkinson’s disease in cataract patients: A population-based cohort study. Parkinsonism Relat. Disord. 2015, 21, 68–71. [Google Scholar] [CrossRef] [PubMed]
- Marcobal, A.; Kashyap, P.C.; Nelson, T.A.; Aronov, P.A.; Donia, M.S.; Spormann, A.; Fischbach, M.A.; Sonnenburg, J.L. A metabolomic view of how the human gut microbiota impacts the host metabolome using humanized and gnotobiotic mice. ISME J. 2013, 7, 1933–1943. [Google Scholar] [CrossRef] [PubMed]
- Vikstrom Bergander, L.; Cai, W.; Klocke, B.; Seifert, M.; Pongratz, I. Tryptamine serves as a proligand of the AHR transcriptional pathway whose activation is dependent of monoamine oxidases. Mol. Endocrinol. 2012, 26, 1542–1551. [Google Scholar] [CrossRef] [PubMed]
- Saraf, M.K.; Piccolo, B.D.; Bowlin, A.K.; Mercer, K.E.; LeRoith, T.; Chintapalli, S.V.; Shankar, K.; Badger, T.M.; Yeruva, L. Formula diet driven microbiota shifts tryptophan metabolism from serotonin to tryptamine in neonatal porcine colon. Microbiome 2017, 5, 77. [Google Scholar] [CrossRef] [PubMed]
- Swanson, K.S.; Grieshop, C.M.; Flickinger, E.A.; Bauer, L.L.; Healy, H.P.; Dawson, K.A.; Merchen, N.R.; Fahey, G.C., Jr. Supplemental fructooligosaccharides and mannanoligosaccharides influence immune function, ileal and total tract nutrient digestibilities, microbial populations and concentrations of protein catabolites in the large bowel of dogs. J. Nutr. 2002, 132, 980–989. [Google Scholar] [CrossRef] [PubMed]
- Crawford, C.; Sepulveda, M.F.; Elliott, J.; Harris, P.A.; Bailey, S.R. Dietary fructan carbohydrate increases amine production in the equine large intestine: Implications for pasture-associated laminitis. J. Anim. Sci. 2007, 85, 2949–2958. [Google Scholar] [CrossRef] [PubMed]
- Mao, S.Y.; Huo, W.J.; Zhu, W.Y. Microbiome-metabolome analysis reveals unhealthy alterations in the composition and metabolism of ruminal microbiota with increasing dietary grain in a goat model. Environ. Microbiol. 2016, 18, 525–541. [Google Scholar] [CrossRef] [PubMed]
- Moshfegh, A.J.; Friday, J.E.; Goldman, J.P.; Ahuja, J.K. Presence of inulin and oligofructose in the diets of Americans. J. Nutr. 1999, 129, 1407S–1411S. [Google Scholar] [CrossRef] [PubMed]
- Ndeh, D.; Rogowski, A.; Cartmell, A.; Luis, A.S.; Basle, A.; Gray, J.; Venditto, I.; Briggs, J.; Zhang, X.; Labourel, A.; et al. Complex pectin metabolism by gut bacteria reveals novel catalytic functions. Nature 2017, 544, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Brito, J.S.; Borges, N.A.; Esgalhado, M.; Magliano, D.C.; Soulage, C.O.; Mafra, D. Aryl hydrocarbon receptor activation in chronic kidney disease: Role of uremic toxins. Nephron 2017, 137, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Gondouin, B.; Cerini, C.; Dou, L.; Sallee, M.; Duval-Sabatier, A.; Pletinck, A.; Calaf, R.; Lacroix, R.; Jourde-Chiche, N.; Poitevin, S.; et al. Indolic uremic solutes increase tissue factor production in endothelial cells by the aryl hydrocarbon receptor pathway. Kidney Int. 2013, 84, 733–744. [Google Scholar] [CrossRef] [PubMed]
- Lamas, B.; Richard, M.L.; Leducq, V.; Pham, H.P.; Michel, M.L.; Da Costa, G.; Bridonneau, C.; Jegou, S.; Hoffmann, T.W.; Natividad, J.M.; et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat. Med. 2016, 22, 598–605. [Google Scholar] [CrossRef] [PubMed]
- Koshima, H.; Honke, S. Chiral bimolecular crystallization of tryptamine and achiral carboxylic acids. J. Org. Chem. 1999, 64, 790–793. [Google Scholar] [CrossRef] [PubMed]
- Martinez, E.; Artigas, F.; Sunol, C.; Tusell, J.M.; Gelpi, E. Liquid-chromatographic determination of indole-3-acetic acid and 5-hydroxyindole-3-acetic acid in human plasma. Clin. Chem. 1983, 29, 1354–1357. [Google Scholar] [PubMed]
- Yokoyama, M.T.; Carlson, J.R. Dissimilation of tryptophan and related indolic compounds by ruminal microorganisms in vitro. Appl. Microbiol. 1974, 27, 540–548. [Google Scholar] [PubMed]
- Nguyen, L.P.; Hsu, E.L.; Chowdhury, G.; Dostalek, M.; Guengerich, F.P.; Bradfield, C.A. D-amino acid oxidase generates agonists of the aryl hydrocarbon receptor from D-tryptophan. Chem. Res. Toxicol. 2009, 22, 1897–1904. [Google Scholar] [CrossRef] [PubMed]
- Behr, C.; Kamp, H.; Fabian, E.; Krennrich, G.; Mellert, W.; Peter, E.; Strauss, V.; Walk, T.; Rietjens, I.M.; van Ravenzwaay, B. Gut microbiome-related metabolic changes in plasma of antibiotic-treated rats. Arch. Toxicol. 2017, 91, 3439–3454. [Google Scholar] [CrossRef] [PubMed]
- Wikoff, W.R.; Anfora, A.T.; Liu, J.; Schultz, P.G.; Lesley, S.A.; Peters, E.C.; Siuzdak, G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc. Natl. Acad. Sci. USA 2009, 106, 3698–3703. [Google Scholar] [CrossRef] [PubMed]
- de Mello, V.D.; Paananen, J.; Lindstrom, J.; Lankinen, M.A.; Shi, L.; Kuusisto, J.; Pihlajamaki, J.; Auriola, S.; Lehtonen, M.; Rolandsson, O.; et al. Indolepropionic acid and novel lipid metabolites are associated with a lower risk of type 2 diabetes in the finnish diabetes prevention study. Sci. Rep. 2017, 7, 46337. [Google Scholar] [CrossRef] [PubMed]
- Kepert, I.; Fonseca, J.; Muller, C.; Milger, K.; Hochwind, K.; Kostric, M.; Fedoseeva, M.; Ohnmacht, C.; Dehmel, S.; Nathan, P.; et al. D-tryptophan from probiotic bacteria influences the gut microbiome and allergic airway disease. J. Allergy Clin. Immunol. 2017, 139, 1525–1535. [Google Scholar] [CrossRef] [PubMed]
- Soutourina, J.; Plateau, P.; Blanquet, S. Metabolism of d-aminoacyl-tRNAs in escherichia coli and saccharomyces cerevisiae cells. J. Biol. Chem. 2000, 275, 32535–32542. [Google Scholar] [CrossRef] [PubMed]
- Hatanaka, T.; Huang, W.; Nakanishi, T.; Bridges, C.C.; Smith, S.B.; Prasad, P.D.; Ganapathy, M.E.; Ganapathy, V. Transport of D-serine via the amino acid transporter ATB0,+ expressed in the colon. Biochem. Biophys. Res. Commun. 2002, 291, 291–295. [Google Scholar] [CrossRef] [PubMed]
- Friedman, M. Chemistry, nutrition, and microbiology of D-amino acids. J. Agric. Food Chem. 1999, 47, 3457–3479. [Google Scholar] [CrossRef] [PubMed]
- Genchi, G. An overview on D-amino acids. Amino Acids 2017, 49, 1521–1533. [Google Scholar] [CrossRef] [PubMed]
- Berka, R.M.; Cui, X.; Yanofsky, C. Genomewide transcriptional changes associated with genetic alterations and nutritional supplementation affecting tryptophan metabolism in bacillus subtilis. Proc. Natl. Acad. Sci. USA 2003, 100, 5682–5687. [Google Scholar] [CrossRef] [PubMed]
- French, S.; Martin, K.; Patterson, T.; Bauerle, R.; Miller, O.L., Jr. Electron microscopic visualization of trp operon expression in Salmonella typhimurium. Proc. Natl. Acad. Sci. USA 1985, 82, 4638–4642. [Google Scholar] [CrossRef] [PubMed]
- Matchett, W.H. Inhibition of tryptophan synthetase by indoleacrylic acid. J. Bacteriol. 1972, 110, 146–154. [Google Scholar] [PubMed]
- Li, A.; Guo, X.; Xie, J.; Liu, X.; Zhang, Z.; Li, Y.; Zhang, Y. Validation of biomarkers in cardiotoxicity induced by periplocin on neonatal rat cardiomyocytes using UPLC-Q-TOF/MS combined with a support vector machine. J. Pharm. Biomed. Anal. 2016, 123, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Wlodarska, M.; Luo, C.; Kolde, R.; d’Hennezel, E.; Annand, J.W.; Heim, C.E.; Krastel, P.; Schmitt, E.K.; Omar, A.S.; Creasey, E.A.; et al. Indoleacrylic acid produced by commensal peptostreptococcus species suppresses inflammation. Cell Host Microbe 2017, 22, 25–37.e26. [Google Scholar] [CrossRef] [PubMed]
- Khodursky, A.B.; Peter, B.J.; Cozzarelli, N.R.; Botstein, D.; Brown, P.O.; Yanofsky, C. DNA microarray analysis of gene expression in response to physiological and genetic changes that affect tryptophan metabolism in escherichia coli. Proc. Natl. Acad. Sci. USA 2000, 97, 12170–12175. [Google Scholar] [CrossRef] [PubMed]
- Jensen, M.T.; Cox, R.P.; Jensen, B.B. 3-methylindole (skatole) and indole production by mixed populations of pig fecal bacteria. Appl. Environ. Microbiol. 1995, 61, 3180–3184. [Google Scholar] [PubMed]
- Whitehead, T.R.; Price, N.P.; Drake, H.L.; Cotta, M.A. Catabolic pathway for the production of skatole and indoleacetic acid by the acetogen clostridium drakei, clostridium scatologenes, and swine manure. Appl. Environ. Microbiol. 2008, 74, 1950–1953. [Google Scholar] [CrossRef] [PubMed]
- Bray, T.M.; Emmerson, K.S. Putative mechanisms of toxicity of 3-methylindole: From free radical to pneumotoxicosis. Annu. Rev. Pharmacol. Toxicol. 1994, 34, 91–115. [Google Scholar] [CrossRef] [PubMed]
- Rasmussen, M.K.; Balaguer, P.; Ekstrand, B.; Daujat-Chavanieu, M.; Gerbal-Chaloin, S. Skatole (3-methylindole) is a partial aryl hydrocarbon receptor agonist and induces CYP1A1/2 and CYP1B1 expression in primary human hepatocytes. PLoS ONE 2016, 11, e0154629. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.W.; Hong, S.L.; Lee, C.H.; Jeon, E.H.; Choi, A.R. Relationship between olfactory function and olfactory neuronal population in C57Bl6 mice injected intraperitoneally with 3-methylindole. Otolaryngol. Head Neck Surg. 2010, 143, 837–842. [Google Scholar] [CrossRef] [PubMed]
- Hammond, A.C.; Carlson, J.R. Inhibition of ruminal degradation of l-tryptophan to 3-methylindole, in vitro. J. Anim. Sci. 1980, 51, 207–214. [Google Scholar] [CrossRef] [PubMed]
- Osborn, M.P.; Park, Y.; Parks, M.B.; Burgess, L.G.; Uppal, K.; Lee, K.; Jones, D.P.; Brantley, M.A., Jr. Metabolome-wide association study of neovascular age-related macular degeneration. PLoS ONE 2013, 8, e72737. [Google Scholar] [CrossRef] [PubMed]
- Sung, C.T.; Chang, S.L.; Entwistle, R.; Ahn, G.; Lin, T.S.; Petrova, V.; Yeh, H.H.; Praseuth, M.B.; Chiang, Y.M.; Oakley, B.R.; et al. Overexpression of a three-gene conidial pigment biosynthetic pathway in aspergillus nidulans reveals the first nrps known to acetylate tryptophan. Fungal Genet. Biol. 2017, 101, 1–6. [Google Scholar] [CrossRef] [PubMed]
- Tammineni, P.; Cai, Q. Defective retrograde transport impairs autophagic clearance in Alzheimer disease neurons. Autophagy 2017, 13, 982–984. [Google Scholar] [CrossRef] [PubMed]
- Kish, S.J.; Mastrogiacomo, F.; Guttman, M.; Furukawa, Y.; Taanman, J.W.; Dozic, S.; Pandolfo, M.; Lamarche, J.; DiStefano, L.; Chang, L.J. Decreased brain protein levels of cytochrome oxidase subunits in Alzheimer’s disease and in hereditary spinocerebellar ataxia disorders: A nonspecific change? J. Neurochem. 1999, 72, 700–707. [Google Scholar] [CrossRef] [PubMed]
- Buchan, J.R.; Stansfield, I. Halting a cellular production line: Responses to ribosomal pausing during translation. Biol. Cell 2007, 99, 475–487. [Google Scholar] [CrossRef] [PubMed]
- Monaco, F.; Fumero, S.; Mondino, A.; Mutani, R. Plasma and cerebrospinal fluid tryptophan in multiple sclerosis and degenerative diseases. J. Neurol. Neurosurg. Psychiatry 1979, 42, 640–641. [Google Scholar] [CrossRef] [PubMed]
- Pratt, J.A.; Jenner, P.; Johnson, A.L.; Shorvon, S.D.; Reynolds, E.H. Anticonvulsant drugs alter plasma tryptophan concentrations in epileptic patients: Implications for antiepileptic action and mental function. J. Neurol. Neurosurg. Psychiatry 1984, 47, 1131–1133. [Google Scholar] [CrossRef] [PubMed]
- Theisen, B.E.; Rumyantseva, A.; Cohen, J.S.; Alcaraz, W.A.; Shinde, D.N.; Tang, S.; Srivastava, S.; Pevsner, J.; Trifunovic, A.; Fatemi, A. Deficiency of WARS2, encoding mitochondrial tryptophanyl tRNA synthetase, causes severe infantile onset leukoencephalopathy. Am. J. Med. Genet. A 2017, 173, 2505–2510. [Google Scholar] [CrossRef] [PubMed]
- Musante, L.; Puttmann, L.; Kahrizi, K.; Garshasbi, M.; Hu, H.; Stehr, H.; Lipkowitz, B.; Otto, S.; Jensen, L.R.; Tzschach, A.; et al. Mutations of the aminoacyl-tRNA-synthetases SARS and WARS2 are implicated in the etiology of autosomal recessive intellectual disability. Hum. Mutat. 2017, 38, 621–636. [Google Scholar] [CrossRef] [PubMed]
- Wortmann, S.B.; Timal, S.; Venselaar, H.; Wintjes, L.T.; Kopajtich, R.; Feichtinger, R.G.; Onnekink, C.; Muhlmeister, M.; Brandt, U.; Smeitink, J.A.; et al. Biallelic variants in WARS2 encoding mitochondrial tryptophanyl-tRNA synthase in six individuals with mitochondrial encephalopathy. Hum. Mutat. 2017, 38, 1786–1795. [Google Scholar] [CrossRef] [PubMed]
- Burke, E.A.; Frucht, S.J.; Thompson, K.; Wolfe, L.A.; Yokoyama, T.; Bertoni, M.; Huang, Y.; Sincan, M.; Adams, D.R.; Taylor, R.W.; et al. Biallelic mutations in mitochondrial tryptophanyl-tRNA synthetase cause Levodopa-rresponsive infantile-onset Parkinsonism. Clin. Genet. 2017, 93, 712–718. [Google Scholar] [CrossRef] [PubMed]
- Tsai, P.C.; Soong, B.W.; Mademan, I.; Huang, Y.H.; Liu, C.R.; Hsiao, C.T.; Wu, H.T.; Liu, T.T.; Liu, Y.T.; Tseng, Y.T.; et al. A recurrent WARS mutation is a novel cause of autosomal dominant distal hereditary motor neuropathy. Brain 2017, 140, 1252–1266. [Google Scholar] [CrossRef] [PubMed]
- Mizuguchi, T.; Nakashima, M.; Kato, M.; Yamada, K.; Okanishi, T.; Ekhilevitch, N.; Mandel, H.; Eran, A.; Toyono, M.; Sawaishi, Y.; et al. PARS2 and NARS2 mutations in infantile-onset neurodegenerative disorder. J. Hum. Genet. 2017, 62, 525–529. [Google Scholar] [CrossRef] [PubMed]
- Cox, P.A.; Davis, D.A.; Mash, D.C.; Metcalf, J.S.; Banack, S.A. Dietary exposure to an environmental toxin triggers neurofibrillary tangles and amyloid deposits in the brain. Proc. Biol. Sci. 2016, 283, 20152397. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, F.H.; Fak, F.; Nookaew, I.; Tremaroli, V.; Fagerberg, B.; Petranovic, D.; Backhed, F.; Nielsen, J. Symptomatic atherosclerosis is associated with an altered gut metagenome. Nat. Commun. 2012, 3, 1245. [Google Scholar] [CrossRef] [PubMed]
- McVeigh, C.; Passmore, P. Vascular dementia: Prevention and treatment. Clin. Interv. Aging 2006, 1, 229–235. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Lucas-Osma, A.M.; Black, S.; Bandet, M.V.; Stephens, M.J.; Vavrek, R.; Sanelli, L.; Fenrich, K.K.; Di Narzo, A.F.; Dracheva, S.; et al. Pericytes impair capillary blood flow and motor function after chronic spinal cord injury. Nat. Med. 2017, 23, 733–741. [Google Scholar] [CrossRef] [PubMed]
- Anwar, M.A.; Ford, W.R.; Broadley, K.J.; Herbert, A.A. Vasoconstrictor and vasodilator responses to tryptamine of rat-isolated perfused mesentery: Comparison with tyramine and beta-phenylethylamine. Br. J. Pharmacol. 2012, 165, 2191–2202. [Google Scholar] [CrossRef] [PubMed]
- Anwar, M.A.; Ford, W.R.; Herbert, A.A.; Broadley, K.J. Signal transduction and modulating pathways in tryptamine-evoked vasopressor responses of the rat isolated perfused mesenteric bed. Vascul. Pharmacol. 2013, 58, 140–149. [Google Scholar] [CrossRef] [PubMed]
- Broadley, K.J. The vascular effects of trace amines and amphetamines. Pharmacol. Ther. 2010, 125, 363–375. [Google Scholar] [CrossRef] [PubMed]
- Repetto, S.; Ambrosetti, P. Changes of urinary tryptamine in angina pectoris. Minerva Med. 1980, 71, 1203. [Google Scholar] [PubMed]
- Mashkovskii, M.D.; Lanskii, V.P. The effect of precursors and various analogs of serotonin on the cerebral circulation. Biull. Eksp. Biol. Med. 1968, 66, 58–61. [Google Scholar] [CrossRef] [PubMed]
- Desai, B.S.; Schneider, J.A.; Li, J.L.; Carvey, P.M.; Hendey, B. Evidence of angiogenic vessels in Alzheimer’s disease. J. Neural. Transm. 2009, 116, 587–597. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.Z.; Nawaz, W. The emerging roles of human trace amines and human trace amine-associated receptors (hTAARs) in central nervous system. Biomed. Pharmacother. 2016, 83, 439–449. [Google Scholar] [CrossRef] [PubMed]
- Zinoviev, V.V.; Rubtsova, N.G.; Lavrik, O.I.; Malygin, E.G.; Akhverdyan, V.Z.; Favorova, O.O.; Kisselev, L.L. Comparison of the atp-[32p]pyrophosphate exchange reactions catalysed by native (two-site) and chemically modified (one-site) tryptophanyl-trna synthetase. FEBS Lett. 1977, 82, 130–134. [Google Scholar] [CrossRef]
- Graves, P.V.; Mazat, J.P.; Juguelin, H.; Labouesse, J.; Labouesse, B. Anticooperative binding of l-tryptophan to tryptophanyl-tRNA synthetase from beef pancreas. Study at equilibrium by dialysis and changes in spectroscopic properties. Eur. J. Biochem. 1979, 96, 509–518. [Google Scholar] [CrossRef] [PubMed]
- Mazat, J.P.; Merle, M.; Graves, P.V.; Merault, G.; Gandar, J.C.; Labouesse, B. Kinetic anticooperativity in pre-steady-state formation of tryptophanyl adenylate by tryptophanyl-tRNA synthetase from beef pancreas. A consequence of the tryptophan anticooperative binding. Eur. J. Biochem. 1982, 128, 389–398. [Google Scholar] [CrossRef] [PubMed]
- Nevinsky, G.A.; Favorova, O.O.; Lavrik, O.I.; Petrova, T.D.; Kochkina, L.L.; Savchenko, T.I. Fluorinated tryptophans as substrates and inhibitors of the ATP--(32p)ppi exchange reaction catalysed by tryptophanyl tRNA synthetase. FEBS Lett. 1974, 43, 135–138. [Google Scholar] [CrossRef]
- Favorova, O.O.; Kochkina, L.L.; Meldrajs, J.A.; Kisselev, L.L.; Zinoviev, V.V. Kinetic parameters of tryptophan: tRNA ligase catalyzed ATP-(32p) pyrophosphate exchange as an approach to extimation of the order of substrate binding. FEBS Lett. 1975, 56, 322–326. [Google Scholar] [CrossRef]
| Inhibitor | Ki (M) |
|---|---|
| 5,7-Difluorotryptophan | 2 × 10−5 ± 0.5 a |
| 4,5,6,7-Tetrafluorotryptophan | 1.2 × 10−5 ± 0.3 a |
| d-Tryptophan | 5 × 10−5 a |
| Tryptamine | 6 × 10−5 a,# |
| β-Indolylacetic acid | 9 × 10−3 b |
| β-Indolylpropionic acid | 8.5 × 10−3 b |
| β-Indolylpyruvic acid | 5 × 10−4 b |
| N-Formyl-l-tryptophan | 4.6 × 10−4 b |
| N-Acetyl-l-tryptophan | 2.5 × 10−4 b |
| Adenine | 1.8 × 10−2 a |
| Adenosine | 3.1 × 10−3 a |
| AD/CN PLASMA | AD/MCI PLASMA | AD/CN CSF | AD/MCI CSF | MCI/CN PLASMA | MCI/CN CSF |
|---|---|---|---|---|---|
| Increase | |||||
| Met His Lys * | Gly His | Pro Pro $ | Tyr Pro § | Ala Leu ## | Pro Pro $ |
| Val Ser Lys | Arg Asn Gln | Tyr Tyr Thr | Asn Gly Ser | Trp Ala Ile © | Gln Pro Lys |
| Phe Ala Arg | Phe Val Val | ||||
| Met Glu Cys | Thr Ser Gln | ||||
| Thr Ser Gln | Glu Ser # | ||||
| Glu Ser # | |||||
| Val Gly | |||||
| Decrease | |||||
| Cys Tyr Cys | Trp Gly Phe ©! | Ala Phe Arg | Pro Lys Pro ** | Ile Ser Lys | Leu Leu Ala |
| Trp Gly Phe ©! | Ala Leu ## | Pro Lys Pro ** | Asp Asn Glu | Asn Gln Gln | |
| Ser Asp Gly | Thr Gly | Leu Glu Gln | Ala Met Lys | ||
| Met Trp Gln ©! | Asp Glu | Met Trp Gln ©! | Glu Ser # | ||
| Met His Lys * | Glu Ser # | Ala Thr Pro | Ala Ala Asp | ||
| Cys Cys Tyr | Tyr Pro § | ||||
| Arg Cys Cys | |||||
| Met Ala His | |||||
| Protein/Peptide | Amino Acids | Function | Database ID |
|---|---|---|---|
| COX subunit VI-c | 75 | electron transfer | Swiss-Prot: P09669.2 |
| tau protein | 758 | microtubule-associated | Swiss-Prot: P10636 |
| islet amyloid peptide | 89 | pro-amylin glycemic | GenBank: AAA52281 |
| beta-amyloid peptide | 40 | not understood | Swiss-Prot: P86906.1 |
| prion protein | 108 | controversial | PDB: 1I4M_A |
| alpha-synuclein | 140 | not understood | GenBank: NP_000336 |
| beta-synuclein | 134 | unknown | GenBank:NP_001001502 |
| gamma-synuclein | 127 | unknown | GenBank: AAL05870 |
| collagen, type XXV | 645 | AD plaque component | GenBank:EAX06240 |
| ubiquitin | 156 | regulation | GenBank: CAA44911 |
| S100B | 92 | regulation | GenBank: CAG46920 |
| histone H2A | 130 | chromatin structure | GenBank: CAA58539 |
| histone H3 | 136 | chromatin structure | GenBank: CAB02546 |
| neurofilament medium | 540 | cytoskeleton | GenBank:NP_001099011 |
| myelin basic protein | 160 | myelination | GenBank: NP_001020263 |
| arrestin | 409 | signal trunsduction | GenBank: CAA77577 |
| TATA box binding | 338 | transcription factor | GenBank: AAI09054 |
| calcitonin | 141 | hormone | GenBank: NP_001029124 |
| thyroid hormone | 138 | stimulating hormone | GenBank: AAH69298 |
| glycoprotein hormones | 116 | hormone | GenBank: NP_000726 |
| oxytocin | 125 | hormone | GenBank: AAI01844 |
| arginine vasopressin | 164 | hormone | GenBank: AAI26197 |
| prothymosin alpha | 111 | immunity | GenBank NP_001092755 |
| snapin | 136 | synaptic transmission | GenBank: AAD11417.1 |
| Interleukin-9 | 144 | cytokine | GenBank: AAH66284.1 |
| interleukin-18 | 189 | Increased in AD | NCBI: NP_001230140.1 |
| epidermal growth factor | 71 | Increased in AD | GenBank: CAA34902.2 |
| interleukin-2 isoform X1 | 131 | lymphokine | NCBI: XP_016863666.1 |
| C-X-C motif chemokine 10 precursor | 98 | Cytokine, elevated in AD | NCBI: NP_001556.2 |
| NADH dehydrogenase (ubiquinone) flavoprotein 3 | 108 | mitochondrial isoform b Renal carcinoma antigen NY-REN-4 | NCBI: NP_001001503.1 |
| NADH dehydrogenase (ubiquinone) flavoprotein 3 | 473 | mitochondrial isoform a precursor | NCBI: NP_066553.3 |
| NADH dehydrogenase (ubiquinone) iron-sulfur | 124 | mitochondrial precursor protein 6 | NCBI: NP_004544.1 |
| NADH dehydrogenase | 119 | NADHDH2 | GenBank: AAP97198.1 |
| NADH dehydrogenase | 210 | Human gut metagenom | GenBank: EKC78685.1 |
| NADH dehydrogenase | 167 | human gut metagenom | GenBank: EKC44884.1 |
| syntaxin | 259 | synaptic vesicles REN31 | PIR: G01485 |
| syntaxin-2 isoform 3 | 277 | synaptic vesicles | NCBI: NP_001337978 |
| syntaxin-3 isoform 1 | 289 | synaptic vesicles | NP_004168.1 |
| GTPase HRas | 189 | regulating cell division | Swiss-Prot: P01112.1 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paley, E.L.; Perry, G. Towards an Integrative Understanding of tRNA Aminoacylation–Diet–Host–Gut Microbiome Interactions in Neurodegeneration. Nutrients 2018, 10, 410. https://doi.org/10.3390/nu10040410
Paley EL, Perry G. Towards an Integrative Understanding of tRNA Aminoacylation–Diet–Host–Gut Microbiome Interactions in Neurodegeneration. Nutrients. 2018; 10(4):410. https://doi.org/10.3390/nu10040410
Chicago/Turabian StylePaley, Elena L., and George Perry. 2018. "Towards an Integrative Understanding of tRNA Aminoacylation–Diet–Host–Gut Microbiome Interactions in Neurodegeneration" Nutrients 10, no. 4: 410. https://doi.org/10.3390/nu10040410






