Carotenoid Biosynthetic and Catabolic Pathways: Gene Expression and Carotenoid Content in Grains of Maize Landraces
Abstract
:1. Introduction
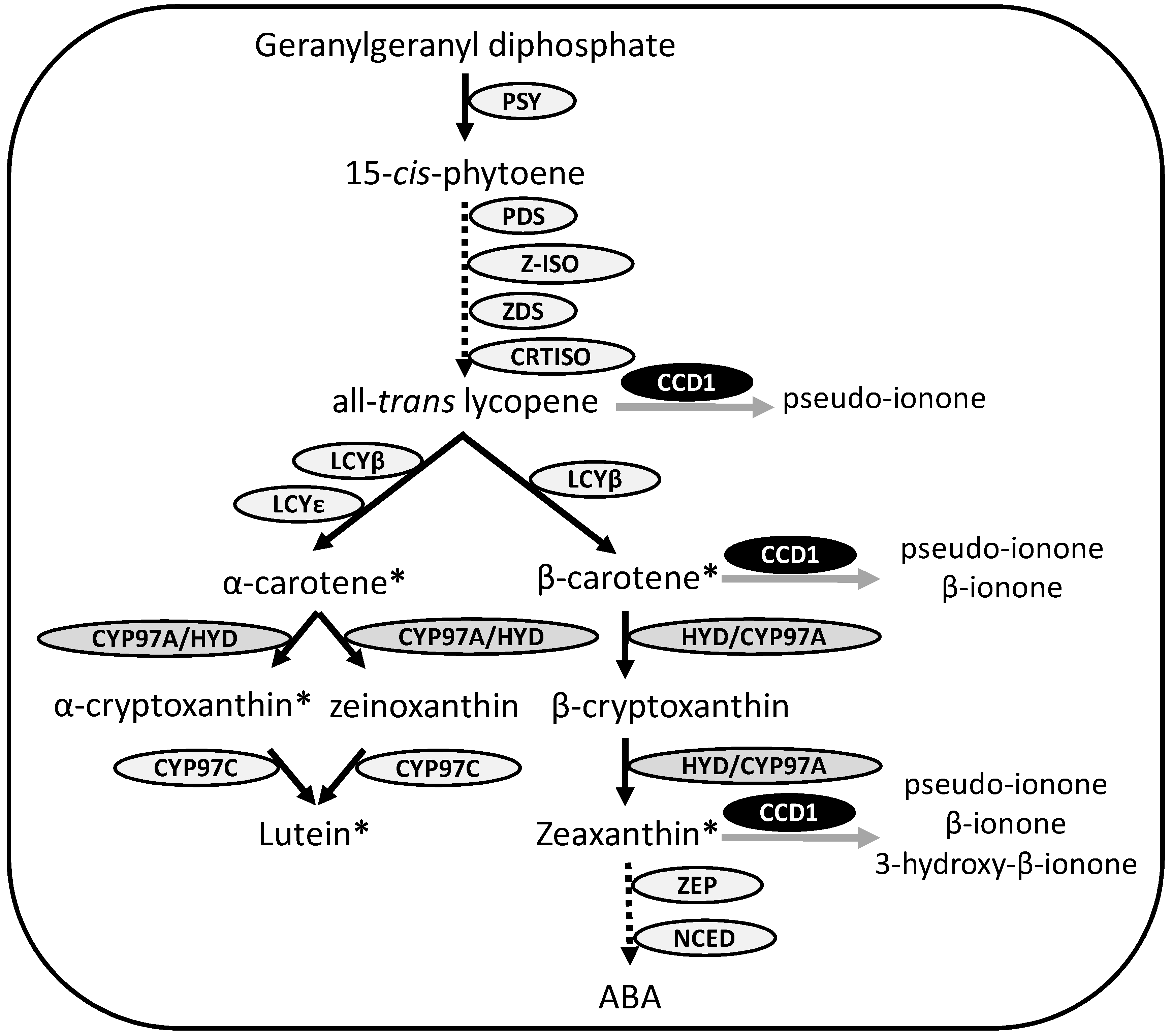
2. Experimental Section
2.1. Experimental Design and Varieties
2.2. Sample Collection and Storage
2.3. Quantification of Carotenoids
2.4. Primer Design and Gene Expression Analysis
| Gene | GenBank Accession | Forward Primer Sequence | Reverse Primer Sequence | Amplicon Length | Product TM °C a |
|---|---|---|---|---|---|
| TUB | NM_001111988.1 | AGAACTGCGACTGCCTCCAAAGG | AGATGAGCAGGGTGCCCATTC | 84 | 82.6 |
| ACT | J01238.1 | CATGGAGAACTGGCATCACACCTT | CTGCGTCATTTTCTCTCTGTTGGC | 118 | 84.3 |
| UBI | M73785.1 | GTTTAAGCTGCCGATGTGCCTG | GACACGACTCATGACACGAACA | 89 | 82.3 |
| PSY1 | FJ971251.1 | GACAGATGAGCTTGTAGATGGGC | TCAGAGAGAGCGGCATCAAGCA | 129 | 83.1 |
| HYD3 | AY844958 | GGGGATTACGCTGTTCGG | GTGGTGTATCTTGTGCGAGG | 130 | 89.5 |
| CYP97C | BE552887 | GTTGACATTGGATGTGATTGG | AACCAACCTTCCAGTATGGC | 150 | 78.7 |
| CCD1 | DQ100346.1 | CTGCTGTGGATTTTCCTCGTG | TATGATGCCAGTCACCTTCGC | 104 | 82.3 |
2.5. Statistical Analysis
3. Results and Discussion
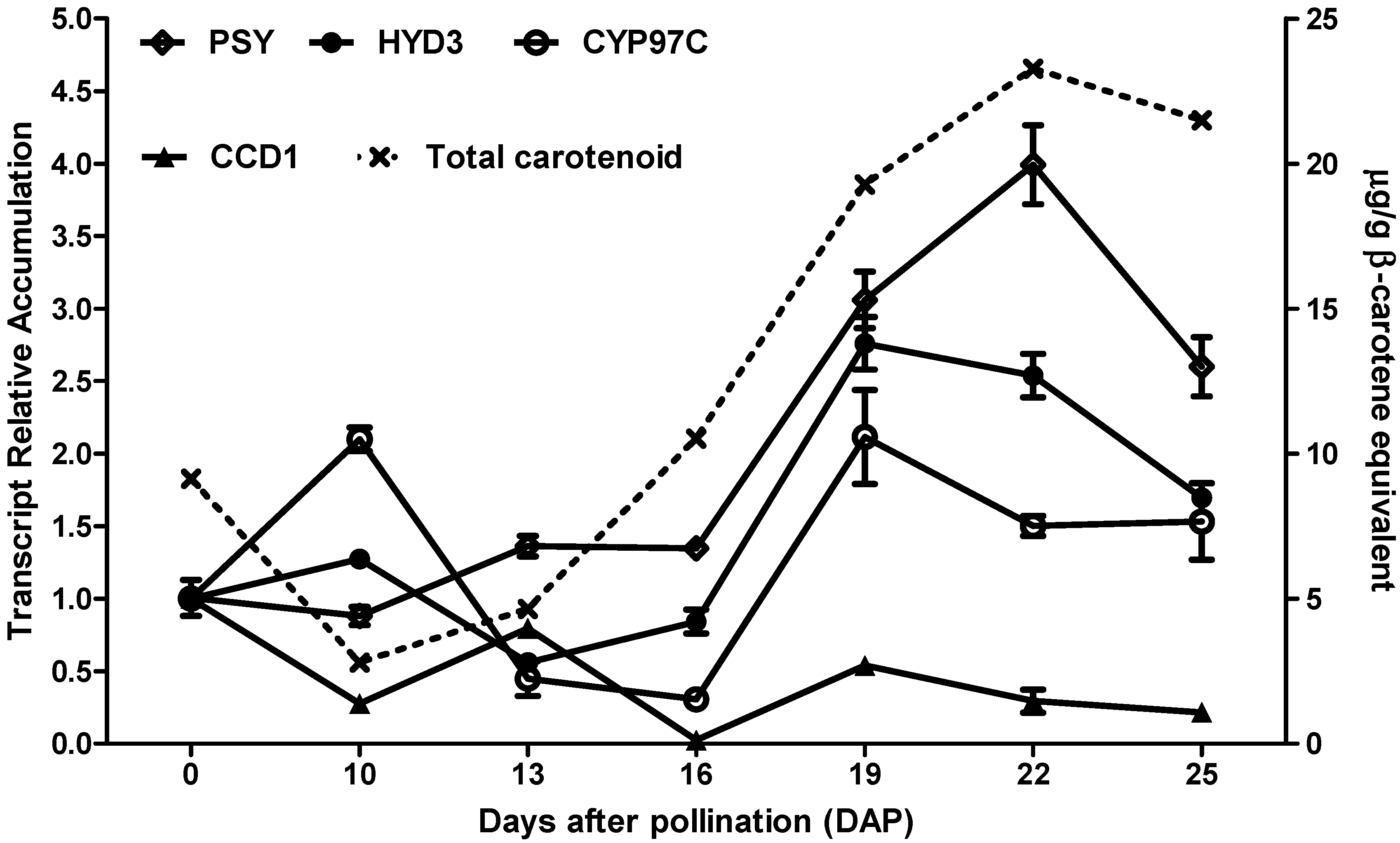
3.1. Time-Series Expression Profiles of Carotenoid-Related Genes during Multiple Grain Developmental Stages of the Hybrid 30F53
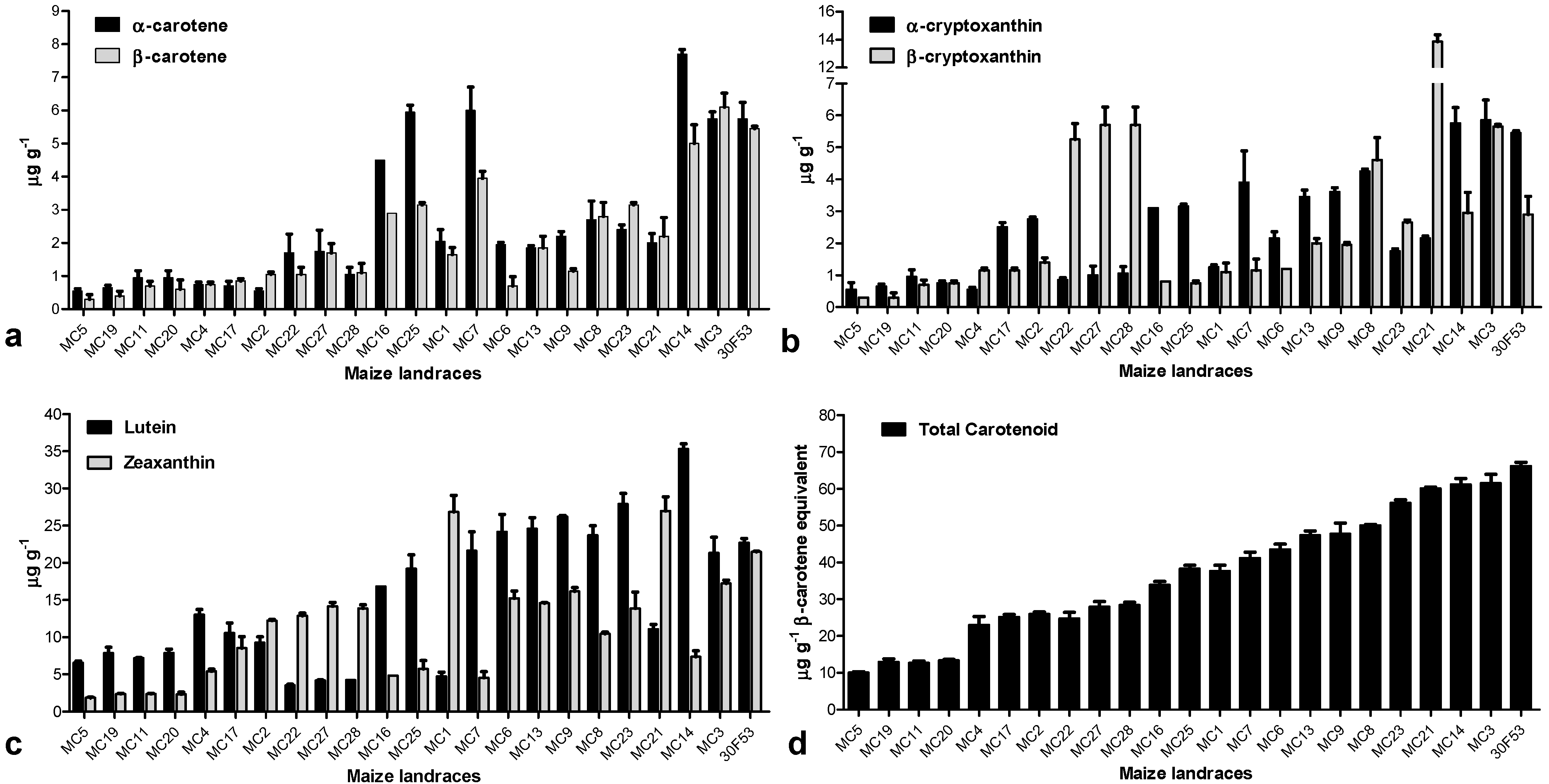
3.2. Carotenoid Profile and the Expression Pattern of Carotenoid-Related Genes in Maize Landraces

| Variables | PSY1 | CYP97C | HYD3 | CCD1 | Total Carotenoids | α-Carotene | β-Carotene | α-Cryptoxanthin | β-Cryptoxanthin | Lutein | Zeaxanthin |
|---|---|---|---|---|---|---|---|---|---|---|---|
| PSY1 | - | 0.49 ** | 0.70 *** | −0.29 | 0.46 ** | 0.13 | 0.12 | 0.07 | 0.41 ** | 0.23 | 0.52 *** |
| CYP97C | 0.49 ** | - | 0.34 * | 0.09 | 0.25 | 0.30 | 0.24 | 0.14 | −0.06 | 0.26 | 0.03 |
| HYD3 | 0.70 *** | 0.34 * | - | 0.10 | 0.25 | −0.16 | −0.05 | −0.25 | 0.72 *** | −0.22 | 0.57 *** |
| CCD1 | −0.29 | 0.09 | 0.10 | - | −0.50 ** | −0.37 * | −0.42 ** | −0.52 *** | 0.00 | −0.42 ** | −0.36 * |
| Total Carotenoids | 0.46 ** | 0.25 | 0.25 | −0.50 ** | - | 0.67 *** | 0.79 *** | 0.79 *** | 0.45 ** | 0.76 *** | 0.63 *** |
| α-Carotene | 0.13 | 0.30 | −0.16 | −0.37 * | 0.67 *** | - | 0.90 *** | 0.81 *** | 0.05 | 0.66 *** | 0.07 |
| β-Carotene | 0.12 | 0.24 | −0.05 | −0.42 ** | 0.79 *** | 0.90 *** | - | 0.85 *** | 0.23 | 0.63 *** | 0.26 |
| α-Cryptoxanthin | 0.07 | 0.14 | −0.25 | −0.52 *** | 0.79 *** | 0.81 *** | 0.85 *** | - | 0.11 | 0.77 *** | 0.23 |
| β-Cryptoxanthin | 0.41 ** | −0.06 | 0.72 *** | 0.00 | 0.45 ** | 0.05 | 0.23 | 0.11 | - | −0.08 | 0.61 *** |
| Lutein | 0.23 | 0.26 | −0.22 | −0.42 ** | 0.76 *** | 0.66 *** | 0.63 *** | 0.77 *** | −0.08 | - | 0.07 |
| Zeaxanthin | 0.52 *** | 0.03 | 0.57 *** | −0.36 * | 0.63 *** | 0.07 | 0.26 | 0.23 | 0.61 *** | 0.07 | - |
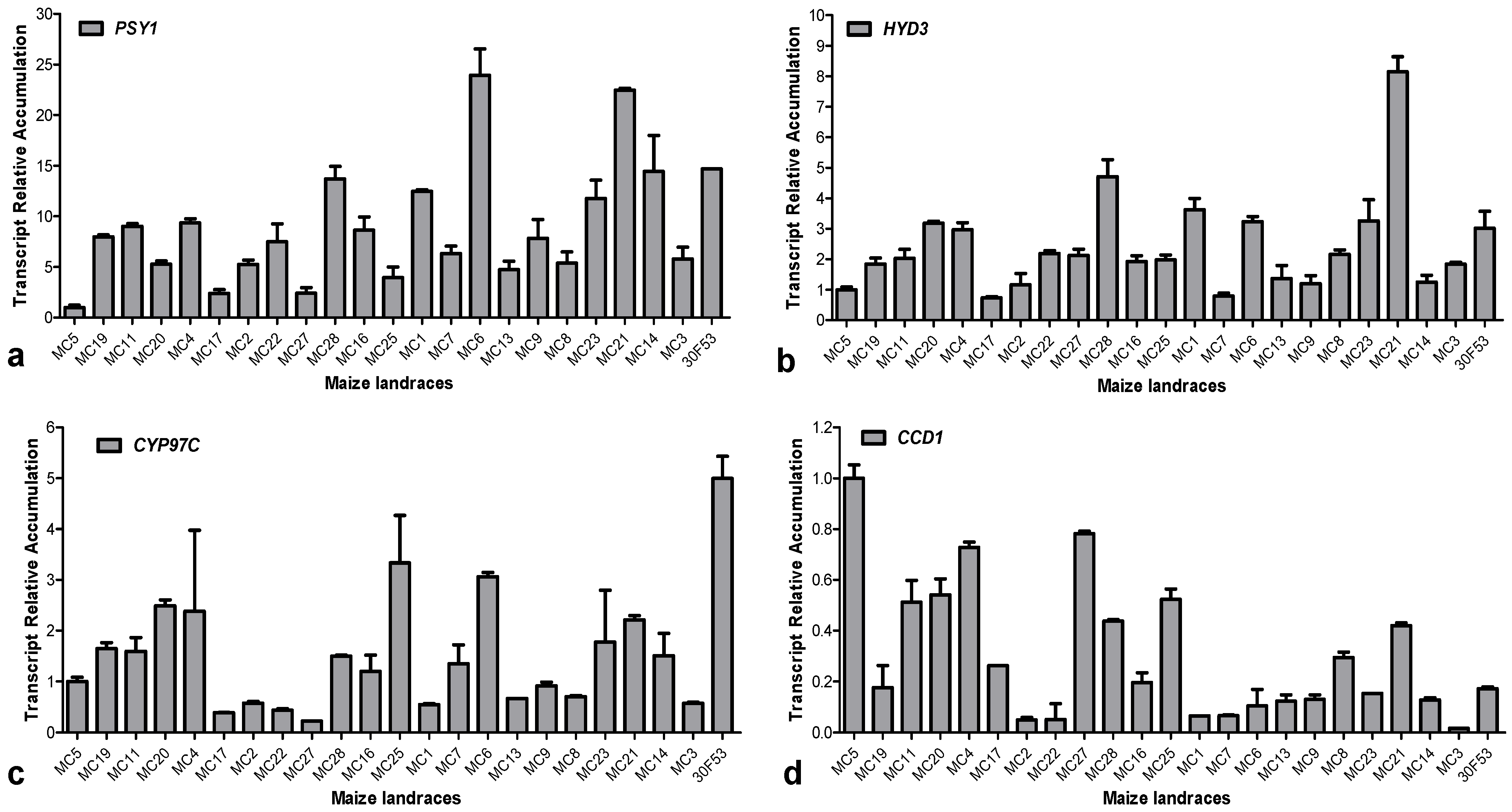
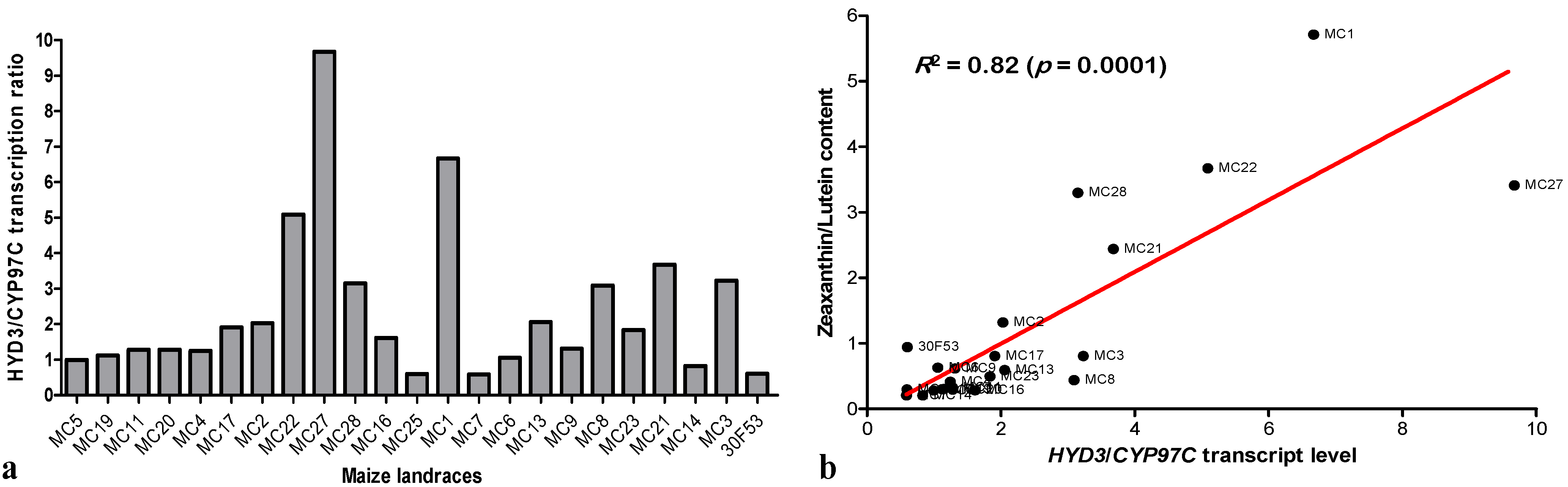
3.3. Expression of Hydroxylases and Their Effects on Carotenoid Content
3.4. Expression Pattern of ZmCCD1 in Maize Landraces and Its Correlation with Carotenoid Content
4. Conclusions
Acknowledgment
Conflicts of Interest
References
- FAOSTATE (Food and Agriculture Organization of the United Nations). Available online: http://www.faostat.fao.org (accessed on 7 June 2013).
- Vallabhaneni, R.; Gallagher, C.E.; Licciardello, N.; Cuttriss, A.J.; Quinlan, R.F.; Wurtzel, E.T. Metabolite sorting of a germplasm collection reveals the Hydroxylase3 locus as a new target for maize provitamin A biofortification. Plant Physiol. 2009, 151, 1635–1645. [Google Scholar] [CrossRef]
- Kuhnen, S.; Lemos, P.M.; Campestrini, L.H.; Ogliari, J.B.; Dias, P.F.; Maraschin, M. Carotenoid and anthocyanin contents of grains of Brazilian maize landraces. J. Sci. Food Agric. 2011, 91, 1548–1553. [Google Scholar] [CrossRef]
- Fraser, P.D.; Bramley, P.M. The biosynthesis and nutritional uses of carotenoids. Prog. Lipid Res. 2004, 43, 228–265. [Google Scholar] [CrossRef]
- WHO (World Health Organization). Global Prevalence of Vitamin A Deficiency in Populations at Risk 1995–2005. In WHO Global Database on Vitamin A Deficiency; WHO: Geneva, Switzerland, 2009; pp. 1–55. [Google Scholar]
- Messias, R.S.; Galli, V.; Silva, S.D.A.; Schirmer, M.A.; Rombaldi, C.V. Micronutrient and functional compounds biofortification of maize grains. Crit. Rev. Food Sci. Nutr. 2013. [Google Scholar] [CrossRef]
- Gallagher, C.E.; Matthews, P.D.; Faqiang, L.; Wurtzel, E.T. Gene duplication in the carotenoid biosynthetic pathway preceded evolution of the grasses. Plant Physiol. 2004, 135, 1776–1783. [Google Scholar] [CrossRef]
- Chen, Y.; Li, F.; Wurtzel, E.T. Isolation and characterization of the Z-ISO gene encoding a missing component of carotenoid biosynthesis in plants. Plant Physiol. 2010, 153, 66–79. [Google Scholar] [CrossRef]
- Sun, Z.; Hans, J.; Walter, M.H.; Matusova, R.; Beekwilder, J.; Verstappen, F.W.A.; Ming, Z.; Echtelt, E.V.; Strack, D.; Bisseling, T.; et al. Cloning and characterisation of a maize carotenoid cleavage dioxygenase (ZmCCD1) and its involvement in the biosynthesis of apocarotenoids with various roles in mutualistic and parasitic interactions. Planta 2008, 228, 789–801. [Google Scholar] [CrossRef]
- Auldridge, M.E.; McCarty, D.R.; Klee, H.J. Plant carotenoid cleavage oxygenases and their apocarotenoid products. Curr. Opin. Plant Biol. 2006, 9, 315–321. [Google Scholar] [CrossRef]
- Ohmiya, A.; Kishimoto, S.; Aida, R.; Yoshioka, S.; Sumitomo, K. Carotenoid cleavage dioxygenase (CmCCD4a) contributes to white color formation in chrysanthemum petals. Plant Physiol. 2006, 142, 1193–1201. [Google Scholar] [CrossRef]
- Walter, M.H.; Strack, D. Carotenoids and their cleavage products: Biosynthesis and functions. Nat. Prod. Rep. 2011, 28, 663–692. [Google Scholar] [CrossRef]
- Davies, W.; Kudoyarova, G.; Hartung, W. Long-distance ABA signaling and its relation to other signaling pathways in the detection of soil drying and the mediation of the plants response to drought. J. Plant Growth Regul. 2005, 24, 285–295. [Google Scholar] [CrossRef]
- Auldridge, M.E.; Block, A.; Vogel, J.T.; Dabney-Smith, C.; Mila, I.; Bouzayen, M.; Magallanes-Lundback, M.; DellaPenna, D.; McCarty, D.R.; Klee, H.J. Characterization of three members of the Arabidopsis carotenoid cleavage dioxygenase family demonstrates the divergent roles of this multifunctional enzyme family. Plant J. 2006, 45, 982–993. [Google Scholar] [CrossRef]
- Wurtzel, E.T.; Cuttriss, A.; Vallabhaneni, R. Maize provitamin a carotenoids, current resources, and future metabolic engineering challenges. Front. Plant Sci. 2012, 3, 29. [Google Scholar]
- Harjes, C.; Rocheford, T.R.; Bai, L.; Brutnell, T.P.; Kandianis, C.B.; Sowinski, S.G.; Stapleton, A.E.; Vallabhaneni, R.; Williams, M.; Wurtzel, E.T.; et al. Natural genetic variation in lycopene epsilon cyclase tapped for maize biofortification. Science 2008, 319, 330–333. [Google Scholar] [CrossRef]
- Vallabhaneni, R.; Wurtzel, E.T. Timing and biosynthetic potential for carotenoid accumulation in genetically diverse germplasm of maize. Plant Physiol. 2009, 150, 562–572. [Google Scholar] [CrossRef]
- Paine, J.A.; Shipton, C.A.; Chaggar, S.; Howells, R.M.; Kennedy, M.J.; Vernon, G.; Wright, S.Y.; Hinchliffe, E.; Adams, J.L.; Silverstone, A.L.; et al. Improving the nutritional value of Golden Rice through increased pro-vitamin A content. Nat. Biotechnol. 2005, 23, 482–487. [Google Scholar] [CrossRef]
- Aluru, M.; Xu, Y.; Guo, R.; Wang, Z.G.; Li, S.S.; White, W.; Wang, K.; Rodermel, S. Generation of transgenic maize with enhanced provitamin A content. J. Exp. Bot. 2008, 59, 3551–3562. [Google Scholar] [CrossRef]
- Giuliano, G.; Tavazza, R.; Diretto, G.; Beyer, P.; Taylor, M.A. Metabolic engineering of carotenoid biosynthesis in plants. Trends Biotechnol. 2008, 26, 139–145. [Google Scholar] [CrossRef]
- Naqvi, S.; Zhu, C.; Farre, G.; Ramessar, K.; Bassie, L.; Breitenbach, J.; Conesa, D.P.; Ros, G.; Sandmann, G.; Capell, T.; et al. Transgenic multivitamin corn through biofortification of endosperm with three vitamins representing three distinct metabolic pathways. Proc. Natl. Acad. Sci. USA 2009, 106, 7762–7767. [Google Scholar] [CrossRef]
- Zhu, C.; Naqvi, S.; Breitenbach, J.; Sandmann, G.; Christou, P.; Capell, T. Combinatorial genetic transformation generates a library of metabolic phenotypes for the carotenoid pathway in maize. Proc. Natl. Acad. Sci. USA 2008, 105, 18232–18237. [Google Scholar] [CrossRef]
- Ann, B.M.; Gothandam, K.M. Overview of genetic manipulation in plant carotenoid biosynthesis pathway. Res. J. Biotechnol. 2012, 7, 3. [Google Scholar]
- Fanning, K.J.; Martin, I.; Wong, L.; Keating, V.; Puna, S.; O’Harec, T. Screening sweetcorn for enhanced zeaxanthin concentration. J. Sci. Food Agric. 2010, 90, 91–96. [Google Scholar] [CrossRef]
- Rodriguez-Amaya, D.B.; Kimura, M. HarvestPlus Handbook for Carotenoid Analysis; International Food Policy Research Institute (IFPRI): Washington, DC, USA, 2004. [Google Scholar]
- Kimura, M.; Kobori, C.N.; Rodriguez-Amaya, D.B.; Nestel, P. Screening and HPLC methods for carotenoids in sweetpotato, cassava and maize for plant breeding trials. Food Chem. 2007, 100, 1734–1746. [Google Scholar] [CrossRef]
- Messias, R.S.; Galli, V.; Silva, S.D.A.; Schirmer, M.A.; Pillon, C.N. Metodologias de Extração e Avaliação Semi Quantitativa da Expressão de Genes de Metabolismo Secundário do Milho (Zea mays L.). Available online: http://ainfo.cnptia.embrapa.br/digital/bitstream/item/30587/1/boletim-117.pdf (accessed on 11 December 2013). (in Portuguese).
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Li, F.; Vallabhaneni, R.; Yu, J.; Rocheford, T.; Wurtzel, E.T. The maize phytoene synthase gene family: Overlapping roles for carotenogenesis in endosperm, photomorphogenesis, and thermal stress tolerance. Plant Physiol. 2008, 147, 1334–1346. [Google Scholar] [CrossRef]
- Simkin, A.J.; Kuntz, M.; Moreau, H.; McCarthy, J. Carotenoid profiling and the expression of carotenoid biosynthetic genes in developing coffee grain. Plant Physiol. Biochem. 2010, 48, 434–442. [Google Scholar] [CrossRef]
- Rodríguez-Ávila, N.L.; Narváez-Zapata, J.A.; Ramírez-Benítez, J.E.; Aguilar-Espinosa, M.L.; Rivera-Madrid, R. Identification and expression pattern of a new carotenoid cleavage dioxygenase gene member from Bixa orellana. J. Exp. Bot. 2011, 62, 5385–5395. [Google Scholar] [CrossRef]
- Vallabhaneni, R.; Bradbury, L.M.; Wurtzel, E.T. The carotenoid dioxygenase gene family in maize, sorghum, and rice. Arch. Biochem. Biophys. 2010, 504, 104–111. [Google Scholar] [CrossRef]
- Bramley, P.M. Regulation of carotenoid formation during tomato fruit ripening and development. J. Exp. Bot. 2002, 53, 2107–2113. [Google Scholar] [CrossRef]
- Ampomah-Dwamena, C.; Dejnoprat, S.; Lewis, D.; Sutherland, P.; Volz, R.K.; Allan, A.C. Metabolic and gene expression analysis of apple (Malus × domestica) carotenogenesis. J. Exp. Bot. 2012, 63, 4497–4511. [Google Scholar] [CrossRef]
- Cazzonelli, C.I.; Pogson, B.J. Source to sink: Regulation of carotenoid biosynthesis in plants. Trends Plant Sci. 2010, 15, 266–274. [Google Scholar] [CrossRef]
- Palaisa, K.A.; Morgante, M.; Williams, M.; Rafalski, A. Contrasting effects of selection on sequence diversity and linkage disequilibrium at two phytoene synthase loci. Plant Cell 2003, 15, 1795–1806. [Google Scholar] [CrossRef]
- Howitt, C.A.; Cavanagh, C.R.; Bowerman, A.F.; Cazzonelli, C.; Rampling, L.; Mimica, J.L.; Pogson, B.J. Alternative splicing, activation of cryptic exons and amino acid substitutions in carotenoid biosynthetic genes are associated with lutein accumulation in wheat endosperm. Funct. Integr. Genomic 2008, 9, 363–376. [Google Scholar]
- Quinlan, R.F.; Shumskaya, M.; Bradbury, L.M.T.; Beltrán, J.; Ma, C.; Kennelly, E.J.; Wurtzel, E.T. Synergistic interactions between carotene ring hydroxylases drive lutein formation in plant carotenoid biosynthesis. Plant Physiol. 2012, 160, 204–214. [Google Scholar] [CrossRef]
- García-Limones, C.; Schnäbele, K.; Blanco-Portales, R.; Luz Bellido, M.; Caballero, J.L.; Schwab, W.; Muñoz-Blanco, J. Functional characterization of FaCCD1: A carotenoid cleavage dioxygenase from strawberry involved in lutein degradation during fruit ripening. J. Agric. Food Chem. 2008, 56, 9277–9285. [Google Scholar] [CrossRef]
- Vogel, J.T.; Tan, B.C.; McCarty, D.R.; Klee, H.J. The carotenoid cleavage dioxygenase 1 enzyme has broad substrate specificity, cleaving multiple carotenoids at two different bond positions. J. Biol. Chem. 2008, 283, 11364–11373. [Google Scholar] [CrossRef]
- Floss, D.S.; Walter, M.H. Role of carotenoid cleavage dioxygenase 1 (CCD1) in apocarotenoid biogenesis revisited. Plant Signal. Behav. 2009, 4, 172–175. [Google Scholar] [CrossRef]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Da Silva Messias, R.; Galli, V.; Dos Anjos e Silva, S.D.; Rombaldi, C.V. Carotenoid Biosynthetic and Catabolic Pathways: Gene Expression and Carotenoid Content in Grains of Maize Landraces. Nutrients 2014, 6, 546-563. https://doi.org/10.3390/nu6020546
Da Silva Messias R, Galli V, Dos Anjos e Silva SD, Rombaldi CV. Carotenoid Biosynthetic and Catabolic Pathways: Gene Expression and Carotenoid Content in Grains of Maize Landraces. Nutrients. 2014; 6(2):546-563. https://doi.org/10.3390/nu6020546
Chicago/Turabian StyleDa Silva Messias, Rafael, Vanessa Galli, Sérgio Delmar Dos Anjos e Silva, and Cesar Valmor Rombaldi. 2014. "Carotenoid Biosynthetic and Catabolic Pathways: Gene Expression and Carotenoid Content in Grains of Maize Landraces" Nutrients 6, no. 2: 546-563. https://doi.org/10.3390/nu6020546
APA StyleDa Silva Messias, R., Galli, V., Dos Anjos e Silva, S. D., & Rombaldi, C. V. (2014). Carotenoid Biosynthetic and Catabolic Pathways: Gene Expression and Carotenoid Content in Grains of Maize Landraces. Nutrients, 6(2), 546-563. https://doi.org/10.3390/nu6020546




