Impact of Maternal Malnutrition on Gut Barrier Defense: Implications for Pregnancy Health and Fetal Development
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Model
2.2. Maternal SI and Fetal Gut Collection
2.3. RNA Extraction and mRNA Expression Analysis
2.4. Expression and Localization of Lysozyme Protein and Quantification of Goblet Cells in Maternal SI
2.5. Data Analysis
3. Results
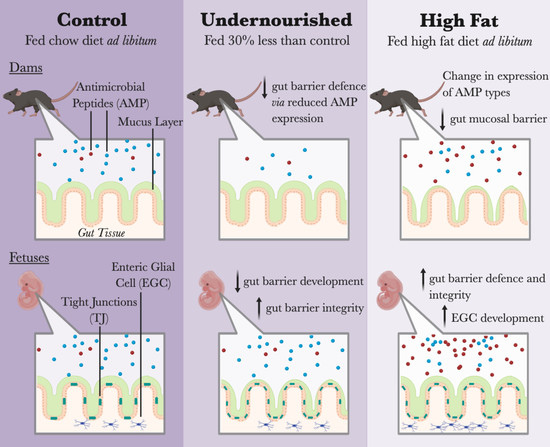
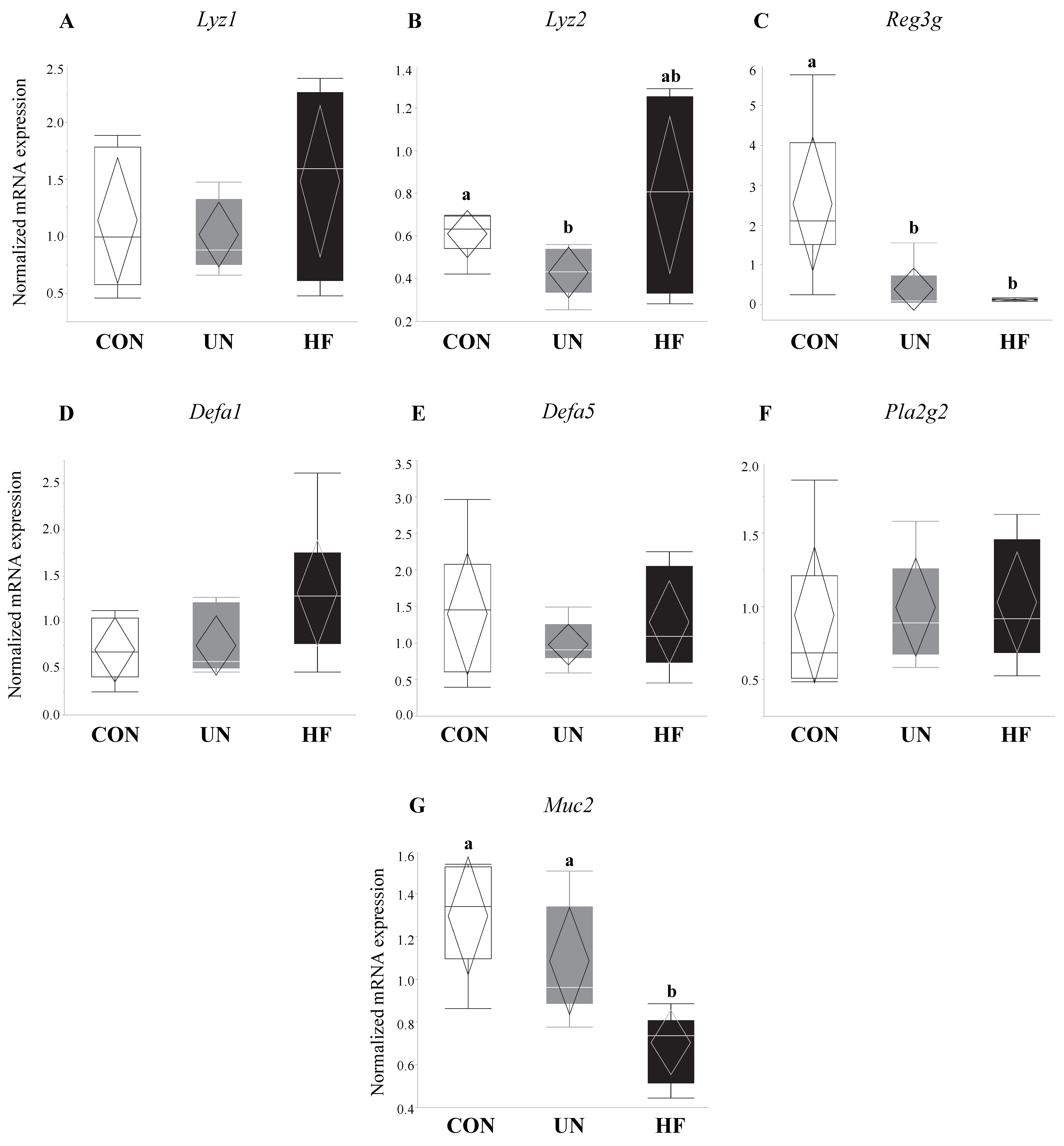
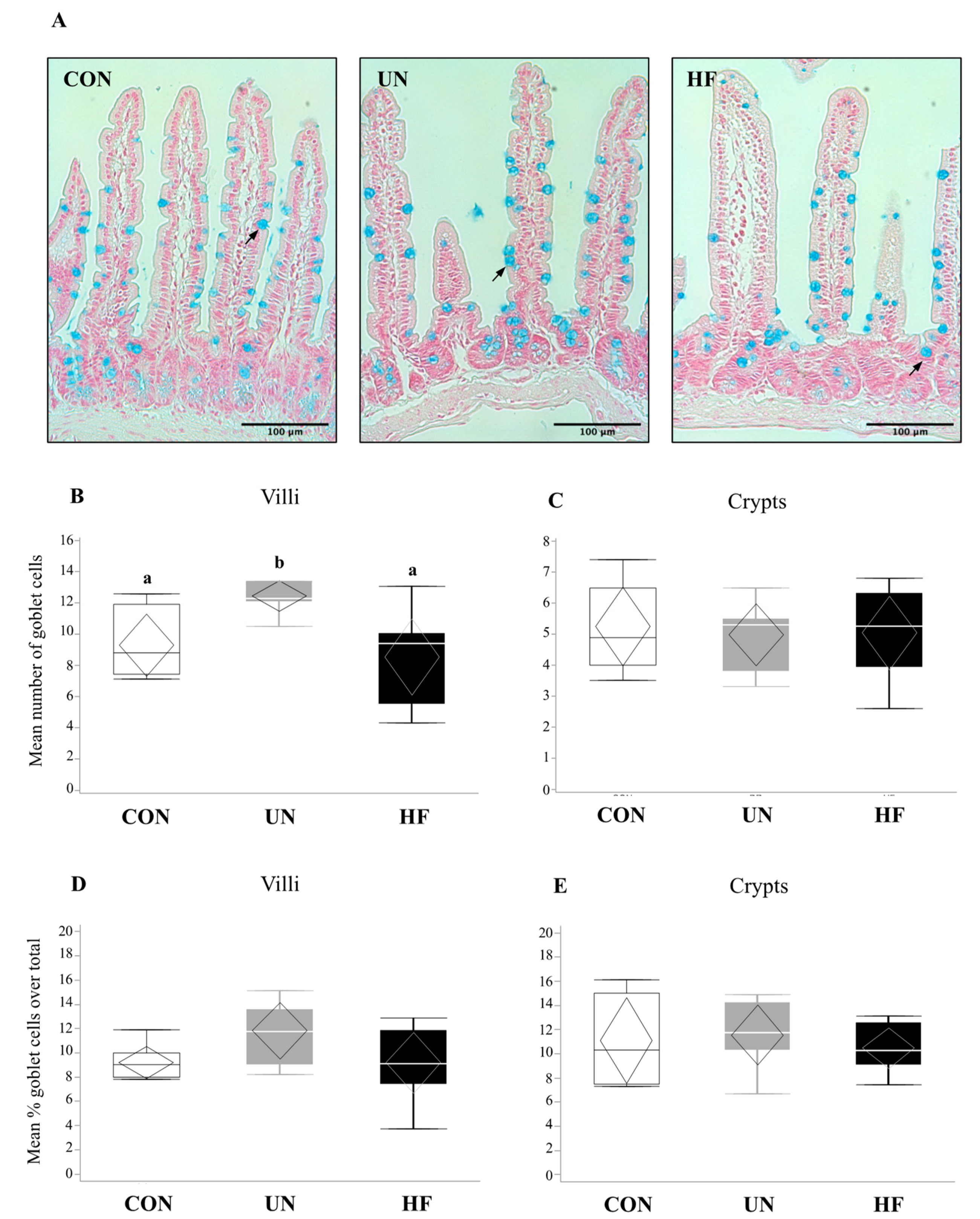
3.1. Malnutrition Was Associated with Reduced Gut Barrier Function and Integrity
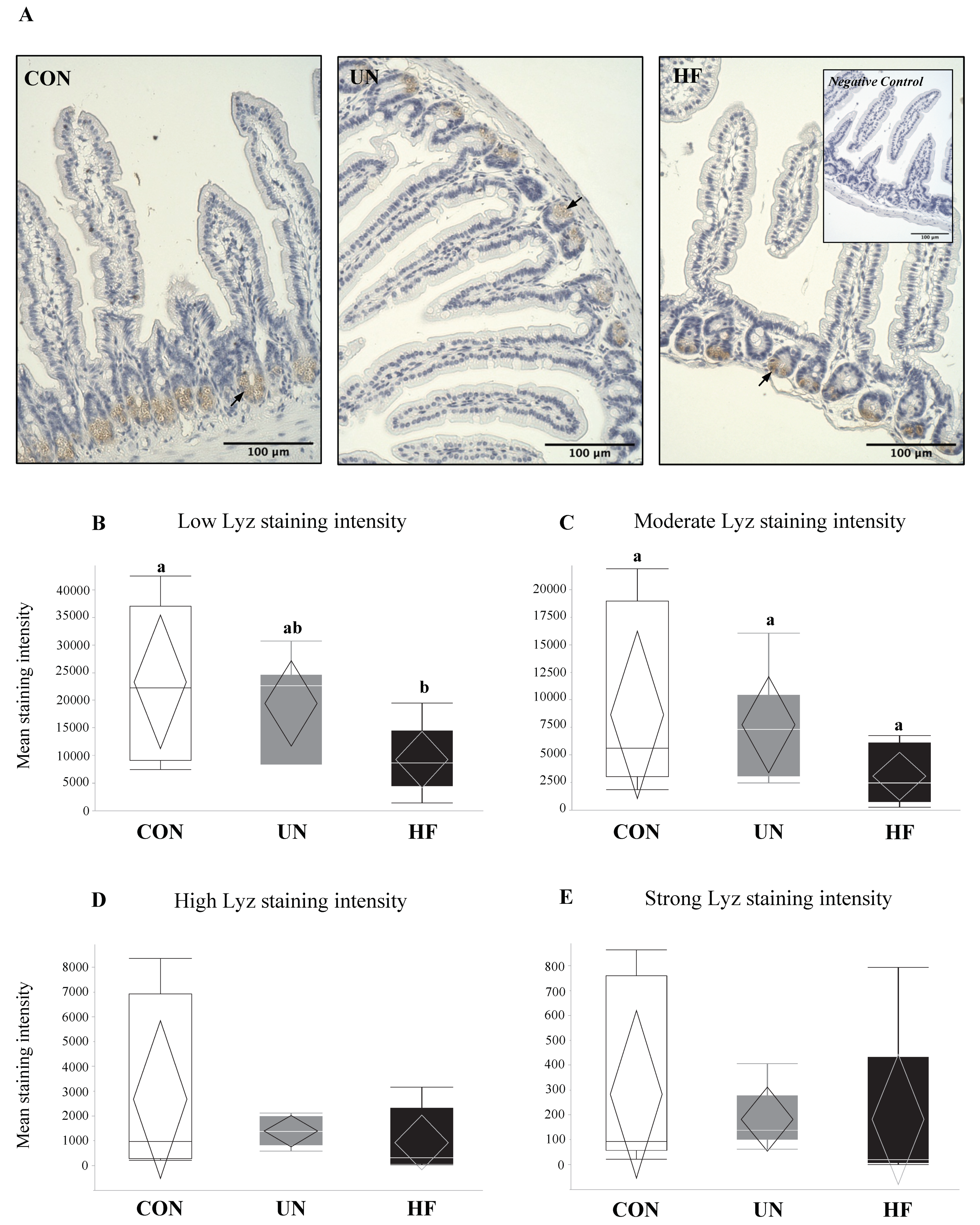
3.2. Maternal HF Diet May Be Associated with Increased Lyz Production
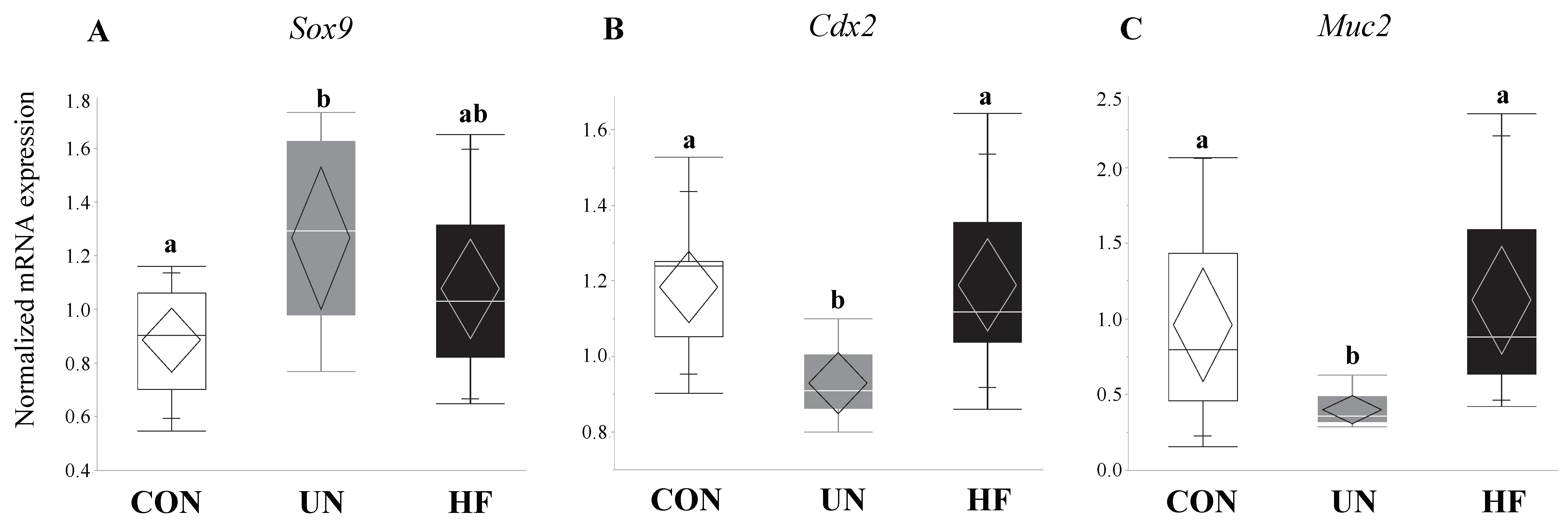
3.3. Maternal UN Was Associated with Delayed Fetal Gut Development and Reduced Mucus Production
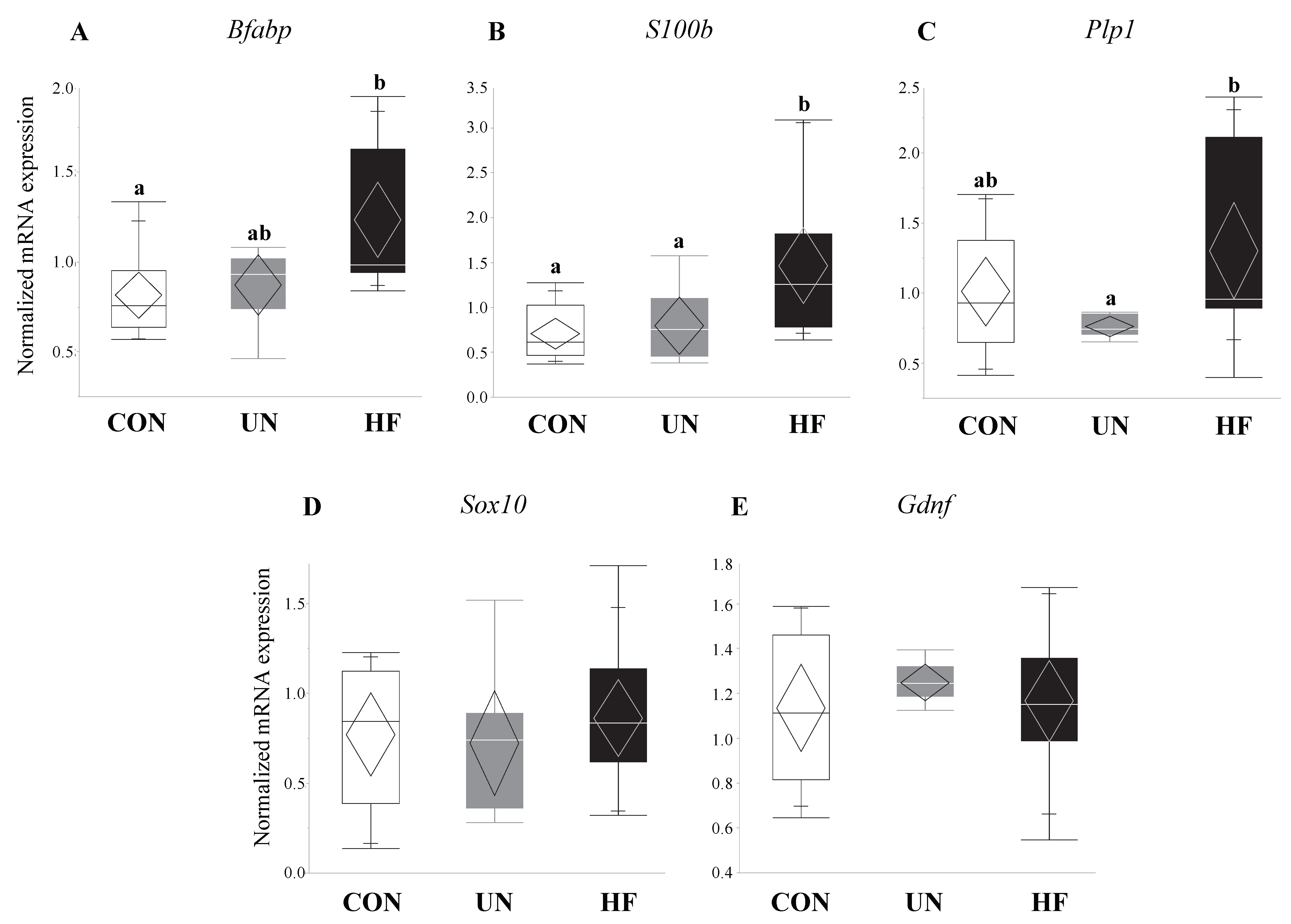
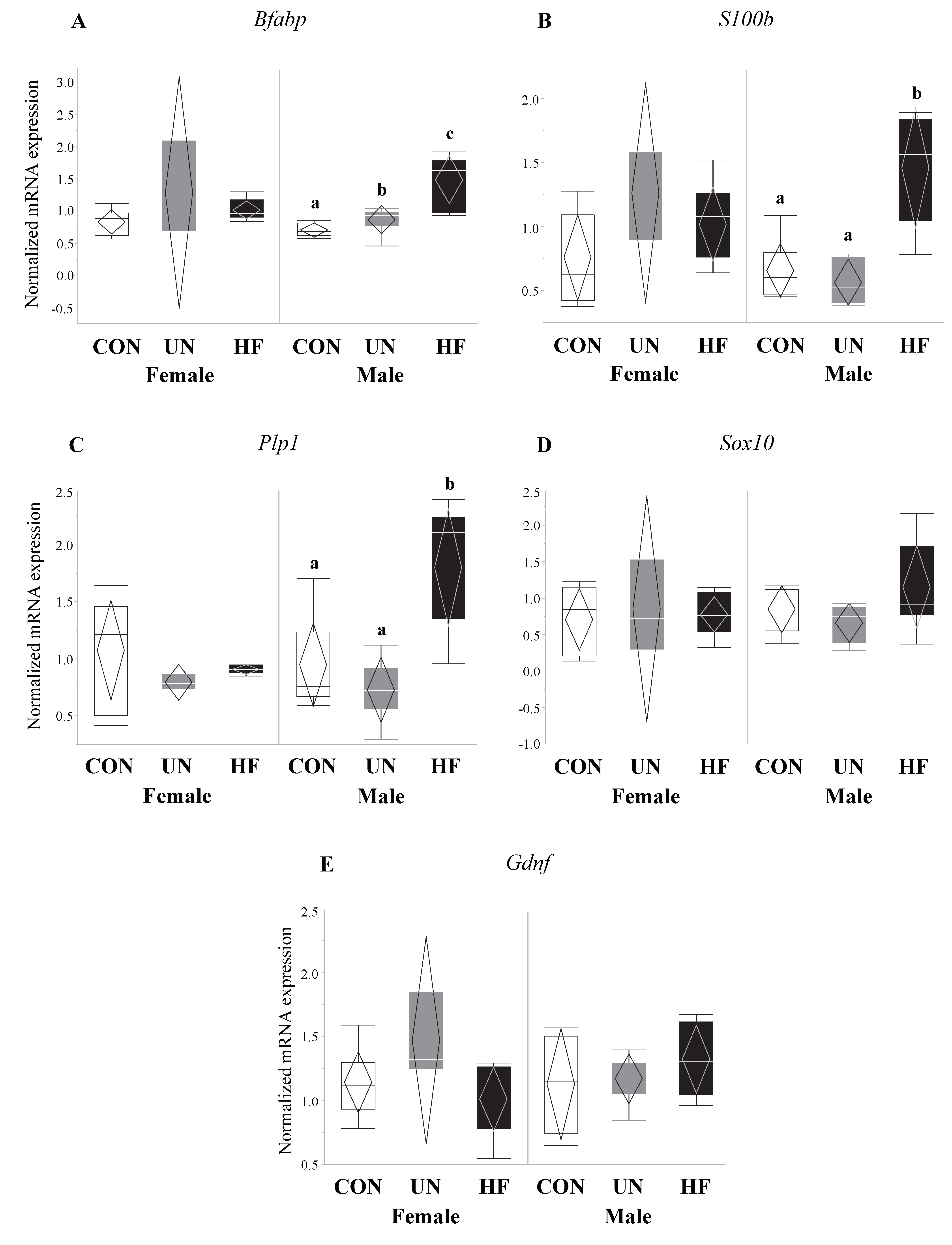
3.4. Maternal HF Diet Was Associated with Increased Fetal EGC Development
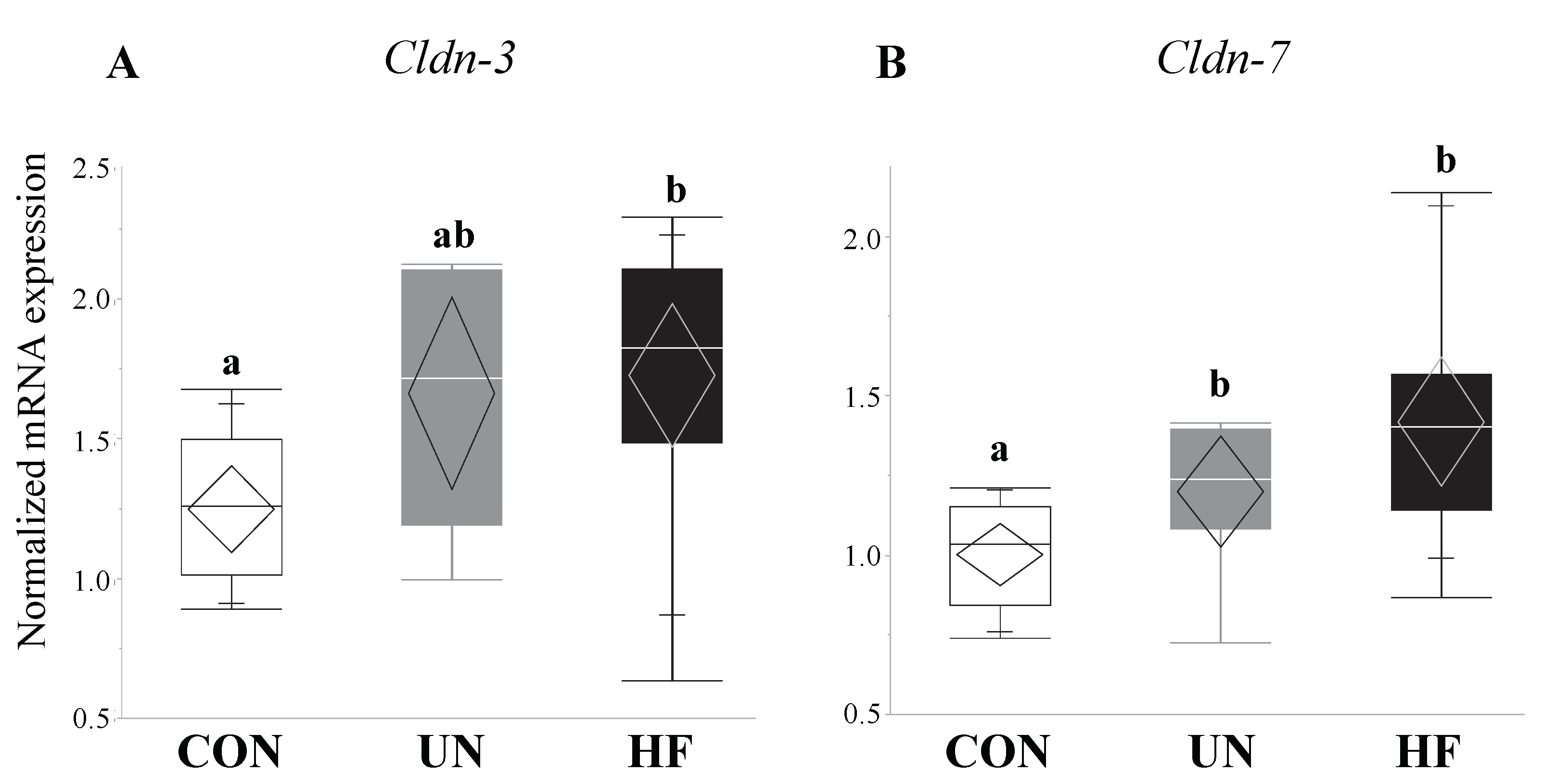
3.5. Maternal HF Diet Was Associated with Increased Fetal Gut Barrier Function and Integrity
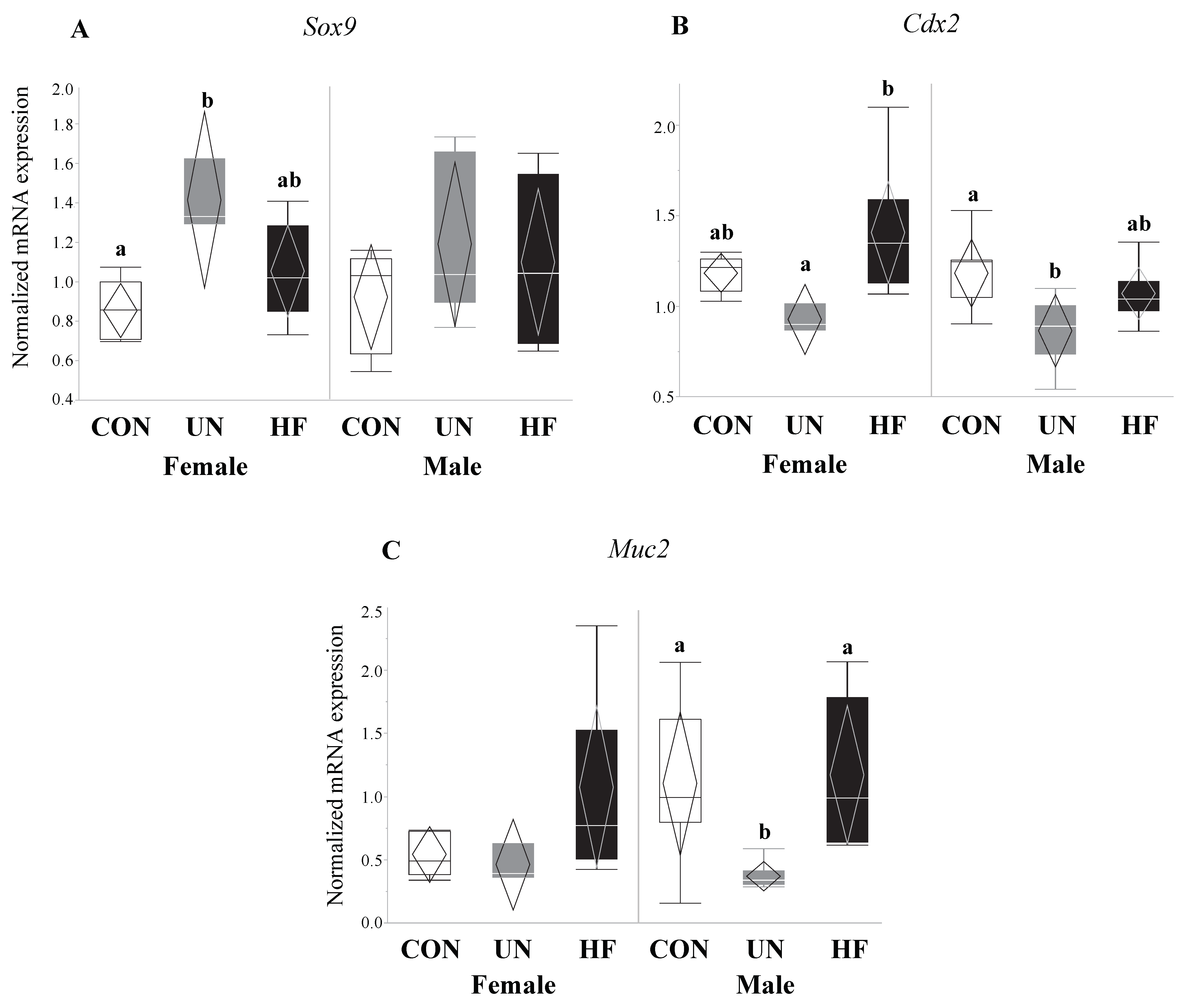
3.6. Maternal Malnutrition Altered Fetal Gut mRNA Expression in Males More than in Females
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Vaishnava, S.; Behrendt, C.L.; Ismail, A.S.; Eckmann, L.; Hooper, L.V. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proc. Natl. Acad. Sci. USA 2008, 105, 20858–20863. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kinross, J.M.; Darzi, A.W.; Nicholson, J.K. Gut microbiome-host interactions in health and disease. Genome Med. 2011, 3, 14. [Google Scholar] [CrossRef] [PubMed]
- Greenwood-Van Meerveld, B.; Johnson, A.C.; Grundy, D. Gastrointestinal physiology and function. In Handbook of Experimental Pharmacology; Springer: Cham, Switzerland, 2017; Volume 239, pp. 1–16. [Google Scholar]
- LeBlanc, J.G.; Milani, C.; de Giori, G.S.; Sesma, F.; van Sinderen, D.; Ventura, M. Bacteria as vitamin suppliers to their host: A gut microbiota perspective. Curr. Opin. Biotechnol. 2013, 24, 160–168. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, J.K.; Holmes, E.; Kinross, J.; Burcelin, R.; Gibson, G.; Jia, W.; Pettersson, S. Host-gut microbiota metabolic interactions. Science 2012, 336, 1262–1267. [Google Scholar] [CrossRef] [PubMed]
- Stecher, B.; Hardt, W.-D. Mechanisms controlling pathogen colonization of the gut. Curr. Opin. Microbiol. 2011, 14, 82–91. [Google Scholar] [CrossRef]
- Carabotti, M.; Scirocco, A.; Maselli, M.A.; Severi, C. The gut-brain axis: Interactions between enteric microbiota, central and enteric nervous systems. Ann. Gastroenterol. 2015, 28, 203–209. [Google Scholar] [PubMed]
- De Vadder, F.; Kovatcheva-Datchary, P.; Goncalves, D.; Vinera, J.; Zitoun, C.; Duchampt, A.; Bäckhed, F.; Mithieux, G. Microbiota-generated metabolites promote metabolic benefits via gut-brain neural circuits. Cell 2014, 156, 84–96. [Google Scholar] [CrossRef]
- Field, B.C.T.; Chaudhri, O.B.; Bloom, S.R. Bowels control brain: Gut hormones and obesity. Nat. Rev. Endocrinol. 2010, 6, 444–453. [Google Scholar] [CrossRef]
- Tracey, K.J. The inflammatory reflex. Nature 2002, 420, 853–859. [Google Scholar] [CrossRef]
- Kane, A.V.; Dinh, D.M.; Ward, H.D. Childhood malnutrition and the intestinal microbiome. Pediatr. Res. 2015, 77, 256–262. [Google Scholar] [CrossRef]
- Pearce, S.C.; Al-Jawadi, A.; Kishida, K.; Yu, S.; Hu, M.; Fritzky, L.F.; Edelblum, K.L.; Gao, N.; Ferraris, R.P. Marked differences in tight junction composition and macromolecular permeability among different intestinal cell types. BMC Biol. 2018, 16, 19. [Google Scholar] [CrossRef] [PubMed]
- Ayabe, T.; Satchell, D.P.; Wilson, C.L.; Parks, W.C.; Selsted, M.E.; Ouellette, A.J. Secretion of microbicidal α-defensins by intestinal Paneth cells in response to bacteria. Nat. Immunol. 2000, 1, 113–118. [Google Scholar] [CrossRef] [PubMed]
- Bry, L.; Falk, P.; Huttner, K.; Ouellette, A.; Midtvedt, T.; Gordon, J.I. Paneth cell differentiation in the developing intestine of normal and transgenic mice. Proc. Natl. Acad. Sci. USA 1994, 91, 10335–10339. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; van Es, J.H.; Snippert, H.J.; Stange, D.E.; Vries, R.G.; van den Born, M.; Barker, N.; Shroyer, N.F.; van de Wetering, M.; Clevers, H. Paneth cells constitute the niche for Lgr5 stem cells in intestinal crypts. Nature 2011, 469, 415–418. [Google Scholar] [CrossRef] [PubMed]
- Said, H.M. Intestinal absorption of water-soluble vitamins in health and disease. Biochem. J. 2011, 437, 357–372. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rühl, A. Glial cells in the gut. Neurogastroenterol. Motil. 2005, 17, 777–790. [Google Scholar] [CrossRef] [PubMed]
- Rühl, A.; Franzke, S.; Stremmel, W. IL-1beta and IL-10 have dual effects on enteric glial cell proliferation. Neurogastroenterol. Motil. 2001, 13, 89–94. [Google Scholar] [CrossRef]
- Murakami, M.; Ohta, T.; Ito, S. Lipopolysaccharides enhance the action of bradykinin in enteric neurons via secretion of interleukin-1beta from enteric glial cells. J. Neurosci. Res. 2009, 87, 2095–2104. [Google Scholar] [CrossRef]
- Savidge, T.C.; Newman, P.; Pothoulakis, C.; Ruhl, A.; Neunlist, M.; Bourreille, A.; Hurst, R.; Sofroniew, M.V. Enteric glia regulate intestinal barrier function and inflammation via release of S-nitrosoglutathione. Gastroenterology 2007, 132, 1344–1358. [Google Scholar] [CrossRef]
- Bernstein, C.N.; Vidrich, A. Isolation, identification, and culture of normal mouse colonic glia. Glia 1994, 12, 108–116. [Google Scholar] [CrossRef]
- Rothman, T.P.; Tennyson, V.M.; Gershon, M.D. Colonization of the bowel by the precursors of enteric glia: Studies of normal and congenitally aganglionic mutant mice. J. Comp. Neurol. 1986, 252, 493–506. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, S.; Huq, S.; Yatsunenko, T.; Haque, R.; Mahfuz, M.; Alam, M.A.; Benezra, A.; DeStefano, J.; Meier, M.F.; Muegge, B.D.; et al. Persistent gut microbiota immaturity in malnourished Bangladeshi children. Nature 2014, 510, 417–421. [Google Scholar] [CrossRef] [PubMed]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444, 1027–1031. [Google Scholar] [CrossRef] [PubMed]
- Tremaroli, V.; Bäckhed, F. Functional interactions between the gut microbiota and host metabolism. Nature 2012, 489, 242–249. [Google Scholar] [CrossRef]
- Araújo, J.R.; Tomas, J.; Brenner, C.; Sansonetti, P.J. Impact of high-fat diet on the intestinal microbiota and small intestinal physiology before and after the onset of obesity. Biochimie 2017, 141, 97–106. [Google Scholar] [CrossRef] [PubMed]
- Genton, L.; Cani, P.D.; Schrenzel, J. Alterations of gut barrier and gut microbiota in food restriction, food deprivation and protein-energy wasting. Clin. Nutr. 2015, 34, 341–349. [Google Scholar] [CrossRef]
- Hodin, C.M.; Verdam, F.J.; Grootjans, J.; Rensen, S.S.; Verheyen, F.K.; Dejong, C.H.C.; Buurman, W.A.; Greve, J.W.; Lenaerts, K. Reduced Paneth cell antimicrobial protein levels correlate with activation of the unfolded protein response in the gut of obese individuals. J. Pathol. 2011, 225, 276–284. [Google Scholar] [CrossRef]
- Lee, J.-C.; Lee, H.-Y.; Kim, T.K.; Kim, M.-S.; Park, Y.M.; Kim, J.; Park, K.; Kweon, M.-N.; Kim, S.-H.; Bae, J.-W.; et al. Obesogenic diet-induced gut barrier dysfunction and pathobiont expansion aggravate experimental colitis. PLoS ONE 2017, 12, e0187515. [Google Scholar] [CrossRef]
- Guo, X.; Tang, R.; Yang, S.; Lu, Y.; Luo, J.; Liu, Z. Rutin and its combination with inulin attenuate gut dysbiosis, the inflammatory status and endoplasmic reticulum stress in Paneth cells of obese mice induced by high-fat diet. Front. Microbiol. 2018, 9, 2651. [Google Scholar] [CrossRef]
- Guo, X.; Li, J.; Tang, R.; Zhang, G.; Zeng, H.; Wood, R.J.; Liu, Z. High fat diet alters gut microbiota and the expression of Paneth cell-antimicrobial peptides preceding changes of circulating inflammatory cytokines. Mediat. Inflamm. 2017, 2017, 9474896. [Google Scholar] [CrossRef] [PubMed]
- Hodin, C.M.; Lenaerts, K.; Grootjans, J.; de Haan, J.J.; Hadfoune, M.; Verheyen, F.K.; Kiyama, H.; Heineman, E.; Buurman, W.A. Starvation compromises Paneth cells. Am. J. Pathol. 2011, 179, 2885–2893. [Google Scholar] [CrossRef] [PubMed]
- Baudry, C.; Reichardt, F.; Marchix, J.; Bado, A.; Schemann, M.; des Varannes, S.B.; Neunlist, M.; Moriez, R. Diet-induced obesity has neuroprotective effects in murine gastric enteric nervous system: Involvement of leptin and glial cell line-derived neurotrophic factor. J. Physiol. 2012, 590, 533–544. [Google Scholar] [CrossRef] [PubMed]
- McMenamin, C.A.; Clyburn, C.; Browning, K.N. High-fat diet during the perinatal period induces loss of myenteric nitrergic neurons and increases enteric glial density, prior to the development of obesity. Neuroscience 2018, 393, 369–380. [Google Scholar] [CrossRef] [PubMed]
- NCD Risk Factor Collaboration (NCD-RisC). Worldwide trends in body-mass index, underweight, overweight, and obesity from 1975 to 2016: A pooled analysis of 2416 population-based measurement studies in 128·9 million children, adolescents, and adults. Lancet (Lond. Engl.) 2017, 390, 2627–2642. [Google Scholar] [CrossRef]
- Ruiter-Ligeti, J.; Vincent, S.; Czuzoj-Shulman, N.; Abenhaim, H.A. Risk factors, incidence, and morbidity associated with obstetric clostridium difficile infection. Obstet. Gynecol. 2018, 131, 387–391. [Google Scholar] [CrossRef] [PubMed]
- Pompei, A.; Cordisco, L.; Amaretti, A.; Zanoni, S.; Matteuzzi, D.; Rossi, M. Folate production by bifidobacteria as a potential probiotic property. Appl. Environ. Microbiol. 2007, 73, 179–185. [Google Scholar] [CrossRef]
- Wikoff, W.R.; Anfora, A.T.; Liu, J.; Schultz, P.G.; Lesley, S.A.; Peters, E.C.; Siuzdak, G. Metabolomics analysis reveals large effects of gut microflora on mammalian blood metabolites. Proc. Natl. Acad. Sci. USA 2009, 106, 3698–3703. [Google Scholar] [CrossRef] [Green Version]
- Reid, J.N.S.; Bisanz, J.E.; Monachese, M.; Burton, J.P.; Reid, G. The rationale for probiotics improving reproductive health and pregnancy outcome. Am. J. Reprod. Immunol. 2013, 69, 558–566. [Google Scholar] [CrossRef]
- Connor, K.L.; Chehoud, C.; Altrichter, A.; Chan, L.; DeSantis, T.Z.; Lye, S.J. Maternal metabolic, immune, and microbial systems in late pregnancy vary with malnutrition in mice. Biol. Reprod. 2018, 98, 579–592. [Google Scholar] [CrossRef]
- Gohir, W.; Whelan, F.J.; Surette, M.G.; Moore, C.; Schertzer, J.D.; Sloboda, D.M. Pregnancy-related changes in the maternal gut microbiota are dependent upon the mother’s periconceptional diet. Gut Microbes 2015, 6, 310–320. [Google Scholar] [CrossRef] [PubMed]
- Million, M.; Diallo, A.; Raoult, D. Gut microbiota and malnutrition. Microb. Pathog. 2017, 106, 127–138. [Google Scholar] [CrossRef] [PubMed]
- Wankhade, U.D.; Zhong, Y.; Kang, P.; Alfaro, M.; Chintapalli, S.V.; Thakali, K.M.; Shankar, K. Enhanced offspring predisposition to steatohepatitis with maternal high-fat diet is associated with epigenetic and microbiome alterations. PLoS ONE 2017, 12, e0175675. [Google Scholar] [CrossRef]
- Peterson, L.W.; Artis, D. Intestinal epithelial cells: Regulators of barrier function and immune homeostasis. Nat. Rev. Immunol. 2014, 14, 141–153. [Google Scholar] [CrossRef]
- Young, H.M.; Bergner, A.J.; Müller, T. Acquisition of neuronal and glial markers by neural crest-derived cells in the mouse intestine. J. Comp. Neurol. 2003, 456, 1–11. [Google Scholar] [CrossRef]
- Burns, A.J.; Thapar, N. Advances in ontogeny of the enteric nervous system. Neurogastroenterol. Motil. 2006, 18, 876–887. [Google Scholar] [CrossRef]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Grubišić, V.; Gulbransen, B.D. Enteric glia: The most alimentary of all glia. J. Physiol. 2017, 595, 557–570. [Google Scholar] [CrossRef]
- Kabouridis, P.S.; Lasrado, R.; McCallum, S.; Chng, S.H.; Snippert, H.J.; Clevers, H.; Pettersson, S.; Pachnis, V. Microbiota controls the homeostasis of glial cells in the gut lamina propria. Neuron 2015, 85, 289–295. [Google Scholar] [CrossRef]
- Stockinger, S.; Hornef, M.W.; Chassin, C. Establishment of intestinal homeostasis during the neonatal period. Cell. Mol. Life Sci. 2011, 68, 3699–3712. [Google Scholar] [CrossRef]
- Santer, R.M.; Conboy, V.B. Prenatal undernutrition permanently decreases enteric neuron number and sympathetic innervation of Auerbach’s plexus in the rat. J. Anat. 1990, 168, 57–62. [Google Scholar] [PubMed]
- Stenkamp-Strahm, C.; Patterson, S.; Boren, J.; Gericke, M.; Balemba, O. High-fat diet and age-dependent effects on enteric glial cell populations of mouse small intestine. Auton. Neurosci. Basic Clin. 2013, 177, 199–210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coad, J.; Al-Rasasi, B.; Morgan, J. Nutrient insult in early pregnancy. Proc. Nutr. Soc. 2002, 61, 51–59. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomas, J.; Mulet, C.; Saffarian, A.; Cavin, J.-B.; Ducroc, R.; Regnault, B.; Tan, C.K.; Duszka, K.; Burcelin, R.; Wahli, W.; et al. High-fat diet modifies the PPAR-γ pathway leading to disruption of microbial and physiological ecosystem in murine small intestine. Proc. Natl. Acad. Sci. USA 2016, 113, E5934–E5943. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.-A.; Gu, W.; Lee, I.-A.; Joh, E.-H.; Kim, D.-H. High fat diet-induced gut microbiota exacerbates inflammation and obesity in mice via the TLR4 signaling pathway. PLoS ONE 2012, 7, e47713. [Google Scholar] [CrossRef] [PubMed]
- de La Serre, C.B.; Ellis, C.L.; Lee, J.; Hartman, A.L.; Rutledge, J.C.; Raybould, H.E. Propensity to high-fat diet-induced obesity in rats is associated with changes in the gut microbiota and gut inflammation. Am. J. Physiol. Gastrointest. Liver Physiol. 2010, 299, G440–G448. [Google Scholar] [CrossRef] [PubMed]
- Ley, R.E.; Bäckhed, F.; Turnbaugh, P.; Lozupone, C.A.; Knight, R.D.; Gordon, J.I. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. USA 2005, 102, 11070–11075. [Google Scholar] [CrossRef] [Green Version]
- Turnbaugh, P.J.; Backhed, F.; Fulton, L.; Gordon, J.I. Marked alterations in the distal gut microbiome linked to diet-induced obesity. Cell Host Microbe 2008, 3, 213–223. [Google Scholar] [CrossRef]
- Suzuki, T.; Hara, H. Dietary fat and bile juice, but not obesity, are responsible for the increase in small intestinal permeability induced through the suppression of tight junction protein expression in LETO and OLETF rats. Nutr. Metab. 2010, 7, 19. [Google Scholar] [CrossRef]
- Everard, A.; Lazarevic, V.; Gaïa, N.; Johansson, M.; Ståhlman, M.; Backhed, F.; Delzenne, N.M.; Schrenzel, J.; François, P.; Cani, P.D. Microbiome of prebiotic-treated mice reveals novel targets involved in host response during obesity. ISME J. 2014, 8, 2116–2130. [Google Scholar] [CrossRef]
- Loonen, L.M.P.; Stolte, E.H.; Jaklofsky, M.T.J.; Meijerink, M.; Dekker, J.; van Baarlen, P.; Wells, J.M. REG3γ-deficient mice have altered mucus distribution and increased mucosal inflammatory responses to the microbiota and enteric pathogens in the ileum. Mucosal Immunol. 2014, 7, 939–947. [Google Scholar] [CrossRef] [PubMed]
- Markart, P.; Faust, N.; Graf, T.; Na, C.-L.; Weaver, T.E.; Akinbi, H.T. Comparison of the microbicidal and muramidase activities of mouse lysozyme M and P. Biochem. J. 2004, 380, 385–392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hansson, G.C. Role of mucus layers in gut infection and inflammation. Curr. Opin. Microbiol. 2012, 15, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Schulz, M.D.; Atay, C.; Heringer, J.; Romrig, F.K.; Schwitalla, S.; Aydin, B.; Ziegler, P.K.; Varga, J.; Reindl, W.; Pommerenke, C.; et al. High-fat-diet-mediated dysbiosis promotes intestinal carcinogenesis independently of obesity. Nature 2014, 514, 508–512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Fouts, D.E.; Stärkel, P.; Hartmann, P.; Chen, P.; Llorente, C.; DePew, J.; Moncera, K.; Ho, S.B.; Brenner, D.A.; et al. Intestinal REG3 lectins protect against alcoholic steatohepatitis by reducing mucosa-associated microbiota and preventing bacterial translocation. Cell Host Microbe 2016, 19, 227–239. [Google Scholar] [CrossRef] [PubMed]
- Vaishnava, S.; Yamamoto, M.; Severson, K.M.; Ruhn, K.A.; Yu, X.; Koren, O.; Ley, R.; Wakeland, E.K.; Hooper, L.V. The antibacterial lectin RegIIIgamma promotes the spatial segregation of microbiota and host in the intestine. Science 2011, 334, 255–258. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, J.; O’Farrelly, C.; Feighery, C.; Murchan, P.; Leonard, N.; Fulton, G.; O’Morain, C.; Keane, F.; Tanner, W. Impaired gut barrier function in malnourished patients. Br. J. Surg. 2005, 83, 1288–1291. [Google Scholar] [CrossRef]
- Welsh, F.K.S.; Farmery, S.M.; MacLennan, K.; Sheridan, M.B.; Barclay, G.R.; Guillou, P.J.; Reynolds, J.V. Gut barrier function in malnourished patients. Gut 1998, 42, 396–401. [Google Scholar] [CrossRef] [Green Version]
- Mondal, D.; Minak, J.; Alam, M.; Liu, Y.; Dai, J.; Korpe, P.; Liu, L.; Haque, R.; Petri, W.A. Contribution of enteric infection, altered intestinal barrier function, and maternal malnutrition to infant malnutrition in Bangladesh. Clin. Infect. Dis. 2012, 54, 185–192. [Google Scholar] [CrossRef]
- Deitch, E.A.; Winterton, J.; Li, M.; Berg, R. The gut as a portal of entry for bacteremia. Role of protein malnutrition. Ann. Surg. 1987, 205, 681–692. [Google Scholar] [CrossRef]
- Oriá, R.B.; Vieira, C.M.G.; Pinkerton, R.C.; de Castro Costa, C.M.; Lopes, M.B.; Hussaini, I.; Shi, W.; Brito, G.A.C.; Lima, A.A.M.; Guerrant, R.L. Apolipoprotein E knockout mice have accentuated malnutrition with mucosal disruption and blunted insulin-like growth factor I responses to refeeding. Nutr. Res. 2006, 26, 427–435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guerrant, R.L.; Oriá, R.B.; Moore, S.R.; Oriá, M.O.; Lima, A.A. Malnutrition as an enteric infectious disease with long-term effects on child development. Nutr. Rev. 2008, 66, 487–505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dock-Nascimento, D.B.; Junqueira, K.; de Aguilar-Nascimento, J.E. Rapid restoration of colonic goblet cells induced by a hydrolyzed diet containing probiotics in experimental malnutrition. Acta Cir. Bras. 2007, 22, 72–76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wehkamp, J.; Schauber, J.; Stange, E.F. Defensins and cathelicidins in gastrointestinal infections. Curr. Opin. Gastroenterol. 2007, 23, 32–38. [Google Scholar] [CrossRef] [PubMed]
- Nieuwenhuis, E.E.S.; Matsumoto, T.; Lindenbergh, D.; Willemsen, R.; Kaser, A.; Simons-Oosterhuis, Y.; Brugman, S.; Yamaguchi, K.; Ishikawa, H.; Aiba, Y.; et al. Cd1d-dependent regulation of bacterial colonization in the intestine of mice. J. Clin. Investig. 2009, 119, 1241–1250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grootjans, J.; Hodin, C.M.; de Haan, J.-J.; Derikx, J.P.M.; Rouschop, K.M.A.; Verheyen, F.K.; van Dam, R.M.; Dejong, C.H.C.; Buurman, W.A.; Lenaerts, K. Level of activation of the unfolded protein response correlates with Paneth cell apoptosis in human small intestine exposed to ischemia/reperfusion. Gastroenterology 2011, 140, 529–539. [Google Scholar] [CrossRef]
- Shi, J.; Aono, S.; Lu, W.; Ouellette, A.J.; Hu, X.; Ji, Y.; Wang, L.; Lenz, S.; van Ginkel, F.W.; Liles, M.; et al. A novel role for defensins in intestinal homeostasis: Regulation of IL-1beta secretion. J. Immunol. 2007, 179, 1245–1253. [Google Scholar] [CrossRef]
- Al-Sadi, R.; Ye, D.; Dokladny, K.; Ma, T.Y. Mechanism of IL-1beta-induced increase in intestinal epithelial tight junction permeability. J. Immunol. 2008, 180, 5653–5661. [Google Scholar] [CrossRef]
- Al-Sadi, R.M.; Ma, T.Y. IL-1beta causes an increase in intestinal epithelial tight junction permeability. J. Immunol. 2007, 178, 4641–4649. [Google Scholar] [CrossRef]
- Cunningham-Rundles, S. Malnutrition and gut immune function. Curr. Opin. Gastroenterol. 1994, 10, 664–670. [Google Scholar] [CrossRef]
- Barker, D.J.P.; Osmond, C.; Kajantie, E.; Eriksson, J.G. Growth and chronic disease: Findings in the Helsinki birth cohort. Ann. Hum. Biol. 2009, 36, 445–458. [Google Scholar] [CrossRef] [PubMed]
- Rao, M.; Nelms, B.D.; Dong, L.; Salinas-Rios, V.; Rutlin, M.; Gershon, M.D.; Corfas, G. Enteric glia express proteolipid protein 1 and are a transcriptionally unique population of glia in the mammalian nervous system. GLIA 2015, 63, 2040–2057. [Google Scholar] [CrossRef] [PubMed]
- Cabarrocas, J.; Savidge, T.C.; Liblau, R.S. Role of enteric glial cells in inflammatory bowel disease. GLIA 2003, 41, 81–93. [Google Scholar] [CrossRef] [PubMed]
- Margolis, K.G.; Gershon, M.D. Enteric neuronal regulation of intestinal inflammation. Trends Neurosci. 2016, 39, 614–624. [Google Scholar] [CrossRef] [PubMed]
- Laranjeira, C.; Sandgren, K.; Kessaris, N.; Richardson, W.; Potocnik, A.; Van den Berghe, P.; Pachnis, V. Glial cells in the mouse enteric nervous system can undergo neurogenesis in response to injury. J. Clin. Investig. 2011, 121, 3412–3424. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Challier, J.C.; Basu, S.; Bintein, T.; Minium, J.; Hotmire, K.; Catalano, P.M.; Hauguel-de Mouzon, S. Obesity in pregnancy stimulates macrophage accumulation and inflammation in the placenta. Placenta 2008, 29, 274–281. [Google Scholar] [CrossRef] [PubMed]
- Bilbo, S.D.; Tsang, V. Enduring consequences of maternal obesity for brain inflammation and behavior of offspring. FASEB J. 2010, 24, 2104–2115. [Google Scholar] [CrossRef]
- Roberts, K.A.; Riley, S.C.; Reynolds, R.M.; Barr, S.; Evans, M.; Statham, A.; Hor, K.; Jabbour, H.N.; Norman, J.E.; Denison, F.C. Placental structure and inflammation in pregnancies associated with obesity. Placenta 2011, 32, 247–254. [Google Scholar] [CrossRef]
- Freeman, L.R.; Small, B.J.; Bickford, P.C.; Umphlet, C.; Granholm, A.-C. A high-fat/high-cholesterol diet inhibits growth of fetal hippocampal transplants via increased inflammation. Cell Transpl. 2011, 20, 1499–1514. [Google Scholar] [CrossRef]
- Wu, Y.-Y.; Hsu, C.-M.; Chen, P.-H.; Fung, C.-P.; Chen, L.-W. Toll-like receptor stimulation induces nondefensin protein expression and reverses antibiotic-induced gut defense impairment. Infect. Immun. 2014, 82, 1994–2005. [Google Scholar] [CrossRef]
- Hu, G.; Gong, A.-Y.; Roth, A.L.; Huang, B.Q.; Ward, H.D.; Zhu, G.; Larusso, N.F.; Hanson, N.D.; Chen, X.-M. Release of luminal exosomes contributes to TLR4-mediated epithelial antimicrobial defense. PLoS Pathog. 2013, 9, e1003261. [Google Scholar] [CrossRef] [PubMed]
- De Oca, E.P.M. Antimicrobial peptide elicitors: New hope for the post-antibiotic era. Innate Immun. 2013, 19, 227–241. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Ding, L.; Lu, Q.; Chen, Y.-H. Claudins in intestines. Tissue Barriers 2013, 1, e24978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holmes, J.L.; Van Itallie, C.M.; Rasmussen, J.E.; Anderson, J.M. Claudin profiling in the mouse during postnatal intestinal development and along the gastrointestinal tract reveals complex expression patterns. Gene Expr. Patterns 2006, 6, 581–588. [Google Scholar] [CrossRef] [PubMed]
- Patel, R.M.; Myers, L.S.; Kurundkar, A.R.; Maheshwari, A.; Nusrat, A.; Lin, P.W. Probiotic bacteria induce maturation of intestinal Claudin 3 expression and barrier function. Am. J. Pathol. 2012, 180, 626–635. [Google Scholar] [CrossRef]
- Ding, L.; Lu, Z.; Foreman, O.; Tatum, R.; Lu, Q.; Renegar, R.; Cao, J.; Chen, Y.-H. Inflammation and disruption of the mucosal architecture in claudin-7-deficient mice. Gastroenterology 2012, 142, 305–315. [Google Scholar] [CrossRef] [PubMed]
- Polak-Charcon, S.; Shoham, J.; Ben-Shaul, Y. Tight junctions in epithelial cells of human fetal hindgut, normal colon, and colon adenocarcinoma. J. Natl. Cancer Inst. 1980, 65, 53–62. [Google Scholar]
- Blache, P.; van de Wetering, M.; Duluc, I.; Domon, C.; Berta, P.; Freund, J.-N.; Clevers, H.; Jay, P. SOX9 is an intestine crypt transcription factor, is regulated by the Wnt pathway, and represses the CDX2 and MUC2 genes. J. Cell Biol. 2004, 166, 37–47. [Google Scholar] [CrossRef]
- Sherman, M.P. New concepts of microbial translocation in the neonatal intestine: Mechanisms and prevention. Clin. Perinatol. 2010, 37, 565–579. [Google Scholar] [CrossRef]
- Mai, V.; Torrazza, R.M.; Ukhanova, M.; Wang, X.; Sun, Y.; Li, N.; Shuster, J.; Sharma, R.; Hudak, M.L.; Neu, J. Distortions in development of intestinal microbiota associated with late onset sepsis in preterm infants. PLoS ONE 2013, 8, e52876. [Google Scholar] [CrossRef]
- Deitch, E.A. Gut-origin sepsis: Evolution of a concept. Surgeon 2012, 10, 350–356. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buddington, R.K.; Diamond, J.M. Ontogenetic development of intestinal nutrient transporters. Annu. Rev. Physiol. 1989, 51, 601–619. [Google Scholar] [CrossRef] [PubMed]
- Rouwet, E.V.; Heineman, E.; Buurman, W.A.; ter Riet, G.; Ramsay, G.; Blanco, C.E. Intestinal permeability and carrier-mediated monosaccharide absorption in preterm neonates during the early postnatal period. Pediatr. Res. 2002, 51, 64–70. [Google Scholar] [CrossRef] [PubMed]
- Clifford, S.H.; Weller, K.F. The absorption of vitamin A in prematurely born infants: With experience in the use of absorbable vitamin A in the prophylaxis of retrolental fibroplasia. Pediatrics 1948, 1, 505–511. [Google Scholar] [PubMed]
- Siggers, R.H.; Siggers, J.; Thymann, T.; Boye, M.; Sangild, P.T. Nutritional modulation of the gut microbiota and immune system in preterm neonates susceptible to necrotizing enterocolitis. J. Nutr. Biochem. 2011, 22, 511–521. [Google Scholar] [CrossRef] [PubMed]
- Gephart, S.M.; McGrath, J.M.; Effken, J.A.; Halpern, M.D. Necrotizing enterocolitis risk: State of the science. Adv. Neonatal Care 2012, 12, 77–87. [Google Scholar] [CrossRef]
- Gregory, K.E.; Deforge, C.E.; Natale, K.M.; Phillips, M.; Van Marter, L.J. Necrotizing enterocolitis in the premature infant: Neonatal nursing assessment, disease pathogenesis, and clinical presentation. Adv. Neonatal Care 2011, 11, 155–164. [Google Scholar] [CrossRef] [PubMed]
- Luig, M.; Lui, K.; NSW & ACT NICUS Group. Epidemiology of necrotizing enterocolitis—Part II: Risks and susceptibility of premature infants during the surfactant era: A regional study. J. Paediatr. Child Health 2005, 41, 174–179. [Google Scholar] [PubMed]
- Di Renzo, G.C.; Rosati, A.; Sarti, R.D.; Cruciani, L.; Cutuli, A.M. Does fetal sex affect pregnancy outcome? Gend. Med. 2007, 4, 19–30. [Google Scholar] [CrossRef]
- Zeitlin, J.; Saurel-Cubizolles, M.-J.; De Mouzon, J.; Rivera, L.; Ancel, P.-Y.; Blondel, B.; Kaminski, M. Fetal sex and preterm birth: Are males at greater risk? Hum. Reprod. 2002, 17, 2762–2768. [Google Scholar] [CrossRef] [PubMed]
- Mayoral, S.R.; Omar, G.; Penn, A.A. Sex differences in a hypoxia model of preterm brain damage. Pediatr. Res. 2009, 66, 248–253. [Google Scholar] [CrossRef] [PubMed]
- Eriksson, J.G.; Kajantie, E.; Osmond, C.; Thornburg, K.; Barker, D.J.P. Boys live dangerously in the womb. Am. J. Hum. Biol. 2010, 22, 330–335. [Google Scholar] [CrossRef] [PubMed]
- Van Abeelen, A.F.M.; de Rooij, S.R.; Osmond, C.; Painter, R.C.; Veenendaal, M.V.E.; Bossuyt, P.M.M.; Elias, S.G.; Grobbee, D.E.; van der Schouw, Y.T.; Barker, D.J.P.; et al. The sex-specific effects of famine on the association between placental size and later hypertension. Placenta 2011, 32, 694–698. [Google Scholar] [CrossRef] [PubMed]
- Aiken, C.E.; Ozanne, S.E. Sex differences in developmental programming models. Reproduction 2013, 145, R1–R13. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Gene | Sequence 5’–3’ | Source |
|---|---|---|
| Gut barrier function markers | ||
| Lyz1 | F-GGGAACCTGTGACCTGTCTT | Accession: NM_013590.4 |
| R-GCCTCATGACACTGGGAACA | ||
| Lyz2 | F-TCTACTGCAGCTCATTCGGT | Accession: NM_017372.3 |
| R-CTTAGAGGGGAAATCGAGGGAA | ||
| Pla2g2 | F-AGGATTCCCCCAAGGATGCCAC | PMID: 19855381 |
| R-CAGCCGTTTCTGACAGGAGTTCTGG | ||
| Defa1 | F-TCAAGAGGCTGCAAAGGAAGAGAAC | PMID: 19855381 |
| R-TGGTCTCCATGTTCAGCGACAGC | ||
| Defa5 | F-CTTGTCCTGCTGGCCTTCC | Accession: NM_007851.2 |
| R-TAGACACAGCCTGGTCCTCT | ||
| Reg3g | F-CCATCTTCACGTAGCAGC | PMID: 22723890 |
| R-CAAGATGTCCTGAGGGC | ||
| Tlr4 | Bio-Rad (Mississauga, ON, Canada) | Assay ID: qMmuCID0023548 |
| Gut barrier integrity markers | ||
| Muc2 | F-ACCTGGAAGGCCCAATCAAG | Accession: NM_023566.3 |
| R-CAGCGTAGTTGGCACTCTCA | ||
| Cldn-3 | F-GCACCCACCAAGATCCTCTA | PMID: 17383680 |
| R-AGGCTGTCTGTCCTCTTCCA | ||
| Cldn-7 | F-CATTGTGGCAGGTCTTGCTG | Accession: NM_016887.6 |
| R-CATGGGCGTCAAGGGGTTAT | ||
| EGC maturation and function markers | ||
| Bfabp | F-GGTTCGGTTGGATGGAGACA | Accession: NM_021272.3 |
| R-AGTCACGACCATCTTGCCAT | ||
| S100b | F-TGGCTGCGGAAGTTGAGATT | Accession: NM_009115.3 |
| R-ATGGCTCCCAGCAGCTAAAG | ||
| Gdnf | F-ACCAGTGACTCCAATATGCCTG | Accession: NM_001301357.1 |
| R-CTGCCGCTTGTTTATCTGGTG | ||
| Sox10 | Bio-Rad (Mississauga, ON, Canada) | Assay ID: qMmuCID0007045 |
| Plp1 | Bio-Rad (Mississauga, ON, Canada) | Assay ID: qMmuCED0061105 |
| Fetal gut development markers | ||
| Cdx2 | F-AGCCAAGTGAAAACCAGGAC | Accession: NM_007673.3 |
| R-AGTGAAACTCCTTCTCCAGCTC | ||
| Sox9 | F-GCCACGGAACAGACTCACAT | Accession: NM_011448.4 |
| R-AGATTGCCCAGAGTGCTCG | ||
| Reference genes | ||
| Ywhaz | F-GCAACGATGTACTGTCTCTTTTGG | Accession: NM_011740.3 |
| R-GTCCACAATTCCTTTCTTGTCATC | ||
| Tbp | F-CGGACAACTGCGTTGATTTTC | Accession: NM_013684.3 |
| R-AGCCCAACTTCTGCACAACTC | ||
| Actb | F-TCGTGCGTGACATCAAAGAGA | Accession: NM_007393.5 |
| R-GAACCGCTCGTTGCCAATA | ||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Srugo, S.A.; Bloise, E.; Nguyen, T.T.-T.N.; Connor, K.L. Impact of Maternal Malnutrition on Gut Barrier Defense: Implications for Pregnancy Health and Fetal Development. Nutrients 2019, 11, 1375. https://doi.org/10.3390/nu11061375
Srugo SA, Bloise E, Nguyen TT-TN, Connor KL. Impact of Maternal Malnutrition on Gut Barrier Defense: Implications for Pregnancy Health and Fetal Development. Nutrients. 2019; 11(6):1375. https://doi.org/10.3390/nu11061375
Chicago/Turabian StyleSrugo, Sebastian A., Enrrico Bloise, Tina Tu-Thu Ngoc Nguyen, and Kristin L. Connor. 2019. "Impact of Maternal Malnutrition on Gut Barrier Defense: Implications for Pregnancy Health and Fetal Development" Nutrients 11, no. 6: 1375. https://doi.org/10.3390/nu11061375