Host Genotype and Weather Effects on Fusarium Head Blight Severity and Mycotoxin Load in Spring Barley
Abstract
:1. Introduction
2. Results
2.1. Content of Fungal DNA in Grain of Spring Barley Genotypes Inoculated with Fusarium spp. and Evaluation of Basal Quantitative FHB Resistance
2.2. Evaluation of Genotype Resistance and Pathogen Abundance after Fusarium spp. Co-Inoculation
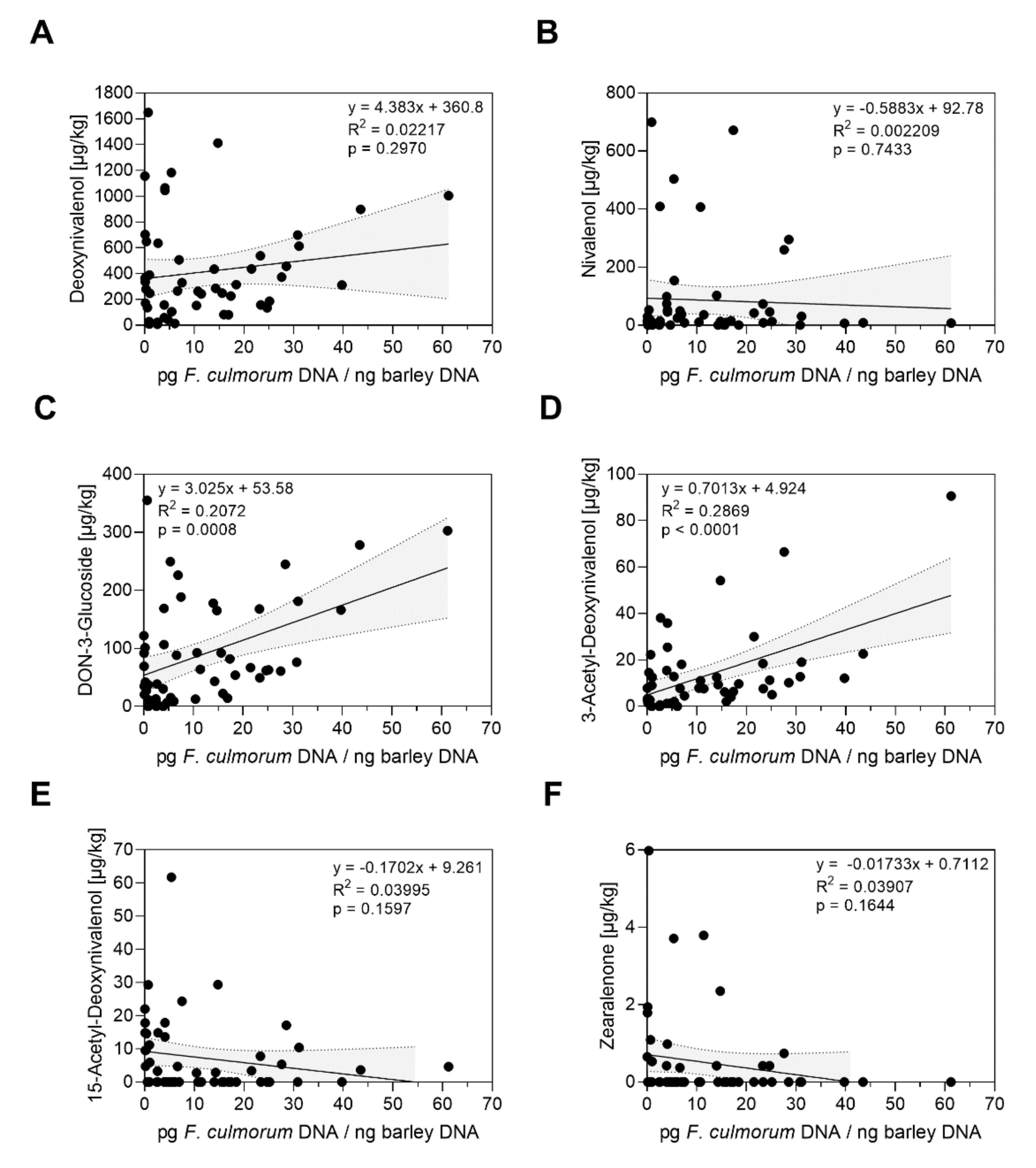
2.3. Correlation of Fusarium spp. DNA Contents and Mycotoxin Contamination in Barley Grain
2.4. Correlation of Plant Height and Grain Infection
2.5. Genotype Resistance under Different Inoculation Methods and Comparison of Bruised Grain Inoculation with Spray Inoculation
2.6. Correlation of Head Symptoms and Fungal Grain Infestation
2.7. Effect of Weather on Yearly Infection Success
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Inoculation Trials
5.2. Preparation of Fungal Inoculum
5.3. Inoculation of Barley Plants with Bruised Grain Material and Spraying of Heads
5.4. Weather Conditions
5.5. Statistical Analysis
5.6. Determination of Plant Height
5.7. Assessment of Symptoms and Sampling of Barley Heads
5.8. Isolation of Genomic DNA from Mature Grain Material
5.9. Quantification of Fusarium spp. DNA
5.10. Quantification of Multiple Fusarium spp. Toxins
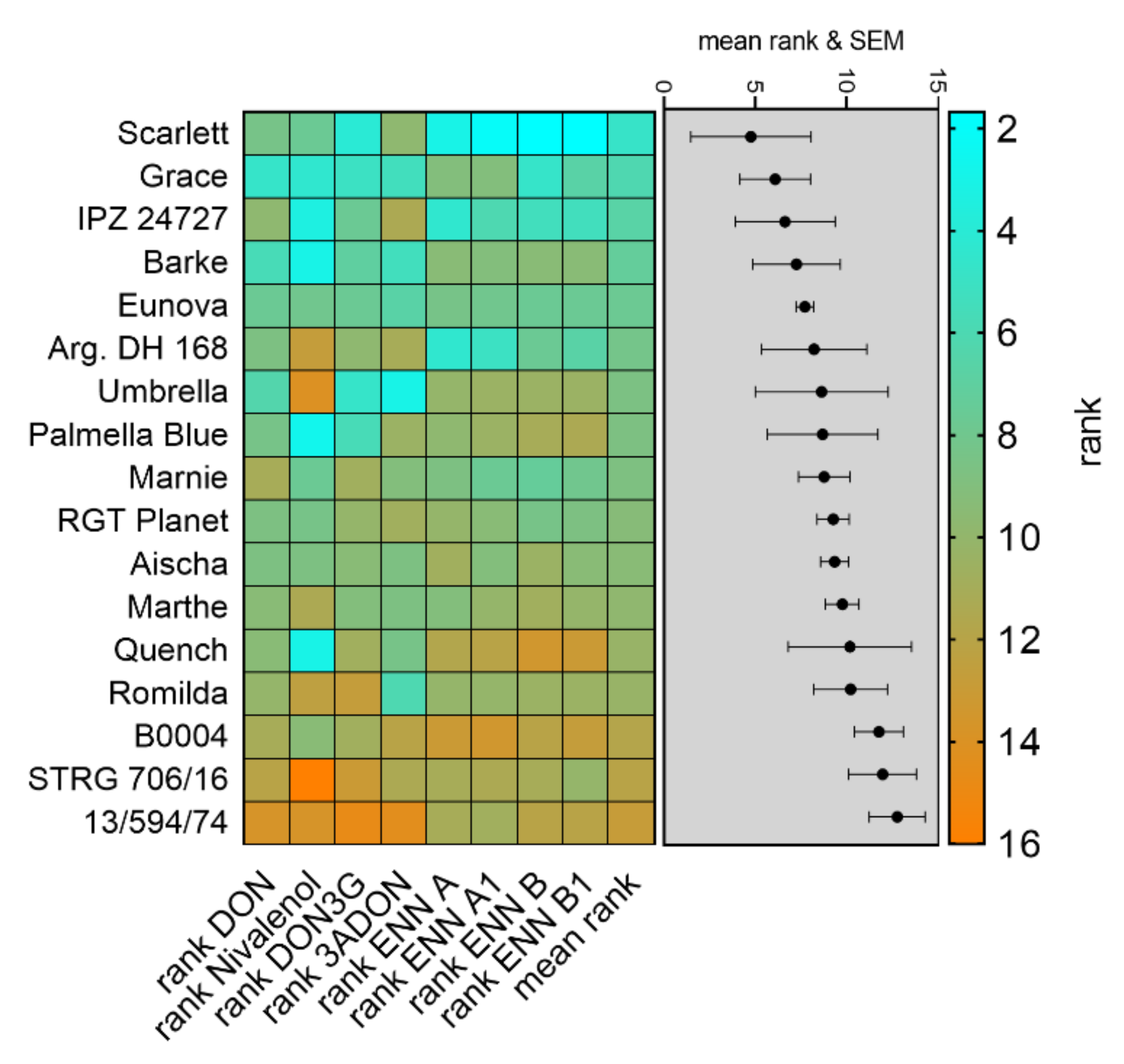
5.11. Calculation of Disease Severity Ranking
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Havis, N.D.; Brown, J.K.; Clemente, G.; Frei, P.; Jedryczka, M.; Kaczmarek, J.; Kaczmarek, M.; Matusinsky, P.; McGrann, G.R.; Pereyra, S.; et al. Ramularia collo-cygni—An emerging pathogen of barley crops. Phytopathology 2015, 105, 895–904. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parry, D.W.; Jenkinson, P.; McLeod, L. Fusarium ear blight (scab) in small grain cereals—A review. Plant Pathol. 1995, 44, 207–238. [Google Scholar] [CrossRef]
- Osborne, L.E.; Stein, J.M. Epidemiology of Fusarium head blight on small-grain cereals. Int. J. Food Microbiol. 2007, 119, 103–108. [Google Scholar] [CrossRef] [PubMed]
- McMullen, M.; Bergstrom, G.; De Wolf, E.; Dill-Macky, R.; Hershman, D.; Shaner, G.; Van Sanford, D. A unified effort to fight an enemy of wheat and barley: Fusarium head blight. Plant Dis. 2012, 96, 1712–1728. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rojas, E.C.; Jørgensen, H.J.; Jensen, B.; Collinge, D.B. Fusarium diseases: Biology and management perspectives. In Integrated Disease Management of Wheat and Barley; Burleigh Dodds Science Publishing: Cambridge, UK, 2018. [Google Scholar]
- Lewandowski, S.M.; Bushnell, W.R.; Evans, C.K. Distribution of mycelial colonies and lesions in field-grown barley inoculated with Fusarium graminearum. Phytopathology 2006, 96, 567–581. [Google Scholar] [CrossRef] [Green Version]
- Linkmeyer, A.; Hofer, K.; Rychlik, M.; Herz, M.; Hausladen, H.; Hückelhoven, R.; Hess, M. Influence of inoculum and climatic factors on the severity of Fusarium head blight in German spring and winter barley. Food Addit. Contam. Part A 2016, 33, 489–499. [Google Scholar] [CrossRef]
- Geißinger, C.; Hofer, K.; Habler, K.; Heß, M.; Hückelhoven, R.; Rychlik, M.; Becker, T.; Gastl, M. Fusarium species on barley malt: Is visual assessment an appropriate tool for detection? Cereal Chem. 2017, 94, 659–669. [Google Scholar] [CrossRef]
- Bottalico, A.; Perrone, G. Toxigenic Fusarium species and mycotoxins associated with head blight in small-grain cereals in Europe. In Mycotoxins in Plant Disease; Springer: Dordrecht, The Netherlands, 2002; pp. 611–624. [Google Scholar]
- Foroud, N.A.; Baines, D.; Gagkaeva, T.Y.; Thakor, N.; Badea, A.; Steiner, B.; Bürstmayr, M.; Bürstmayr, H. Trichothecenes in cereal grains–An update. Toxins 2019, 11, 634. [Google Scholar] [CrossRef] [Green Version]
- Sarlin, T.; Laitila, A.; Pekkarinen, A.; Haikara, A. Effects of three Fusarium species on the quality of barley and malt. J. Am. Soc. Brew. Chem. 2005, 63, 43–49. [Google Scholar]
- Hofer, K.; Geißinger, C.; König, C.; Gastl, M.; Hückelhoven, R.; Hess, M.; Coleman, A.D. Influence of Fusarium isolates on the expression of barley genes related to plant defense and malting quality. J. Cereal. Sci. 2016, 69, 17–24. [Google Scholar] [CrossRef]
- Oliveira, P.M.; Mauch, A.; Jacob, F.; Waters, D.M.; Arendt, E.K. Fundamental study on the influence of Fusarium infection on quality and ultrastructure of barley malt. Int. J. Food Microbiol. 2012, 156, 32–43. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.M.; Nicholson, P.; Thomsett, M.A.; Simpson, D.; Cooke, B.M.; Doohan, F.M.; Brennan, J.; Monaghan, S.; Moretti, A.; Mule, G.; et al. Relationship between the fungal complex causing Fusarium head blight of wheat and environmental conditions. Phytopathology 2018, 98, 69–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van der Lee, T.; Zhang, H.; van Diepeningen, A.; Waalwijk, C. Biogeography of Fusarium graminearum species complex and chemotypes: A review. Food Addit. Contam. Part A 2015, 32, 453–460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schöneberg, T.; Musa, T.; Forrer, H.R.; Mascher, F.; Bucheli, T.D.; Bertossa, M.; Keller, B.; Vogelgsang, S. Infection conditions of Fusarium graminearum in barley are variety specific and different from those in wheat. Eur. J. Plant Pathol. 2018, 151, 975–989. [Google Scholar] [CrossRef] [Green Version]
- Hofer, K.; Hückelhoven, R.; Hess, M. Analysis of archive samples of spring and winter barley support an increase in individual Fusarium species in Bavarian barley grain over the last decades. J. Plant Dis. Prot. 2019, 126, 247–254. [Google Scholar] [CrossRef]
- Habler, K.; Hofer, K.; Geißinger, C.; Schüler, J.; Hückelhoven, R.; Hess, M.; Gastl, M.; Rychlik, M. Fate of Fusarium toxins during the malting process. J. Agric. Food Chem. 2016, 64, 1377–1384. [Google Scholar] [CrossRef]
- Drakopoulos, D.; Sulyok, M.; Jenny, E.; Kägi, A.; Bänziger, I.; Logrieco, A.F.; Krska, R.; Vogelgsang, S. Fusarium Head Blight and Associated Mycotoxins in Grains and Straw of Barley: Influence of Agricultural Practices. Agronomy 2021, 11, 801. [Google Scholar] [CrossRef]
- Fernando, W.D.; Oghenekaro, A.O.; Tucker, J.R.; Badea, A. Building on a foundation: Advances in epidemiology, resistance breeding, and forecasting research for reducing the impact of Fusarium head blight in wheat and barley. Can. J. Plant Pathol. 2021, 43, 1–32. [Google Scholar] [CrossRef]
- Valverde-Bogantes, E.; Bianchini, A.; Herr, J.R.; Rose, D.J.; Wegulo, S.N.; Hallen-Adams, H.E. Recent population changes of Fusarium head blight pathogens: Drivers and implications. Can. J. Plant Pathol. 2020, 42, 315–329. [Google Scholar] [CrossRef]
- Ward, T.J.; Clear, R.M.; Rooney, A.P.; O’Donnell, K.; Gaba, D.; Patrick, S.; Starkey, D.E.; Gilbert, J.; Geiser, D.M.; Nowicki, T.W. An adaptive evolutionary shift in Fusarium head blight pathogen populations is driving the rapid spread of more toxigenic Fusarium graminearum in North America. Fungal Genet. Biol. 2008, 45, 473–484. [Google Scholar] [CrossRef]
- Waalwijk, C.; Kastelein, P.; de Vries, I.; Kerényi, Z.; van der Lee, T.; Hesselink, T.; Köhl, J.; Kema, G. Major changes in Fusarium spp. in wheat in the Netherlands. Eur. J. Plant Pathol. 2003, 109, 743–754. [Google Scholar] [CrossRef]
- Xue, A.G.; Chen, Y.; Seifert, K.; Guo, W.; Blackwell, B.A.; Harris, L.J.; Overy, D.P. Prevalence of Fusarium species causing head blight of spring wheat, barley and oat in Ontario during 2001–2017. Can. J. Plant Pathol. 2019, 41, 392–402. [Google Scholar] [CrossRef]
- Parikka, P.; Hakala, K.; Tiilikkala, K. Expected shifts in Fusarium species’ composition on cereal grain in Northern Europe due to climatic change. Food Addit. Contam. Part A 2012, 29, 1543–1555. [Google Scholar] [CrossRef] [PubMed]
- Madgwick, J.W.; West, J.S.; White, R.P.; Semenov, M.A.; Townsend, J.A.; Turner, J.A.; Fitt, B.D. Impacts of climate change on wheat anthesis and Fusarium ear blight in the UK. Eur. J. Plant Pathol. 2011, 130, 117–131. [Google Scholar] [CrossRef] [Green Version]
- Trnka, M.; Rötter, R.P.; Ruiz-Ramos, M.; Kersebaum, K.C.; Olesen, J.E.; Žalud, Z.; Semenov, M.A. Adverse weather conditions for European wheat production will become more frequent with climate change. Nat. Clim. Chang. 2014, 4, 637–643. [Google Scholar] [CrossRef]
- Chakraborty, S.; Newton, A.C. Climate change, plant diseases and food security: An overview. Plant Pathol. 2011, 60, 2–14. [Google Scholar] [CrossRef]
- Delgado-Baquerizo, M.; Guerra, C.A.; Cano-Díaz, C.; Egidi, E.; Wang, J.T.; Eisenhauer, N.; Singh, B.K.; Maestre, F.T. The proportion of soil-borne pathogens increases with warming at the global scale. Nat. Clim. Chang. 2020, 10, 1–5. [Google Scholar] [CrossRef]
- Pinotti, L.; Ottoboni, M.; Giromini, C.; Dell’Orto, V.; Cheli, F. Mycotoxin contamination in the EU feed supply chain: A focus on cereal byproducts. Toxins 2016, 8, 45. [Google Scholar] [CrossRef]
- Hückelhoven, R.; Hofer, K.; Coleman, A.; Heß, M. Fusarium infection of malting barley has to be managed over the entire value chain. J. Plant Dis. Prot. 2018, 125, 1–4. [Google Scholar] [CrossRef]
- Moretti, A.; Pascale, M.; Logrieco, A.F. Mycotoxin risks under a climate change scenario in Europe. Trends Food Sci. Technol. 2019, 84, 38–40. [Google Scholar] [CrossRef]
- Ingvordsen, C.H.; Backes, G.; Lyngkjaer, M.F.; Peltonen-Sainio, P.; Jensen, J.D.; Jalli, M.; Jahoor, A.; Rassmussen, M.; Mikkelsen, T.N.; Stockmarr, A.; et al. Significant decrease in yield under future climate conditions: Stability and production of 138 spring barley accessions. Eur. J. Agron. 2015, 63, 105–113. [Google Scholar] [CrossRef] [Green Version]
- Sallam, A.; Alqudah, A.M.; Dawood, M.F.; Baenziger, P.S.; Börner, A. Drought stress tolerance in wheat and barley: Advances in physiology, breeding and genetics research. Int. J. Mol. Sci. 2019, 20, 3137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pandey, P.; Ramegowda, V.; Senthil-Kumar, M. Shared and unique responses of plants to multiple individual stresses and stress combinations: Physiological and molecular mechanisms. Front. Plant Sci. 2015, 6, 723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramegowda, V.; Senthil-Kumar, M. The interactive effects of simultaneous biotic and abiotic stresses on plants: Mechanistic understanding from drought and pathogen combination. J. Plant Physiol. 2015, 176, 47–54. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Sonnewald, U. Differences and commonalities of plant responses to single and combined stresses. Plant J. 2017, 90, 839–855. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buerstmayr, M.; Steiner, B.; Buerstmayr, H. Breeding for Fusarium head blight resistance in wheat—Progress and challenges. Plant Breed. 2020, 139, 429–454. [Google Scholar] [CrossRef]
- De la Pena, R.C.; Smith, K.P.; Capettini, F.; Muehlbauer, G.J.; Gallo-Meagher, M.; Dill-Macky, R.; Somers, D.A.; Rasmusson, D.C. Quantitative trait loci associated with resistance to Fusarium head blight and kernel discoloration in barley. Theor. Appl. Genet. 1999, 99, 561–569. [Google Scholar] [CrossRef]
- Zhu, H.; Gilchrist, L.; Hayes, P.; Kleinhofs, A.; Kudrna, D.; Liu, Z.; Prom, L.; Steffenson, B.; Toojinda, T.; Vivar, H. Does function follow form? Principal QTLs for Fusarium head blight (FHB) resistance are coincident with QTLs for inflorescence traits and plant height in a doubled-haploid population of barley. Theor. Appl. Genet. 1999, 99, 1221–1232. [Google Scholar] [CrossRef]
- Rudd, J.C.; Horsley, R.D.; McKendry, A.L.; Elias, E.M. Host plant resistance genes for Fusarium head blight: Sources, mechanisms, and utility in conventional breeding systems. Crop Sci. 2001, 41, 620–627. [Google Scholar] [CrossRef] [Green Version]
- Mesfin, A.; Smith, K.P.; Dill-Macky, R.; Evans, C.K.; Waugh, R.; Gustus, C.D.; Muehlbauer, G.J. Quantitative trait loci for Fusarium head blight resistance in barley detected in a two-rowed by six-rowed population. Crop Sci. 2003, 43, 307–318. [Google Scholar] [CrossRef]
- Legge, W.G.; Therrien, M.C.; Tucker, J.R.; Banik, M.; Tekauz, A.; Somers, D.; Savard, M.E.; Rossnagel, B.G.; Lefol, E.; Voth, D.; et al. Progress in breeding for resistance to Fusarium head blight in barley. Can. J. Plant Pathol. 2004, 26, 436–442. [Google Scholar] [CrossRef]
- Linkmeyer, A.; Götz, M.; Hu, L.; Asam, S.; Rychlik, M.; Hausladen, H.; Hess, M.; Hückelhoven, R. Assessment and introduction of quantitative resistance to Fusarium head blight in elite spring barley. Phytopathology 2013, 103, 1252–1259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ogrodowicz, P.; Kuczyńska, A.; Mikołajczak, K.; Adamski, T.; Surma, M.; Krajewski, P.; Ćwiek-Kupczyńska, H.; Kempa, M.; Rokicki, M.; Jasińska, D. Mapping of quantitative trait loci for traits linked to Fusarium head blight in barley. PLoS ONE 2020, 15, e0222375. [Google Scholar] [CrossRef] [PubMed]
- Khanal, R.; Choo, T.M.; Xue, A.G.; Vigier, B.; Savard, M.E.; Blackwell, B.; Wang, J.; Yang, J.; Martin, R.A. Response of Barley Genotypes to Fusarium Head Blight under Natural Infection and Artificial Inoculation Conditions. Plant Pathol. J. 2021, 37, 455–464. [Google Scholar] [CrossRef] [PubMed]
- Schroeder, H.W.; Christensen, J.J. Factors affecting resistance of wheat to scab caused by Gibberella zeae. Phytopathology 1963, 53, 831–838. [Google Scholar]
- Mesterházy, Á. Types and components of resistance to Fusarium head blight of wheat. Plant Breed. 1995, 114, 377–386. [Google Scholar] [CrossRef]
- Boutigny, A.L.; Richard-Forget, F.; Barreau, C. Natural mechanisms for cereal resistance to the accumulation of Fusarium trichothecenes. Eur. J. Plant Pathol. 2008, 121, 411–423. [Google Scholar] [CrossRef]
- Martin, C.; Schöneberg, T.; Vogelgsang, S.; Vincenti, J.; Bertossa, M.; Mauch-Mani, B.; Mascher, F. Factors of wheat grain resistance to Fusarium head blight. Phytopathol. Mediterr. 2017, 56, 154–166. [Google Scholar]
- Martin, C.; Schöneberg, T.; Vogelgsang, S.; Morisoli, R.; Bertossa, M.; Mauch-Mani, B.; Mascher, F. Resistance against Fusarium graminearum and the relationship to β-glucan content in barley grains. Eur. J. Plant Pathol. 2018, 152, 621–634. [Google Scholar] [CrossRef]
- Jenkinson, P.; Parry, D.W. Splash dispersal of conidia of Fusarium culmorum and Fusarium avenaceum. Mycol. Res. 1994, 98, 506–510. [Google Scholar] [CrossRef]
- Hörberg, H.M. Patterns of splash dispersed conidia of Fusarium poae and Fusarium culmorum. Eur. J. Plant Pathol. 2002, 108, 73–80. [Google Scholar] [CrossRef]
- Bai, G.; Shaner, G. Management and resistance in wheat and barley to Fusarium head blight. Annu. Rev. Phytopathol. 2004, 42, 135–161. [Google Scholar] [CrossRef] [PubMed]
- Nicolaisen, M.; Supronienė, S.; Nielsen, L.K.; Lazzaro, I.; Spliid, N.H.; Justesen, A.F. Real-time PCR for quantification of eleven individual Fusarium species in cereals. J. Microbiol. Methods 2009, 76, 234–240. [Google Scholar] [CrossRef]
- Hoheneder, F.; Hofer, K.; Groth, J.; Herz, M.; Heß, M.; Hückelhoven, R. Ramularia leaf spot disease of barley is highly host genotype-dependent and suppressed by continuous drought stress in the field. J. Plant Dis. Prot. 2021, 128, 1–19. [Google Scholar] [CrossRef]
- Salas, B.; Steffenson, B.J.; Casper, H.H.; Tacke, B.; Prom, L.K.; Fetch, T.G., Jr.; Schwarz, P.B. Fusarium species pathogenic to barley and their associated mycotoxins. Plant Dis. 1999, 83, 667–674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mesterázy, Á.; Bartók, T.; Kászonyi, G.; Varga, M.; Tóth, B.; Varga, J. Common resistance to different Fusarium spp. causing Fusarium head blight in wheat. Eur. J. Plant Pathol. 2005, 112, 267–281. [Google Scholar] [CrossRef]
- Xue, A.; Ho, K.; Butler, G.; Vigier, B.; Babcock, C. Pathogenicity of Fusarium species causing head blight in barley. Phytoprotection 2006, 87, 55–61. [Google Scholar] [CrossRef] [Green Version]
- Geddes, J.; Eudes, F.; Tucker, J.R.; Legge, W.G.; Selinger, L.B. Evaluation of inoculation methods on infection and deoxynivalenol production by Fusarium graminearum on barley. Can. J. Plant Pathol. 2008, 30, 66–73. [Google Scholar] [CrossRef]
- Imathiu, S.; Edwards, S.; Ray, R.; Back, M. Artificial inoculum and inoculation techniques commonly used in the investigation of Fusarium head blight in cereals. Acta Phytopathol. Entomol. Hung. 2014, 49, 129–139. [Google Scholar] [CrossRef]
- Yan, W.; Li, H.B.; Cai, S.B.; Ma, H.X.; Rebetzke, G.J.; Liu, C.J. Effects of plant height on type I and type II resistance to Fusarium head blight in wheat. Plant Pathol. 2011, 60, 506–512. [Google Scholar] [CrossRef]
- Buerstmayr, H.; Legzdina, L.; Steiner, B.; Lemmens, M. Variation for resistance to Fusarium head blight in spring barley. Euphytica 2004, 137, 279–290. [Google Scholar] [CrossRef]
- Ma, Z.; Steffenson, B.J.; Prom, L.K.; Lapitan, N.L. Mapping of quantitative trait loci for Fusarium head blight resistance in barley. Phytopathology 2000, 90, 1079–1088. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, G.T.; Franckowiak, J.D.; Neate, S.M.; Zhang, B.; Horsley, R.D. A native QTL for Fusarium head blight resistance in North American barley (Hordeum vulgare L.) independent of height, maturity, and spike type loci. Genome 2010, 53, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Spanic, V.; Katanic, Z.; Sulyok, M.; Krska, R.; Puskas, K.; Vida, G.; Drezner, G.; Šarkanj, B. Multiple fungal metabolites including mycotoxins in naturally infected and Fusarium-inoculated wheat samples. Microorganisms 2020, 8, 578. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Habler, K.; Rychlik, M. Multi-mycotoxin stable isotope dilution LC-MS/MS method for Fusarium toxins in cereals. Anal. Bioanal. Chem. 2016, 408, 307–317. [Google Scholar] [CrossRef] [PubMed]
- Mesterházy, Á.; Bartók, T.; Mirocha, C.G.; Komoroczy, R. Nature of wheat resistance to Fusarium head blight and the role of deoxynivalenol for breeding. Plant Breed. 1999, 118, 97–110. [Google Scholar] [CrossRef]
- Poppenberger, B.; Berthiller, F.; Lucyshyn, D.; Sieberer, T.; Schuhmacher, R.; Krska, R.; Kuchler, K.; Glössl, J.; Luschnig, C.; Adam, G. Detoxification of the Fusarium mycotoxin deoxynivalenol by a UDP-glucosyltransferase from Arabidopsis thaliana. J. Biol. Chem. 2003, 278, 47905–47914. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xing, L.; Gao, L.; Chen, Q.; Pei, H.; Di, Z.; Xiao, J.; Wang, H.; Ma, L.; Chen, P.; Cao, A.; et al. Over-expressing a UDP-glucosyltransferase gene (Ta-UGT 3) enhances Fusarium Head Blight resistance of wheat. Plant Growth Regul. 2018, 84, 561–571. [Google Scholar] [CrossRef]
- Yoshida, M.; Kawada, N.; Nakajima, T. Effect of infection timing on Fusarium head blight and mycotoxin accumulation in open-and closed-flowering barley. Phytopathology 2007, 97, 1054–1062. [Google Scholar] [CrossRef] [Green Version]
- Musa, T.; Hecker, A.; Vogelgsang, S.; Forrer, H.R. Forecasting of Fusarium head blight and deoxynivalenol content in winter wheat with FusaProg. EPPO Bull. 2007, 37, 283–289. [Google Scholar] [CrossRef]
- Brustolin, R.; Zoldan, S.M.; Reis, E.M.; Zanatta, T.; Carmona, M. Weather requirements and rain forecast to time fungicide application for Fusarium head blight control in wheat. Summa Phytopathol. 2013, 39, 248–251. [Google Scholar] [CrossRef] [Green Version]
- Hofgaard, I.S.; Seehusen, T.; Aamot, H.U.; Riley, H.; Razzaghian, J.; Le, V.H.; Hjelkrem, A.R.; Dill-Macky, R.; Brodal, G. Inoculum potential of Fusarium spp. relates to tillage and straw management in Norwegian fields of spring oats. Front. Microbiol. 2016, 7, 556. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beccari, G.; Prodi, A.; Tini, F.; Bonciarelli, U.; Onofri, A.; Oueslati, S.; Limayma, M.; Covarelli, L. Changes in the Fusarium head blight complex of malting barley in a three-year field experiment in Italy. Toxins 2017, 9, 120. [Google Scholar] [CrossRef] [Green Version]
- Nielsen, L.K.; Cook, D.J.; Edwards, S.G.; Ray, R.V. The prevalence and impact of Fusarium head blight pathogens and mycotoxins on malting barley quality in UK. Int. J. Food Microbiol. 2014, 179, 38–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hietaniemi, V.; Rämö, S.; Yli-Mattila, T.; Jestoi, M.; Peltonen, S.; Kartio, M.; Sieviläinen, E.; Koivisto, T.; Parikka, P. Updated survey of Fusarium species and toxins in Finnish cereal grains. Food Addit. Contam. Part A 2016, 33, 831–848. [Google Scholar] [CrossRef]
- Agrarmeteorologie Bayern. 2021. Available online: https://www.am.rlp.de/Internet/AM/NotesBAM.nsf/bamweb/89952241408487f9c12573920045299b?OpenDocument&TableRow=3.10#3 (accessed on 17 February 2021).
- Joint Research Centre. Maize Seed Sampling and DNA Extraction. Document CRLV04/05XP. 2007. Available online: http://gmo-crl.jrc.ec.europa.eu/summaries/MIR604_DNAExtr.pdf (accessed on 20 December 2021).
- Mandel, J. The Statistical Analysis of Experimental Data; Interscience Publishers: Geneva, Switzerland, 1964; pp. 160–192. [Google Scholar]
- Asam, S.; Rychlik, M. Synthesis of four carbon-13-labeled type A trichothecene mycotoxins and their application as internal standards in stable isotope dilution assays. J. Agric. Food Chem. 2006, 54, 6535–6546. [Google Scholar] [CrossRef]
- Hu, L.; Rychlik, M. Biosynthesis of 15N3-labeled enniatins and beauvericin and their application to stable isotope dilution assays. J. Agric. Food Chem. 2012, 60, 7129–7136. [Google Scholar] [CrossRef]
- Ward, J.H. Hierarchical grouping to optimize an objective function. J. Am. Stat. Assoc. 1963, 58, 236–244. [Google Scholar] [CrossRef]
- Comadran, J.; Kilian, B.; Russell, J.; Ramsay, L.; Stein, N.; Ganal, M.; Shaw, P.; Bayer, M.; Thomas, W.; Marshall, D.; et al. Natural variation in a homolog of Antirrhinum CENTRORADIALIS contributed to spring growth habit and environmental adaptation in cultivated barley. Nat. Genet. 2012, 44, 1388–1392. [Google Scholar] [CrossRef]
- Wimmer, V.; Albrecht, T.; Auinger, H.J.; Schön, C.C. synbreed: A framework for the analysis of genomic prediction data using R. Bioinformatics 2012, 28, 2086–2087. [Google Scholar] [CrossRef] [Green Version]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2021; Available online: https://www.R-project.org/ (accessed on 20 December 2021).






Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hoheneder, F.; Biehl, E.M.; Hofer, K.; Petermeier, J.; Groth, J.; Herz, M.; Rychlik, M.; Heß, M.; Hückelhoven, R. Host Genotype and Weather Effects on Fusarium Head Blight Severity and Mycotoxin Load in Spring Barley. Toxins 2022, 14, 125. https://doi.org/10.3390/toxins14020125
Hoheneder F, Biehl EM, Hofer K, Petermeier J, Groth J, Herz M, Rychlik M, Heß M, Hückelhoven R. Host Genotype and Weather Effects on Fusarium Head Blight Severity and Mycotoxin Load in Spring Barley. Toxins. 2022; 14(2):125. https://doi.org/10.3390/toxins14020125
Chicago/Turabian StyleHoheneder, Felix, Eva Maria Biehl, Katharina Hofer, Johannes Petermeier, Jennifer Groth, Markus Herz, Michael Rychlik, Michael Heß, and Ralph Hückelhoven. 2022. "Host Genotype and Weather Effects on Fusarium Head Blight Severity and Mycotoxin Load in Spring Barley" Toxins 14, no. 2: 125. https://doi.org/10.3390/toxins14020125




