Secondary Metabolite Gene Regulation in Mycotoxigenic Fusarium Species: A Focus on Chromatin
Abstract
:1. Introduction
2. Chromatin: The Basic Packaging Form of DNA
3. The Many Shades of Histone Post-Translational Modifications
3.1. Histone Lysine Methylation—A Ballett of Transcriptional Homeostasis
3.1.1. H3K4me3 Facilitated by Set1/Kmt2 Is a Hallmark of Euchromatin
3.1.2. Kmt1 Is Involved in the Establishment of H3K9me3-Dependent Heterochromatin
3.1.3. H3K27me3 Is a Bona Fide Mark for Facultative Heterochromatin Installed by Kmt6
3.1.4. Two Enzymes, Kmt3 and Ash1, Are Involved in H3K36me3
3.1.5. Kmt5-Mediated H4K20me3—A Histone Mark to Be Explored
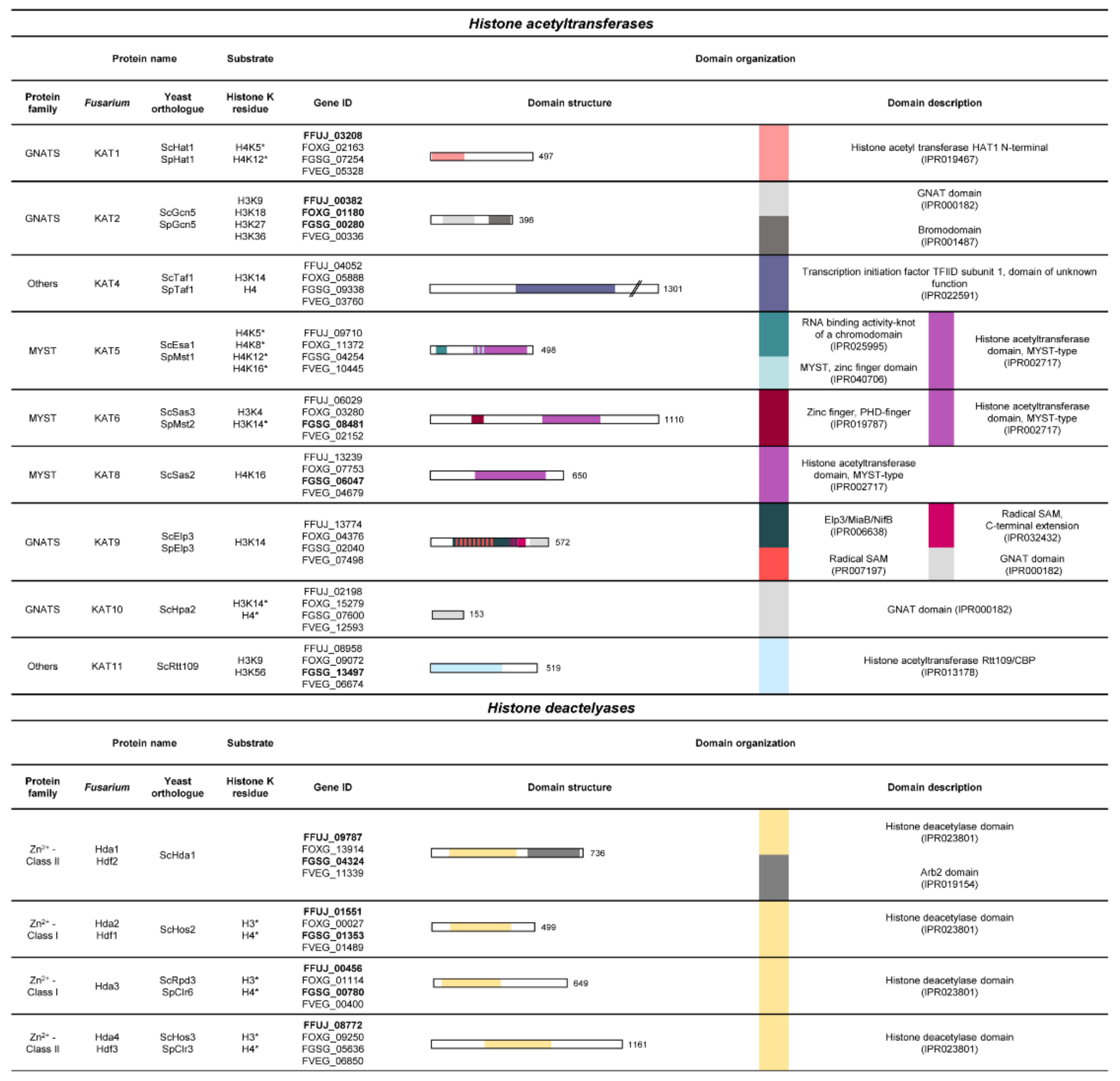
3.2. Histone Lysine Acetylation
3.2.1. The SAGA Complex
3.2.2. The NuA4 and NuA3 Complex
3.2.3. The Roles of So Far Less Characterized Histone Acetyltransferases
3.2.4. HAT1—A Standalone Histone Acetyltransferase
3.2.5. The Group of Zn2+-Dependent HDACs: Class I and II
4. The Dynamic Interplay of Histone-Modifying Enzymes, Global and Narrow Regulators
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- Leslie, J.F.; Summerell, B.A. The Fusarium Laboratory Manual; Blackwell Publishing: Iowa, IA, USA, 2006; Volume 6, ISBN 9780813819198. [Google Scholar]
- Dean, R.; van Kan, J.A.L.; Pretorius, Z.A.; Hammond-Kosack, K.E.; di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; et al. The Top 10 Fungal Pathogens in Molecular Plant Pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef] [Green Version]
- Nucci, F.; Nouér, S.A.; Capone, D.; Anaissie, E.; Nucci, M. Fusariosis. Semin. Respir. Crit. Care Med. 2015, 36, 706–714. [Google Scholar] [CrossRef] [PubMed]
- O’Donnell, K.; Al-Hatmi, A.M.S.; Aoki, T.; Brankovics, B.; Cano-Lira, J.F.; Coleman, J.J.; de Hoog, G.S.; di Pietro, A.; Frandsen, R.J.N.; Geiser, D.M.; et al. No to Neocosmospora: Phylogenomic and Practical Reasons for Continued Inclusion of the Fusarium Solani Species Complex in the Genus Fusarium. mSphere 2020, 5, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Aoki, T.; O’Donnell, K.; Geiser, D.M. Systematics of Key Phytopathogenic Fusarium Species: Current Status and Future Challenges. J. Gen. Plant Pathol. 2014, 80, 189–201. [Google Scholar] [CrossRef]
- Summerell, B.A. Resolving Fusarium: Current Status of the Genus. Annu. Rev. Phytopathol. 2019, 57, 323–339. [Google Scholar] [CrossRef] [PubMed]
- Van der Lee, T.; Zhang, H.; van Diepeningen, A.; Waalwijk, C. Biogeography of Fusarium Graminearum Species Complex and Chemotypes: A Review. Food Addit. Contam.-Part A Chem. Anal. Control Expo. Risk Assess 2015, 32, 453–460. [Google Scholar] [CrossRef] [Green Version]
- Ma, L.; Geiser, D.M.; Proctor, R.H.; Rooney, A.P.; Donnell, K.O.; Trail, F.; Gardiner, D.M.; Manners, J.M.; Kazan, K. Fusarium Pathogenomics. Annu. Rev. Microbiol. 2013, 67, 399–416. [Google Scholar] [CrossRef] [Green Version]
- Niehaus, E.-M.; Münsterkötter, M.; Proctor, R.H.; Brown, D.W.; Sharon, A.; Idan, Y.; Oren-Young, L.; Sieber, C.M.; Novak, O.; Pencik, A.; et al. Comparative “Omics” of the Fusarium Fujikuroi Species Complex Highlights Differences in Genetic Potential and Metabolite Synthesis. Genome Biol. Evol. 2016, 8, 3574–3599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Proctor, R.H.; Brown, D.W.; Plattner, R.D.; Desjardins, A.E. Co-Expression of 15 Contiguous Genes Delineates a Fumonisin Biosynthetic Gene Cluster in Gibberella Moniliformis. Fungal Genet. Biol. 2003, 38, 237–249. [Google Scholar] [CrossRef]
- Proctor, R.H.; Plattner, R.D.; Desjardins, A.E.; Busman, M.; Butchko, R.A.E. Fumonisin Production in the Maize Pathogen Fusarium Verticillioides: Genetic Basis of Naturally Occurring Chemical Variation. J. Agric. Food Chem. 2006, 54, 2424–2430. [Google Scholar] [CrossRef] [Green Version]
- Steogonpień, L.; Koczyk, G.; Waśkiewicz, A. FUM Cluster Divergence in Fumonisins-Producing Fusarium Species. Fungal Biol. 2011, 115, 112–123. [Google Scholar] [CrossRef]
- Pasquali, M.; Beyer, M.; Logrieco, A.; Audenaert, K.; Balmas, V.; Basler, R.; Boutigny, A.L.; Chrpová, J.; Czembor, E.; Gagkaeva, T.; et al. A European Database of Fusarium Graminearum and F. Culmorum Trichothecene Genotypes. Front. Microbiol. 2016, 7, 406. [Google Scholar] [CrossRef] [Green Version]
- Gautier, C.; Pinson-Gadais, L.; Richard-Forget, F. Fusarium Mycotoxins Enniatins: An Updated Review of Their Occurrence, the Producing Fusarium Species, and the Abiotic Determinants of Their Accumulation in Crop Harvests. J. Agric. Food Chem. 2020, 68, 4788–4798. [Google Scholar] [CrossRef] [PubMed]
- Urbaniak, M.; Stępień, Ł.; Uhlig, S. Evidence for Naturally Produced Beauvericins Containing N-Methyl-Tyrosine in Hypocreales Fungi. Toxins 2019, 11, 182. [Google Scholar] [CrossRef] [Green Version]
- Munkvold, G.P. Fusarium Species and Their Associated Mycotoxins. In Methods in Molecular Biology; Humana Press Inc.: New York, NY, USA, 2017; Volume 1542, pp. 51–106. [Google Scholar]
- Nesic, K.; Ivanovic, S.; Nesic, V. Fusarial Toxins: Secondary Metabolites of Fusarium Fungi. Rev. Environ. Contam. Toxicol. Vol. 2014, 228, 101–120. [Google Scholar] [CrossRef]
- Wiemann, P.; Sieber, C.M.K.; von Bargen, K.W.; Studt, L.; Niehaus, E.M.; Espino, J.J.; Huß, K.; Michielse, C.B.; Albermann, S.; Wagner, D.; et al. Deciphering the Cryptic Genome: Genome-Wide Analyses of the Rice Pathogen Fusarium Fujikuroi Reveal Complex Regulation of Secondary Metabolism and Novel Metabolites. PLoS Pathog. 2013, 9, e1003475. [Google Scholar] [CrossRef] [Green Version]
- Niehaus, E.M.; Kim, H.K.; Münsterkötter, M.; Janevska, S.; Arndt, B.; Kalinina, S.A.; Houterman, P.M.; Ahn, I.-P.; Alberti, I.; Tonti, S.; et al. Comparative Genomics of Geographically Distant Fusarium Fujikuroi Isolates Revealed Two Distinct Pathotypes Correlating with Secondary Metabolite Profiles. PLoS Pathog. 2017, 13, e1006670. [Google Scholar] [CrossRef] [Green Version]
- Macheleidt, J.; Mattern, D.J.; Fischer, J.; Netzker, T.; Weber, J.; Schroeckh, V.; Valiante, V.; Brakhage, A.A. Regulation and Role of Fungal Secondary Metabolites. Annu. Rev. Genet. 2016, 50, 371–392. [Google Scholar] [CrossRef] [PubMed]
- Keller, N.P. Fungal Secondary Metabolism: Regulation, Function and Drug Discovery. Nat. Rev. Microbiol. 2019, 17, 167–180. [Google Scholar] [CrossRef] [PubMed]
- Cuzick, A.; Urban, M.; Hammond-Kosack, K. Fusarium Graminearum Gene Deletion Mutants Map1 and Tri5 Reveal Similarities and Differences in the Pathogenicity Requirements to Cause Disease on Arabidopsis and Wheat Floral Tissue. New Phytol. 2008, 177, 990–1000. [Google Scholar] [CrossRef]
- Proctor, R.H.; Hohn, T.M.; McCormick, S.P. Reduced Virulence of Gibberella Zeae Caused by Disruption of a Trichothecene Toxin Biosynthetic Gene. MPMI 1994, 8, 593–601. [Google Scholar] [CrossRef] [Green Version]
- Trail, F. For Blighted Waves of Grain: Fusarium Graminearum in the Postgenomics Era. Plant Physiol. 2009, 149, 103–110. [Google Scholar] [CrossRef] [Green Version]
- Bayram, Ö.; Braus, G.H. Coordination of Secondary Metabolism and Development in Fungi: The Velvet Family of Regulatory Proteins. FEMS Microbiol. Rev. 2012, 36, 1–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiemann, P.; Brown, D.W.; Kleigrewe, K.; Bok, J.W.; Keller, N.P.; Humpf, H.U.; Tudzynski, B. FfVel1 and Fflae1, Components of a Velvet-like Complex in Fusarium Fujikuroi, Affect Differentiation, Secondary Metabolism and Virulence. Mol. Microbiol. 2010, 77, 972–994. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- López-Berges, M.S.; Schäfer, K.; Hera, C.; di Pietro, A. Combinatorial Function of Velvet and AreA in Transcriptional Regulation of Nitrate Utilization and Secondary Metabolism. Fungal Genet. Biol. 2014, 62, 78–84. [Google Scholar] [CrossRef] [PubMed]
- López-Berges, M.S.; Hera, C.; Sulyok, M.; Schäfer, K.; Capilla, J.; Guarro, J.; di Pietro, A. The Velvet Complex Governs Mycotoxin Production and Virulence of Fusarium Oxysporum on Plant and Mammalian Hosts. Mol. Microbiol. 2013, 87, 49–65. [Google Scholar] [CrossRef] [PubMed]
- Merhej, J.; Urban, M.; Dufresne, M.; Hammond-Kosack, K.E.; Richard-Forget, F.; Barreau, C. The Velvet Gene, FgVe1, Affects Fungal Development and Positively Regulates Trichothecene Biosynthesis and Pathogenicity in Fusarium Graminearum. Mol. Plant Pathol. 2012, 13, 363–374. [Google Scholar] [CrossRef]
- Jiang, J.; Liu, X.; Yin, Y.; Ma, Z. Involvement of a Velvet Protein FgVeA in the Regulation of Asexual Development, Lipid and Secondary Metabolisms and Virulence in Fusarium Graminearum. PLoS ONE 2011, 6, e28291. [Google Scholar] [CrossRef] [Green Version]
- Myung, K.; Zitomer, N.C.; Duvall, M.; Glenn, A.E.; Riley, R.T.; Calvo, A.M. The Conserved Global Regulator VeA Is Necessary for Symptom Production and Mycotoxin Synthesis in Maize Seedlings by Fusarium Verticillioides. Plant Pathol. 2012, 61, 152–160. [Google Scholar] [CrossRef] [Green Version]
- Jiang, J.; Yun, Y.; Liu, Y.; Ma, Z. FgVELB Is Associated with Vegetative Differentiation, Secondary Metabolism and Virulence in Fusarium Graminearum. Fungal Genet. Biol. 2012, 49, 653–662. [Google Scholar] [CrossRef]
- Lee, J.; Myong, K.; Kim, J.E.; Kim, H.K.; Yun, S.H.; Lee, Y.W. FgVelB Globally Regulates Sexual Reproduction, Mycotoxin Production and Pathogenicity in the Cereal Pathogen Fusarium Graminearum. Microbiology 2012, 158, 1723–1733. [Google Scholar] [CrossRef] [Green Version]
- Niehaus, E.M.; Rindermann, L.; Janevska, S.; Münsterkötter, M.; Güldener, U.; Tudzynski, B. Analysis of the Global Regulator Lae1 Uncovers a Connection between Lae1 and the Histone Acetyltransferase HAT1 in Fusarium Fujikuroi. Appl. Microbiol. Biotechnol. 2018, 102, 279–295. [Google Scholar] [CrossRef] [PubMed]
- Butchko, R.A.E.; Brown, D.W.; Busman, M.; Tudzynski, B.; Wiemann, P. Lae1 Regulates Expression of Multiple Secondary Metabolite Gene Clusters in Fusarium Verticillioides. Fungal Genet. Biol. 2012, 49, 602–612. [Google Scholar] [CrossRef] [Green Version]
- Niehaus, E.M.; Schumacher, J.; Burkhardt, I.; Rabe, P.; Spitzer, E.; Münsterkötter, M.; Güldener, U.; Sieber, C.M.K.; Dickschat, J.S.; Tudzynski, B. The GATA-Type Transcription Factor Csm1 Regulates Conidiation and Secondary Metabolism in Fusarium Fujikuroi. Front. Microbiol. 2017, 8, 1175. [Google Scholar] [CrossRef]
- Tudzynski, B.; Homann, V.; Feng, B.; Marzluf, G.A. Isolation, Characterization and Disruption of the AreA Nitrogen Regulatory Gene of Gibberella Fujikuroi. Mol. Gen. Genet. 1999, 261, 106–114. [Google Scholar] [CrossRef]
- Mihlan, M.; Homann, V.; Liu, T.-W.D.; Tudzynski, B. AREA Directly Mediates Nitrogen Regulation of Gibberellin Biosynthesis in Gibberella Fujikuroi, but Its Activity Is Not Affected by NMR. Mol. Microbiol. 2003, 47, 975–991. [Google Scholar] [CrossRef] [PubMed]
- Pfannmüller, A.; Leufken, J.; Studt, L.; Michielse, C.B.; Sieber, C.M.K.; Güldener, U.; Hawat, S.; Hippler, M.; Fufezan, C.; Tudzynski, B. Comparative Transcriptome and Proteome Analysis Reveals a Global Impact of the Nitrogen Regulators AreA and AreB on Secondary Metabolism in Fusarium Fujikuroi. PLoS ONE 2017, 12, e0176194. [Google Scholar] [CrossRef] [PubMed]
- Bannister, A.J.; Kouzarides, T. Regulation of Chromatin by Histone Modifications. Cell Res. 2011, 21, 381–395. [Google Scholar] [CrossRef]
- Chen, Z.; Ponts, N. H2A.Z and Chromatin Remodelling Complexes: A Focus on Fungi. Crit. Rev. Microbiol. 2020, 46, 321–337. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Zehraoui, E.; Atanasoff-Kardjalieff, A.K.; Strauss, J.; Studt, L.; Ponts, N. Effect of H2A.Z Deletion Is Rescued by Compensatory Mutations in Fusarium Graminearum. PLoS Genet. 2020, 16, e1009125. [Google Scholar] [CrossRef]
- Yang, X.J.; Seto, E. The Rpd3/Hda1 Family of Lysine Deacetylases: From Bacteria and Yeast to Mice and Men. Nat. Rev. Mol. Cell. Biol. 2008, 9, 206–218. [Google Scholar] [CrossRef] [Green Version]
- Hyun, K.; Jeon, J.; Park, K.; Kim, J. Writing, Erasing and Reading Histone Lysine Methylations. Exp. Mol. Med. 2017, 49, e324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nicholson, T.B.; Veland, N.; Chen, T. Writers, Readers, and Erasers of Epigenetic Marks. In Epigenetic Cancer Therapy; Elsevier Inc.: Amsterdam, The Netherlands, 2015; pp. 31–66. ISBN 9780128002247. [Google Scholar]
- Allfrey, V.G.; Faulkner, R.; Mirsky, A.E. Acetylation and methylation of histones and their possible role in the regulation of rna synthesis*. Proc. Natl. Acad. Sci. USA 1964, 51, 786–794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verrier, L.; Vandromme, M.; Trouche, D. Histone Demethylases in Chromatin Cross-Talks. Biol. Cell 2011, 103, 381–401. [Google Scholar] [CrossRef] [Green Version]
- Blum, M.; Chang, H.Y.; Chuguransky, S.; Grego, T.; Kandasaamy, S.; Mitchell, A.; Nuka, G.; Paysan-Lafosse, T.; Qureshi, M.; Raj, S.; et al. The InterPro Protein Families and Domains Database: 20 Years On. Nucleic Acids Res. 2021, 49, D344–D354. [Google Scholar] [CrossRef]
- Keats Shwab, E.; Bok, J.W.; Galehr, J.; Shwab, E.K.; Tribus, M.; Keller, N.P.; Graessle, S. Histone Deacetylase Activity Regulates Chemical Diversity in Aspergillus. Eukaryot. Cell 2007, 6, 1656–1664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reyes-Dominguez, Y.; Boedi, S.; Sulyok, M.; Wiesenberger, G.; Stoppacher, N.; Krska, R.; Strauss, J. Heterochromatin Influences the Secondary Metabolite Profile in the Plant Pathogen Fusarium Graminearum. Fungal Genet. Biol. 2012, 49, 39–47. [Google Scholar] [CrossRef] [Green Version]
- Studt, L.; Wiemann, P.; Kleigrewe, K.; Humpf, H.; Tudzynski, B. Biosynthesis of Fusarubins Accounts for Pigmentation of Fusarium Fujikuroi Perithecia. Appl. Environ. Microbiol. 2012, 78, 4468–4480. [Google Scholar] [CrossRef] [Green Version]
- Westphal, K.R.; Bachleitner, S.; Severinsen, M.M.; Brundtø, M.L.; Hansen, F.T.; Sørensen, T.; Wollenberg, R.D.; Lysøe, E.; Studt, L.; Sørensen, J.L.; et al. Cyclic, Hydrophobic Hexapeptide Fusahexin Is the Product of a Nonribosomal Peptide Synthetase in Fusarium Graminearum. J. Nat. Prod. 2021, 84, 2070–2080. [Google Scholar] [CrossRef]
- Tobiasen, C.; Aahman, J.; Ravnholt, K.S.; Bjerrum, M.J.; Grell, M.N.; Giese, H. Nonribosomal Peptide Synthetase (NPS) Genes in Fusarium Graminearum, F. Culmorum and F. Pseudograminearium and Identification of NPS2 as the Producer of Ferricrocin. Curr. Genet. 2007, 51, 43–58. [Google Scholar] [CrossRef] [PubMed]
- Varga, J.; Kocsube, S.; Toth, B.; Mesterhazy, A. Nonribosomal peptide synthetase genes in the genome of Fusarium graminearum, causative agent of wheat head blight. Acta Biol. Hung. 2005, 56, 375–388. [Google Scholar] [CrossRef]
- Janevska, S.; Arndt, B.; Niehaus, E.M.; Burkhardt, I.; Rösler, S.M.; Brock, N.L.; Humpf, H.U.; Dickschat, J.S.; Tudzynski, B. Gibepyrone Biosynthesis in the Rice Pathogen Fusarium Fujikuroi Is Facilitated by a Small Polyketide Synthase Gene Cluster. J. Biol. Chem. 2016, 291, 27403–27420. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bömke, C.; Tudzynski, B. Diversity, Regulation, and Evolution of the Gibberellin Biosynthetic Pathway in Fungi Compared to Plants and Bacteria. Phytochemistry 2009, 70, 1876–1893. [Google Scholar] [CrossRef]
- Rhouma, M.B.; Kriaa, M.; Nasr, Y.B.; Mellouli, L.; Kammoun, R. A New Endophytic Fusarium Oxysporum Gibberellic Acid: Optimization of Production Using Combined Strategies of Experimental Designs and Potency on Tomato Growth under Stress Condition. BioMed Res. Int. 2020, 2020, 4587148. [Google Scholar] [CrossRef] [Green Version]
- Tsavkelova, E.A.; Bömke, C.; Netrusov, A.I.; Weiner, J.; Tudzynski, B. Production of Gibberellic Acids by an Orchid-Associated Fusarium Proliferatum Strain. Fungal Genet. Biol. 2008, 45, 1393–1403. [Google Scholar] [CrossRef] [PubMed]
- Munkvold, G.P.; Proctor, R.H.; Moretti, A. Mycotoxin Production in Fusarium According to Contemporary Species Concepts. Annu. Rev. Phytopathol. 2021, 59. [Google Scholar] [CrossRef] [PubMed]
- Ceranic, A.; Svoboda, T.; Berthiller, F.; Sulyok, M.; Samson, J.M.; Güldener, U.; Schuhmacher, R.; Adam, G. Identification and Functional Characterization of the Gene Cluster Responsible for Fusaproliferin Biosynthesis in Fusarium Proliferatum. Toxins 2021, 13, 468. [Google Scholar] [CrossRef]
- Oide, S.; Berthiller, F.; Wiesenberger, G.; Adam, G.; Turgeon, B.G. Individual and Combined Roles of Malonichrome, Ferricrocin, and TAFC Siderophores in Fusarium Graminearum Pathogenic and Sexual Development. Front. Microbiol. 2014, 5, 759. [Google Scholar] [CrossRef] [PubMed]
- Westphal, K.R.; Nielsen, K.A.H.; Wollenberg, R.D.; Møllehøj, M.B.; Bachleitner, S.; Studt, L.; Lysøe, E.; Giese, H.; Wimmer, R.; Sørensen, J.L.; et al. Fusaoctaxin A, an Example of a Two-Step Mechanism for Non-Ribosomal Peptide Assembly and Maturation in Fungi. Toxins 2019, 11, 277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shiono, Y.; Tsuchinari, M.; Shimanuki, K.; Miyajima, T.; Murayama, T.; Koseki, T.; Laatsch, H.; Funakoshi, T.; Takanami, K.; Suzuki, K. Fusaristatins A and B, Two New Cyclic Lipopeptides from an Endophytic Fusarium sp. J. Antibiot. 2007, 60, 309–316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sørensen, J.L.; Sondergaard, T.E.; Covarelli, L.; Fuertes, P.R.; Hansen, F.T.; Frandsen, R.J.N.; Saei, W.; Lukassen, M.B.; Wimmer, R.; Nielsen, K.F.; et al. Identification of the Biosynthetic Gene Clusters for the Lipopeptides Fusaristatin A and W493 B in Fusarium Graminearum and F. Pseudograminearum. J. Nat. Prod. 2014, 77, 2619–2625. [Google Scholar] [CrossRef] [PubMed]
- Bahadoor, A.; Brauer, E.K.; Bosnich, W.; Schneiderman, D.; Johnston, A.; Aubin, Y.; Blackwell, B.; Melanson, J.E.; Harris, L.J. Gramillin A and B: Cyclic Lipopeptides Identified as the Nonribosomal Biosynthetic Products of Fusarium Graminearum. J. Am. Chem. Soc. 2018, 140, 16783–16791. [Google Scholar] [CrossRef] [PubMed]
- Wollenberg, R.D.; Saei, W.; Westphal, K.R.; Sloth Klitgaard, C.; Nielsen, K.L.; Lysøe, E.; Gardiner, D.M.; Wimmer, R.; Sondergaard, T.E.; Sørensen, J.L. Chrysogine Biosynthesis Is Mediated by a Two Module Non- Ribosomal Peptide Synthetase. J. Nat. Prod. 2017, 80, 2131–2135. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Orozco, R.; Wijeratne, E.M.K.; Gunatilaka, A.A.L.; Stock, S.P.; Molnár, I. Biosynthesis of the Cyclooligomer Depsipeptide Beauvericin, a Virulence Factor of the Entomopathogenic Fungus Beauveria Bassiana. Chem. Biol. 2008, 15, 898–907. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.; Zhuo, Y.; Jia, X.P.; Liu, J.T.; Gao, H.; Song, F.H.; Liu, M.; Zhang, L.X. Cloning and Characterization of the Gene Cluster Required for Beauvericin Biosynthesis in Fusarium Proliferatum. Sci. China Life Sci. 2013, 56, 628–637. [Google Scholar] [CrossRef] [Green Version]
- Niehaus, E.-M.; Studt, L.; von Bargen, K.W.; Kummer, W.; Humpf, H.-U.; Reuter, G.; Tudzynski, B. Sound of Silence: The Beauvericin Cluster in Fusarium Fujikuroi Is Controlled by Cluster-Specific and Global Regulators Mediated by H3K27 Modification. Environ. Microbiol. 2016, 18, 4282–4302. [Google Scholar] [CrossRef]
- Niehaus, E.M.; Janevska, S.; von Bargen, K.W.; Sieber, C.M.K.; Harrer, H.; Humpf, H.U.; Tudzynski, B. Apicidin F: Characterization and Genetic Manipulation of a New Secondary Metabolite Gene Cluster in the Rice Pathogen Fusarium Fujikuroi. PLoS ONE 2014, 9, e103336. [Google Scholar] [CrossRef] [Green Version]
- Munawar, A.; Marshall, J.W.; Cox, R.J.; Bailey, A.M.; Lazarus, C.M. Isolation and Characterisation of a Ferrirhodin Synthetase Gene from the Sugarcane Pathogen Fusarium Sacchari. ChemBioChem 2013, 14, 388–394. [Google Scholar] [CrossRef]
- Brown, D.W.; Butchko, R.A.E.; Busman, M.; Proctor, R.H. Identification of Gene Clusters Associated with Fusaric Acid, Fusarin, and Perithecial Pigment Production in Fusarium Verticillioides. Fungal Genet. Biol. 2012, 49, 521–532. [Google Scholar] [CrossRef] [Green Version]
- Niehaus, E.M.; von Bargen, K.W.; Espino, J.J.; Pfannmüller, A.; Humpf, H.U.; Tudzynski, B. Characterization of the Fusaric Acid Gene Cluster in Fusarium Fujikuroi. Appl. Microbiol. Biotechnol. 2014, 98, 1749–1762. [Google Scholar] [CrossRef]
- Studt, L.; Janevska, S.; Niehaus, E.M.; Burkhardt, I.; Arndt, B.; Sieber, C.M.K.; Humpf, H.U.; Dickschat, J.S.; Tudzynski, B. Two Separate Key Enzymes and Two Pathway-Specific Transcription Factors Are Involved in Fusaric Acid Biosynthesis in Fusarium Fujikuroi. Environ. Microbiol. 2016, 18, 936–956. [Google Scholar] [CrossRef]
- Hansen, F.T.; Gardiner, D.M.; Lysøe, E.; Fuertes, P.R.; Tudzynski, B.; Wiemann, P.; Sondergaard, T.E.; Giese, H.; Brodersen, D.E.; Sørensen, J.L. An Update to Polyketide Synthase and Non-Ribosomal Synthetase Genes and Nomenclature in Fusarium. Fungal Genet. Biol. 2015, 75, 20–29. [Google Scholar] [CrossRef] [PubMed]
- Linnemannstöns, P.; Schulte, J.; del Mar Prado, M.; Proctor, R.H.; Avalos, J.; Tudzynski, B. The Polyketide Synthase Gene Pks4 from Gibberella Fujikuroi Encodes a Key Enzyme in the Biosynthesis of the Red Pigment Bikaverin. Fungal Genet. Biol. 2002, 37, 134–148. [Google Scholar] [CrossRef]
- Von Bargen, K.W.; Niehaus, E.M.; Krug, I.; Bergander, K.; Würthwein, E.U.; Tudzynski, B.; Humpf, H.U. Isolation and Structure Elucidation of Fujikurins A-D: Products of the PKS19 Gene Cluster in Fusarium Fujikuroi. J. Nat. Prod. 2015, 78, 1809–1815. [Google Scholar] [CrossRef]
- Kim, Y.T.; Lee, Y.R.; Jin, J.; Han, K.H.; Kim, H.; Kim, J.C.; Lee, T.; Yun, S.H.; Lee, Y.W. Two Different Polyketide Synthase Genes Are Required for Synthesis of Zearalenone in Gibberella Zeae. Mol. Microbiol. 2005, 58, 1102–1113. [Google Scholar] [CrossRef] [PubMed]
- Sørensen, J.L.; Akk, E.; Thrane, U.; Giese, H.; Sondergaard, T.E. Production of Fusarielins by Fusarium. Int. J. Food Microbiol. 2013, 160, 206–211. [Google Scholar] [CrossRef] [PubMed]
- Gai, Y.; Zhao, L.L.; Hu, C.Q.; Zhang, H.P. Fusarielin E, a New Antifungal Antibiotic from Fusarium sp. Chin. Chem. Lett. 2007, 18, 954–956. [Google Scholar] [CrossRef]
- Kobayashi, H.; Sljnaga, R.; Furihatat, K.; Morisaki, N.; Iwasaki, S. Isolation and Structures of an Antifungal Antibiotic, Fusarielin A, and Related Compounds Produced by a Fusarium sp. J. Antibiot. 1995, 48, 42–52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Namikoshi, M.; Kobayashi, H.; Yoshimoto, T.; Meguro, S.; Akano, K. Isolation and Characterization of Bioactive Metabolites from Marine-Derived Filamentous Fungi Collected from Tropical and Sub-Tropical Coral Reefs. Chem. Pharm. Bull. 2000, 48, 1452–1457. [Google Scholar] [CrossRef] [Green Version]
- Malz, S.; Grell, M.N.; Thrane, C.; Maier, F.J.; Rosager, P.; Felk, A.; Albertsen, K.S.; Salomon, S.; Bohn, L.; Schäfer, W.; et al. Identification of a Gene Cluster Responsible for the Biosynthesis of Aurofusarin in the Fusarium Graminearum Species Complex. Fungal Genet. Biol. 2005, 42, 420–433. [Google Scholar] [CrossRef]
- Jørgensen, S.H.; Frandsen, R.J.N.; Nielsen, K.F.; Lysøe, E.; Sondergaard, T.E.; Wimmer, R.; Giese, H.; Sørensen, J.L. Fusarium Graminearum PKS14 Is Involved in Orsellinic Acid and Orcinol Synthesis. Fungal Genet. Biol. 2014, 70, 24–31. [Google Scholar] [CrossRef] [Green Version]
- Reynolds, H.T.; Slot, J.C.; Divon, H.H.; Lysøe, E.; Proctor, R.H.; Brown, D.W. Differential Retention of Gene Functions in a Secondary Metabolite Cluster. Mol. Biol. Evol. 2017, 34, 2002–2015. [Google Scholar] [CrossRef]
- Atanasoff-Kardjalieff, A.K.; Lünne, F.; Kalinina, S.; Strauss, J.; Humpf, H.-U.; Studt, L. Biosynthesis of Fusapyrone Depends on the H3K9 Methyltransferase, FmKmt1, in Fusarium Mangiferae. Front. Fungal Biol. 2021, 1, 671796. [Google Scholar] [CrossRef]
- Evidente, A.; Conti, L.; Altomare, C.; Bottalico, A.; Sindona, G.; Segre, A.L.; Logrieco, A. Fusapyrone and Deoxyfusapyrone, Two Antifungal α-Pyrones From Fusarium Semitectum. Nat. Toxins 1994, 2, 4–13. [Google Scholar] [CrossRef]
- Arndt, B.; Janevska, S.; Schmid, R.; Hübner, F.; Tudzynski, B.; Humpf, H.-U. A Fungal N-Dimethylallyltryptophan Metabolite from Fusarium Fujikuroi. ChemBioChem 2017, 18, 899–904. [Google Scholar] [CrossRef]
- Tudzynski, B.; Hölter, K. Gibberellin Biosynthetic Pathway in Gibberella Fujikuroi: Evidence for a Gene Cluster. Fungal Genet. Biol. 1998, 25, 157–170. [Google Scholar] [CrossRef] [PubMed]
- Linnemannstöns, P.; Prado, M.M.; Fernández-Martín, R.; Tudzynski, B.; Avalos, J. A Carotenoid Biosynthesis Gene Cluster in Fusarium Fujikuroi: The Genes CarB and CarRA. Mol. Genet. Genom. 2002, 267, 593–602. [Google Scholar] [CrossRef]
- Burkhardt, I.; Siemon, T.; Henrot, M.; Studt, L.; Rösler, S.; Tudzynski, B.; Christmann, M.; Dickschat, J.S. Mechanistic Characterisation of Two Sesquiterpene Cyclases from the Plant Pathogenic Fungus Fusarium Fujikuroi. Angew. Chem. 2016, 128, 8890–8893. [Google Scholar] [CrossRef]
- Brock, N.L.; Huss, K.; Tudzynski, B.; Dickschat, J.S. Genetic Dissection of Sesquiterpene Biosynthesis by Fusarium Fujikuroi. ChemBioChem 2013, 14, 311–315. [Google Scholar] [CrossRef] [PubMed]
- McCormick, S.P.; Alexander, N.J.; Harris, L.J. CLM1 of Fusarium Graminearum Encodes a Longiborneol Synthase Required for Culmorin Production. Appl. Environ. Microbiol. 2010, 76, 136–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schotta, G.; Ebert, A.; Reuter, G. SU(VAR)3-9 Is a Conserved Key Function in Heterochromatic Gene Silencing. Genetica 2003, 117, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Farooq, Z.; Banday, S.; Pandita, T.K.; Altaf, M. The Many Faces of Histone H3K79 Methylation. Mutat. Res. /Rev. Mutat. Res. 2016, 768, 46–52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nguyen, A.T.; Zhang, Y. The Diverse Functions of Dot1 and H3K79 Methylation. Genes Dev. 2011, 25, 1345–1358. [Google Scholar] [CrossRef] [Green Version]
- Dillon, S.C.; Zhang, X.; Trievel, R.C.; Cheng, X. The SET-Domain Protein Superfamily: Protein Lysine Methyltransferases. Genome Biol. 2005, 6, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsukada, Y.I.; Fang, J.; Erdjument-Bromage, H.; Warren, M.E.; Borchers, C.H.; Tempst, P.; Zhang, Y. Histone Demethylation by a Family of JmjC Domain-Containing Proteins. Nature 2006, 439, 811–816. [Google Scholar] [CrossRef]
- Lan, F.; Zaratiegui, M.; Villén, J.; Vaughn, M.W.; Verdel, A.; Huarte, M.; Shi, Y.; Gygi, S.P.; Moazed, D.; Martienssen, R.A.; et al. S. Pombe LSD1 Homologs Regulate Heterochromatin Propagation and Euchromatic Gene Transcription. Mol. Cell 2007, 26, 89–101. [Google Scholar] [CrossRef]
- Klose, R.J.; Kallin, E.M.; Zhang, Y. JmjC-Domain-Containing Proteins and Histone Demethylation. Nat. Rev. Genet. 2006, 7, 715–727. [Google Scholar] [CrossRef] [PubMed]
- Accari, S.L.; Fisher, P.R. Emerging Roles of JmjC Domain-Containing Proteins. In International Review of Cell and Molecular Biology; Elsevier Inc.: Amsterdam, The Netherlands, 2015; Volume 319, pp. 165–220. ISBN 9780128022788. [Google Scholar]
- Janevska, S.; Baumann, L.; Sieber, C.M.K.; Münsterkötter, M.; Ulrich, J.; Kämper, J.; Güldener, U.; Tudzynski, B. Elucidation of the Two H3K36me3 Histone Targets and a Major Impact of Ash1 on Genome Stability. Genetics 2018, 208, 153–171. [Google Scholar] [CrossRef] [Green Version]
- Janevska, S.; Güldener, U.; Sulyok, M.; Tudzynski, B.; Studt, L. Set1 and Kdm5 Are Antagonists for H3K4 Methylation and Regulators of the Major Conidiation-Specific Transcription Factor Gene ABA1 in Fusarium Fujikuroi. Environ. Microbiol. 2018, 20, 3343–3362. [Google Scholar] [CrossRef] [Green Version]
- Bachleitner, S.; Sørensen, J.L.; Gacek-Matthews, A.; Sulyok, M.; Studt, L.; Strauss, J. Evidence of a Demethylase- Independent Role for the H3K4- Specific Histone Demethylases in Aspergillus Nidulans and Fusarium Graminearum Secondary Metabolism. Front. Microbiol. 2019, 10, 1759. [Google Scholar] [CrossRef] [Green Version]
- Tang, G.; Yuan, J.; Wang, J.; Zhang, Y.-Z.; Xie, S.-S.; Wang, H.; Tao, Z.; Liu, H.; Kistler, H.C.; Zhao, Y.; et al. Fusarium BP1 Is a Reader of H3K27 Methylation. Nucleic Acids Res. 2021, 49, 10448–10464. [Google Scholar] [CrossRef]
- Takahashi, Y.-H.; Shilatifard, A. Structural Basis for H3K4 Trimethylation by Yeast Set1/ COMPASS. Adv. Enzym. Regul. 2011, 50, 104–110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allis, C.D.; Berger, S.L.; Cote, J.; Dent, S.; Jenuwien, T.; Kouzarides, T.; Pillus, L.; Reinberg, D.; Shi, Y.; Shiekhattar, R.; et al. New Nomenclature for Chromatin-Modifying Enzymes. Cell 2007, 131, 633–636. [Google Scholar] [CrossRef] [Green Version]
- Shilatifard, A. The COMPASS Family of Histone H3K4 Methylases: Mechanisms of Regulation in Development and Disease Pathogenesis. Annu. Rev. Biochem. 2012, 81, 65–95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shilatifard, A. Molecular Implementation and Physiological Roles for Histone H3 Lysine 4 (H3K4) Methylation. Curr. Opin. Cell Biol. 2008, 20, 341–348. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Liu, N.; Yin, Y.; Chen, Y.; Jiang, J.; Ma, Z. Histone H3K4 Methylation Regulates Hyphal Growth, Secondary Metabolism and Multiple Stress Responses in Fusarium Graminearum. Environ. Microbiol. 2015, 17, 4615–4630. [Google Scholar] [CrossRef] [PubMed]
- Studt, L.; Janevska, S.; Arndt, B.; Boedi, S.; Sulyok, M.; Humpf, H.U.; Tudzynski, B.; Strauss, J. Lack of the COMPASS Component Ccl1 Reduces H3K4 Trimethylation Levels and Affects Transcription of Secondary Metabolite Genes in Two Plant-Pathogenic Fusarium Species. Front. Microbiol. 2017, 7, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Connolly, L.R.; Smith, K.M.; Freitag, M. The Fusarium Graminearum Histone H3 K27 Methyltransferase KMT6 Regulates Development and Expression of Secondary Metabolite Gene Clusters. PLoS Genet. 2013, 9, e1003916. [Google Scholar] [CrossRef] [Green Version]
- Gu, Q.; Tahir, H.A.S.; Zhang, H.; Huang, H.; Ji, T.; Sun, X.; Wu, L.; Wu, H.; Gao, X. Involvement of FvSet1 in Fumonisin B1 Biosynthesis, Vegetative Growth, Fungal Virulence, and Environmental Stress Responses in Fusarium Verticillioides. Toxins 2017, 9, 43. [Google Scholar] [CrossRef] [Green Version]
- Gacek-Matthews, A.; Berger, H.; Sasaki, T.; Wittstein, K.; Gruber, C.; Lewis, Z.A.; Strauss, J. KdmB, a Jumonji Histone H3 Demethylase, Regulates Genome-Wide H3K4 Trimethylation and Is Required for Normal Induction of Secondary Metabolism in Aspergillus Nidulans. PLoS Genet. 2016, 12, e1006222. [Google Scholar] [CrossRef]
- Nakayama, J.; Rice, J.C.; Strahl, B.D.; Allis, C.D.; Grewal, S.I.S. Role of Histone H3 Lysine 9 Methylation in Epigenetic Control of Heterochromatin Assembly. Science 2001, 292, 110–113. [Google Scholar] [CrossRef] [Green Version]
- Tamaru, H.; Selker, E.U. A Histone H3 Methyltransferase Controls DNA Methylation in Neurospora Crassa. Nature 2001, 414, 277–283. [Google Scholar] [CrossRef]
- Selker, E.U. Genome Defense and DNA Methylation in Neurospora. Cold Spring Harb. Symp. Quant. Biol. 2004, 69, 119–124. [Google Scholar] [CrossRef]
- Lewis, Z.A.; Adhvaryu, K.K.; Honda, S.; Shiver, A.L.; Knip, M. DNA Methylation and Normal Chromosome Behavior in Neurospora Depend on Five Components of a Histone Methyltransferase Complex, DCDC. PLoS Genet 2010, 6, e1001196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gessaman, J.D.; Selker, E.U. Induction of H3K9me3 and DNA Methylation by Tethered Heterochromatin Factors in Neurospora Crassa. Proc. Natl. Acad. Sci. USA 2017, 114, E9598–E9607. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hiragami, K.; Festenstein, R. Heterochromatin Protein 1: A Pervasive Controlling Influence. Cell. Mol. Life Sci. 2005, 62, 2711–2726. [Google Scholar] [CrossRef]
- Freitag, M.; Hickey, P.C.; Khlafallah, T.K.; Read, N.D.; Selker, E.U. HP1 Is Essential for DNA Methylation in Neurospora. Mol. Cell 2004, 13, 427–434. [Google Scholar] [CrossRef]
- Gu, Q.; Ji, T.; Sun, X.; Huang, H.; Zhang, H.; Lu, X.; Wu, L.; Huo, R.; Wu, H.; Gao, X. Histone H3 Lysine 9 Methyltransferase FvDim5 Regulates Fungal Development, Pathogenicity and Osmotic Stress Responses in Fusarium Verticillioides. FEMS Microbiol. Lett. 2017, 364, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Bonner, C.; Sproule, A.; Rowland, O.; Overy, D.; Subramaniam, R. DNA Methylation Is Responsive to the Environment and Regulates the Expression of Biosynthetic Gene Clusters, Metabolite Production, and Virulence in Fusarium Graminearum. Front. Fungal Biol. 2021, 1. [Google Scholar] [CrossRef]
- Freitag, M. Histone Methylation by SET Domain Proteins in Fungi. Annu. Rev. Microbiol. 2017, 71, 413–439. [Google Scholar] [CrossRef] [PubMed]
- Müller, J.; Hart, C.M.; Francis, N.J.; Vargas, M.L.; Sengupta, A.; Wild, B.; Miller, E.L.; O’Connor, M.B.; Kingston, R.E.; Simon, J.A. Histone Methyltransferase Activity of a Drosophila Polycomb Group Repressor Complex. Cell 2002, 111, 197–208. [Google Scholar] [CrossRef] [Green Version]
- Jamieson, K.; Rountree, M.R.; Lewis, Z.A.; Stajich, J.E.; Selker, E.U. Regional Control of Histone H3 Lysine 27 Methylation in Neurospora. Proc. Natl. Acad. Sci. USA 2013, 110, 6027–6032. [Google Scholar] [CrossRef] [Green Version]
- Cao, R.; Wang, L.; Wang, H.; Xia, L.; Erdjument-Bromage, H.; Tempst, P.; Jones, R.S.; Zhang, Y. Role of Histone H3 Lysine 27 Methylation in Polycomb-Group Silencing. Science 2002, 298, 1039–1043. [Google Scholar] [CrossRef] [Green Version]
- Studt, L.; Rösler, S.M.; Burkhardt, I.; Arndt, B.; Freitag, M.; Humpf, H.U.; Dickschat, J.S.; Tudzynski, B. Knock-down of the Methyltransferase Kmt6 Relieves H3K27me3 and Results in Induction of Cryptic and Otherwise Silent Secondary Metabolite Gene Clusters in Fusarium Fujikuroi. Environ. Microbiol. 2016, 18, 4037–4054. [Google Scholar] [CrossRef] [Green Version]
- Adpressa, D.A.; Connolly, L.R.; Konkel, Z.M.; Neuhaus, G.F.; Chang, X.L.; Pierce, B.R.; Smith, K.M.; Freitag, M.; Loesgen, S. A Metabolomics-Guided Approach to Discover Fusarium Graminearum Metabolites after Removal of a Repressive Histone Modification. Fungal Genet. Biol. 2019, 132, 103256. [Google Scholar] [CrossRef] [PubMed]
- Shao, Z.; Raible, F.; Mollaaghababa, R.; Guyon, J.R.; Wu, C.-T.; Bender, W.; Kingston, R.E. Stabilization of Chromatin Structure by PRC1, a Polycomb Complex. Cell 1999, 98, 37–46. [Google Scholar] [CrossRef] [Green Version]
- Lewis, Z.A. Polycomb Group Systems in Fungi: New Models for Understanding Polycomb Repressive Complex 2. Trends Genet. 2017, 33, 220–231. [Google Scholar] [CrossRef]
- Strahl, B.D.; Grant, P.A.; Briggs, S.D.; Sun, Z.-W.; Bone, J.R.; Caldwell, J.A.; Mollah, S.; Cook, R.G.; Shabanowitz, J.; Hunt, D.F.; et al. Set2 Is a Nucleosomal Histone H3-Selective Methyltransferase That Mediates Transcriptional Repression. Mol. Cell. Biol. 2002, 22, 1298–1306. [Google Scholar] [CrossRef] [Green Version]
- Krogan, N.J.; Kim, M.; Tong, A.; Golshani, A.; Cagney, G.; Canadien, V.; Richards, D.P.; Beattie, B.K.; Emili, A.; Boone, C.; et al. Methylation of Histone H3 by Set2 in Saccharomyces Cerevisiae Is Linked to Transcriptional Elongation by RNA Polymerase II. Mol. Cell. Biol. 2003, 23, 4207–4218. [Google Scholar] [CrossRef] [Green Version]
- Kizer, K.O.; Phatnani, H.P.; Shibata, Y.; Hall, H.; Greenleaf, A.L.; Strahl, B.D. A Novel Domain in Set2 Mediates RNA Polymerase II Interaction and Couples Histone H3 K36 Methylation with Transcript Elongation. Mol. Cell. Biol. 2005, 25, 3305–3316. [Google Scholar] [CrossRef] [Green Version]
- Gu, Q.; Wang, Z.; Sun, X.; Ji, T.; Huang, H.; Yang, Y.; Zhang, H.; Tahir, H.A.S.; Wu, L.; Wu, H.; et al. FvSet2 Regulates Fungal Growth, Pathogenicity, and Secondary Metabolism in Fusarium Verticillioides. Fungal Genet. Biol. 2017, 107, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Klose, R.J.; Gardner, K.E.; Liang, G.; Erdjument-Bromage, H.; Tempst, P.; Zhang, Y. Demethylation of Histone H3K36 and H3K9 by Rph1: A Vestige of an H3K9 Methylation System in Saccharomyces Cerevisiae ? Mol. Cell. Biol. 2007, 27, 3951–3961. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tu, S.; Bulloch, E.M.M.; Yang, L.; Ren, C.; Huang, W.C.; Hsu, P.H.; Chen, C.H.; Liao, C.L.; Yu, H.M.; Lo, W.S.; et al. Identification of Histone Demethylases in Saccharomyces Cerevisiae. J. Biol. Chem. 2007, 282, 14262–14271. [Google Scholar] [CrossRef] [Green Version]
- Schotta, G.; Lachner, M.; Sarma, K.; Ebert, A.; Sengupta, R.; Reuter, G.; Reinberg, D.; Jenuwein, T. A Silencing Pathway to Induce H3-K9 and H4-K20 Trimethylation at Constitutive Heterochromatin. Genes Dev. 2004, 18, 1251–1262. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Jia, S. Degrees Make All the Difference: The Multifunctionality of Histone H4 Lysine 20 Methylation. Epigenetics 2009, 4, 273–276. [Google Scholar] [CrossRef] [Green Version]
- Sanders, S.L.; Portoso, M.; Mata, J.; Bähler, J.; Allshire, R.C.; Kouzarides, T. Methylation of Histone H4 Lysine 20 Controls Recruitment of Crb2 to Sites of DNA Damage. Cell 2004, 119, 603–614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bachleitner, S.; Sulyok, M.; Sørensen, J.L.; Strauss, J.; Studt, L. The H4K20 Methyltransferase Kmt5 Is Involved in Secondary Metabolism and Stress Response in Phytopathogenic Fusarium Species. Fungal Genet. Biol. 2021, 155, 103602. [Google Scholar] [CrossRef]
- Barnes, C.E.; English, D.M.; Cowley, S.M. Acetylation and Co: An Expanding Repertoire of Histone Acylations Regulates Chromatin and Transcription. Essays Biochem. 2019, 63, 97–107. [Google Scholar] [CrossRef] [Green Version]
- Roth, S.Y.; Denu, J.M.; Allis, C.D. Histone Acetyltransferases. Annu. Rev. Biochem. 2001, 70, 81–120. [Google Scholar] [CrossRef] [PubMed]
- Marmorstein, R.; Roth, S.Y. Histone Acetyltransferases: Function, Structure, and Catalysis. Curr. Opin. Genet. Dev. 2001, 11, 155–161. [Google Scholar] [CrossRef]
- Elías-Villalobos, A.; Toullec, D.; Faux, C.; Séveno, M.; Helmlinger, D. Chaperone-Mediated Ordered Assembly of the SAGA and NuA4 Transcription Co-Activator Complexes in Yeast. Nat. Commun. 2019, 10, 5237. [Google Scholar] [CrossRef] [Green Version]
- Rösler, S.M.; Kramer, K.; Finkemeier, I.; Humpf, H.U.; Tudzynski, B. The SAGA Complex in the Rice Pathogen Fusarium Fujikuroi: Structure and Functional Characterization. Mol. Microbiol. 2016, 102, 951–974. [Google Scholar] [CrossRef]
- Liu, J.; An, B.; Luo, H.; He, C.; Wang, Q. The Histone Acetyltransferase FocGCN5 Regulates Growth, Conidiation, and Pathogenicity of the Banana Wilt Disease Causal Agent Fusarium Oxysporum f.sp. Cubense Tropical Race 4. Res. Microbiol. 2021, 103902. [Google Scholar] [CrossRef]
- Kong, X.; van Diepeningen, A.D.; van der Lee, T.A.J.; Waalwijk, C.; Xu, J.; Xu, J.; Zhang, H.; Chen, W.; Feng, J. The Fusarium Graminearum Histone Acetyltransferases Are Important for Morphogenesis, DON Biosynthesis, and Pathogenicity. Front. Microbiol. 2018, 9, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eberharter, A.; Becker, P.B. Histone Acetylation: A Switch between Repressive and Permissive Chromatin. Second in Review on Chromatin Dynamics. EMBO Rep. 2002, 3, 224–229. [Google Scholar] [CrossRef]
- Seto, E.; Yoshida, M. Erasers of Histone Acetylation: The Histone Deacetylase Enzymes. Cold Spring Harb. Perspect. Biol. 2014, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soffers, J.H.M.; Workman, J.L. The SAGA Chromatin-Modifying Complex: The Sum of Its Parts Is Greater than the Whole. Genes Dev. 2020, 34, 1287–1303. [Google Scholar] [CrossRef] [PubMed]
- Helmlinger, D.; Tora, L. Sharing the SAGA. Trends Biochem. Sci. 2017, 42, 850–861. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Huang, P.; Zhou, S. Functional Characterization of the Catalytic and Bromodomain of FgGCN5 in Development, DON Production and Virulence of Fusarium Graminearum. J. Integr. Agric. 2020, 19, 2477–2487. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, J.; Yang, N.; Wen, Z.; Sun, X.; Chai, Y.; Ma, Z. Wheat Microbiome Bacteria Can Reduce Virulence of a Plant Pathogenic Fungus by Altering Histone Acetylation. Nat. Commun. 2018, 9, 3429. [Google Scholar] [CrossRef] [Green Version]
- Setiaputra, D.; Ahmad, S.; Dalwadi, U.; Steunou, A.-L.; Lu, S.; Ross, J.D.; Dong, M.-Q.; Côté, J.; Yip, C.K. Molecular Architecture of the Essential Yeast Histone Acetyltransferase Complex NuA4 Redefines Its Multimodularity. Mol. Cell. Biol. 2018, 38, e00570-17. [Google Scholar] [CrossRef] [Green Version]
- Clarke, A.S.; Lowell, J.E.; Jacobson, S.J.; Pillus, L. Esa1p Is an Essential Histone Acetyltransferase Required for Cell Cycle Progression. Mol. Cell. Biol. 1999, 19, 2515–2526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soukup, A.A.; Chiang, Y.M.; Bok, J.W.; Reyes-Dominguez, Y.; Oakley, B.R.; Wang, C.C.C.; Strauss, J.; Keller, N.P. Overexpression of the Aspergillus Nidulans Histone 4 Acetyltransferase EsaA Increases Activation of Secondary Metabolite Production. Mol. Microbiol. 2012, 86, 314–330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, H.; Xia, A.; Ye, M.; Ren, J.; Li, D.; Liu, H.; Wang, Q.; Lu, P.; Wu, C.; Xu, J.R.; et al. Opposing Functions of Fng1 and the Rpd3 HDAC Complex in H4 Acetylation in Fusarium Graminearum. PLoS Genet. 2020, 16, e1009185. [Google Scholar] [CrossRef] [PubMed]
- Sterner, D.E.; Berger, S.L. Acetylation of Histones and Transcription-Related Factors. Microbiol. Mol. Biol. Rev. 2000, 64, 435–459. [Google Scholar] [CrossRef] [Green Version]
- Ye, M.; Jiang, H.; Fu, X.; Xu, J.R.; Jiang, C. Fng1 Is Involved in Crosstalk between Histone Acetylation and Methylation. Curr. Genet. 2021, 67, 535–538. [Google Scholar] [CrossRef]
- Vicente-Muñoz, S.; Romero, P.; Magraner-Pardo, L.; Martinez-Jimenez, C.P.; Tordera, V.; Pamblanco, M. Comprehensive Analysis of Interacting Proteins and Genome-Wide Location Studies of the Sas3-Dependent NuA3 Histone Acetyltransferase Complex. FEBS Open Bio. 2014, 4, 996–1006. [Google Scholar] [CrossRef] [Green Version]
- Luan, Y.; Ngo, L.; Han, Z.; Wang, X.; Qu, M.; Zheng, Y.G. Histone Acetyltransferases: Enzymes, Assays, and Inhibitors. In Epigenetic Technological Applications; Academic Press: Cambridge, MA, USA, 2015; pp. 291–317. ISBN 9780128013274. [Google Scholar]
- Qin, J.; Wu, M.; Zhou, S. FgEaf6 Regulates Virulence, Asexual/Sexual Development and Conidial Septation in Fusarium Graminearum. Curr. Genet. 2020, 66, 517–529. [Google Scholar] [CrossRef]
- Cavero, S.; Herruzo, E.; Ontoso, D.; San-Segundo, P.A. Impact of Histone H4K16 Acetylation on the Meiotic Recombination Checkpoint in Saccharomyces Cerevisiae. Microb. Cell 2016, 3, 606–620. [Google Scholar] [CrossRef] [Green Version]
- Winkler, S.G.; Kristjuhan, A.; Erdjument-Bromage, H.; Tempst, P.; Svejstrup, J.Q. Elongator Is a Histone H3 and H4 Acetyltransferase Important for Normal Histone Acetylation Levels in Vivo. Proc. Natl. Acad. Sci. USA 2002, 99, 3517–3522. [Google Scholar] [CrossRef] [Green Version]
- Dalwadi, U.; Yip, C.K. Structural Insights into the Function of Elongator. Cell. Mol. Life Sci. 2018, 75, 1613–1622. [Google Scholar] [CrossRef] [PubMed]
- Lee, Y.; Min, K.; Son, H.; Park, A.R.; Kim, J.C.; Choi, G.J.; Lee, Y.W. ELP3 Is Involved in Sexual and Asexual Development, Virulence, and the Oxidative Stress Response in Fusarium Graminearum. Mol. Plant-Microbe Interact. 2014, 27, 1344–1355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Tang, Y.; Cole, P.A.; Marmorstein, R. Structure and Chemistry of the P300/CBP and Rtt109 Histone Acetyltransferases: Implications for Histone Acetyltransferase Evolution and Function. Curr. Opin. Struct. Biol. 2008, 18, 741–747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, R.; Wen, M.; Wu, L.; Lan, H.; Yuan, J.; Wang, S. The Fungi-Specific Histone Acetyltransferase Rtt109 Mediates Morphogenesis, Aflatoxin Synthesis and Pathogenicity in Aspergillus Flavus by Acetylating H3K9. IMA Fungus 2021, 12, 9. [Google Scholar] [CrossRef]
- Cai, Q.; Wang, J.J.; Shao, W.; Ying, S.H.; Feng, M.G. Rtt109-Dependent Histone H3 K56 Acetylation and Gene Activity Are Essential for the Biological Control Potential of Beauveria Bassiana. Pest Manag. Sci. 2018, 74, 2626–2635. [Google Scholar] [CrossRef] [PubMed]
- Parthun, M.R.; Widom, J.; Gottschling, D.E. The Major Cytoplasmic Histone Acetyltransferase in Yeast: Links to Chromatin Replication and Histone Metabolism. Cell 1996, 87, 85–94. [Google Scholar] [CrossRef] [Green Version]
- Parthun, M.R. Histone Acetyltransferase 1: More than Just an Enzyme? Biochim. Biophys. Acta-Gene Regul. Mech. 2012, 1819, 256–263. [Google Scholar] [CrossRef] [Green Version]
- Ruiz-García, A.B.; Sendra, R.; Galiana, M.; Pamblanco, M.; Pérez-Ortín, J.E.; Tordera, V. HAT1 and HAT2 Proteins Are Components of a Yeast Nuclear Histone Acetyltransferase Enzyme Specific for Free Histone H4. J. Biol. Chem. 1998, 273, 12599–12605. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Wang, C.; Liu, W.; Wang, G.; Kang, Z.; Kistler, H.C.; Xu, J.R. The HDF1 Histone Deacetylase Gene Is Important for Conidiation, Sexual Reproduction, and Pathogenesis in Fusarium Graminearum. Mol. Plant-Microbe Interact. 2011, 24, 487–496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Studt, L.; Schmidt, F.J.; Jahn, L.; Sieber, C.M.K.; Connolly, L.R.; Niehaus, E.M.; Freitag, M.; Humpf, H.U.; Tudzynski, B. Two Histone Deacetylases, FfHda1 and FfHda2, Are Important for Fusarium Fujikuroi Secondary Metabolism and Virulence. Appl. Environ. Microbiol. 2013, 79, 7719–7734. [Google Scholar] [CrossRef] [Green Version]
- Maeda, K.; Izawa, M.; Nakajima, Y.; Jin, Q.; Hirose, T.; Nakamura, T.; Koshino, H.; Kanamaru, K.; Ohsato, S.; Kamakura, T.; et al. Increased Metabolite Production by Deletion of an HDA1-Type Histone Deacetylase in the Phytopathogenic Fungi, Magnaporthe Oryzae (Pyricularia Oryzae) and Fusarium Asiaticum. Lett. Appl. Microbiol. 2017, 65, 446–452. [Google Scholar] [CrossRef]
- Tudzynski, B. Nitrogen Regulation of Fungal Secondary Metabolism in Fungi. Front. Microbiol. 2014, 5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Michielse, C.B.; Pfannmüller, A.; Macios, M.; Rengers, P.; Dzikowska, A.; Tudzynski, B. The Interplay between the GATA Transcription Factors AreA, the Global Nitrogen Regulator and AreB in Fusarium Fujikuroi. Mol. Microbiol. 2014, 91, 472–493. [Google Scholar] [CrossRef] [PubMed]
- Wiemann, P.; Willmann, A.; Straeten, M.; Kleigrewe, K.; Beyer, M.; Humpf, H.U.; Tudzynski, B. Biosynthesis of the Red Pigment Bikaverin in Fusarium Fujikuroi: Genes, Their Function and Regulation. Mol. Microbiol. 2009, 72, 931–946. [Google Scholar] [CrossRef] [PubMed]
- Rösler, S.M.; Sieber, C.M.K.; Humpf, H.U.; Tudzynski, B. Interplay between Pathway-Specific and Global Regulation of the Fumonisin Gene Cluster in the Rice Pathogen Fusarium Fujikuroi. Appl. Microbiol. Biotechnol. 2016, 100, 5869–5882. [Google Scholar] [CrossRef]
- Lan, N.; Zhang, H.; Hu, C.; Wang, W.; Calvo, A.M.; Harris, S.D.; Chen, S.; Li, S. Coordinated and Distinct Functions of Velvet Proteins in Fusarium Verticillioides. Eukaryot. Cell 2014, 13, 909–918. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.K.; Lee, S.; Jo, S.M.; McCormick, S.P.; Butchko, R.A.E.; Proctor, R.H.; Yun, S.H. Functional Roles of FgLaeA in Controlling Secondary Metabolism, Sexual Development, and Virulence in Fusarium Graminearum. PLoS ONE 2013, 8, e68441. [Google Scholar] [CrossRef]
- Peñalva, M.A.; Tilburn, J.; Bignell, E.; Arst, H.N. Ambient PH Gene Regulation in Fungi: Making Connections. Trends Microbiol. 2008, 16, 291–300. [Google Scholar] [CrossRef] [Green Version]
- Michielse, C.B.; van Wijk, R.; Reijnen, L.; Manders, E.M.M.; Boas, S.; Olivain, C.; Alabouvette, C.; Rep, M. The Nuclear Protein Sge1 of Fusarium Oxysporum Is Required for Parasitic Growth. PLoS Pathog. 2009, 5, e1000637. [Google Scholar] [CrossRef] [Green Version]
- Michielse, C.B.; Studt, L.; Janevska, S.; Sieber, C.M.K.; Arndt, B.; Espino, J.J.; Humpf, H.U.; Güldener, U.; Tudzynski, B. The Global Regulator FfSge1 Is Required for Expression of Secondary Metabolite Gene Clusters but Not for Pathogenicity in Fusarium Fujikuroi. Environ. Microbiol. 2015, 17, 2690–2708. [Google Scholar] [CrossRef]



| InterPro Accession Number | F. fujikuroi | F. oxysporum | F. graminearum | F. solani | |
|---|---|---|---|---|---|
| Histone lysine methyltransferases | |||||
| SET-domain | IPR001214 | 25 | 37 | 26 | 23 |
| DOT-domain | 1 | 1 | 1 | 1 | |
| Histone lysine demethylases | |||||
| JmjC-domain | IPR003347 | 10 | 28 | 16 | 10 |
| Histone reader proteins | |||||
| Chromo/ Chromoshadow- domain | IPR000953 | 11 | 49 | 13 | 8 |
| BAH-domain | IPR001025 | 4 | 15 | 8 | 4 |
| Histone acetyltransferases | |||||
| GNAT-domain | IPR000182 | 63 | 99 | 63 | 72 |
| MYST-domain | IPR040706 | 3 | 6 | 4 | 3 |
| Other * | IPR022591 IPR013178 | 2 | 8 | 3 | 2 |
| Histone deacetylases | |||||
| Zn2+-dependent | IPR023801 | 4 | 7 | 6 | 4 |
| Sirtuins | IPR026590 | 8 | 15 | 9 | 9 |
| Gene Name | Product | Fusarium spp. | Literature | |||
|---|---|---|---|---|---|---|
| F. fujikuroi | F. graminearum | F. verticillioides | F. mangiferae | |||
| NRPSs | ||||||
| NRPS 1 | Malonichrom | n.p. | Kmt6 | ? | ? | [61] |
| NRPS 2 | Ferricrocin | Gcn5, Hda1 | Kmt6 | ? | ? | [53,61] |
| NRPS 4 | Fusahexin | Kmt2, Kmt3, Ash1, Kmt6 Kdm5, Gcn5 | Kmt6 | ? | ? | [52] |
| NRPS 5 | Fusaoctaxin | n.p. | Kmt6 | n.p. | n.p. | [62] |
| NRPS 6 | Fusarinine | Kmt2, Kmt3 | Kmt6, Gcn5, Sas3, RID/Dim-2 | ? | ? | [54] |
| NRPS7 | Fusaristatins | n.p. | Kmt6, RID/Dim-2 | n.p. | n.p. | [63,64] |
| NRPS 8 | Gramilin | n.p. | Kmt6, Gcn5 RID/Dim-2 | n.p. | n.p. | [65] |
| NRPS 9 | Fusaoctaxin | n.p. | Kmt6 | n.p. | n.p. | [62] |
| NRPS 14 | Chrysogine | n.p. | Kmt6, Kdm5 | n.p. | n.p. | [66] |
| NRPS 17 | Ferrichrome | Gcn5 | ? | ? | n.p. | [54] |
| NRPS 22 | Beauvericin | Ash1, Kmt6, Gcn5, Hda1 | n.p. | ? | Kmt1 | [67,68,69] |
| NRPS 31 | Apicidin F | Kmt2, Kmt3, Ash1, Kmt6 Kdm5, Gcn5 | n.p. | n.p. | n.p. | [70] |
| NRPS 32 | Ferrirhodin | n.p. | n.p. | n.p. | n.p. | [71] |
| NRPS 34 | Fusaric acid | Kmt2, Kmt3, Ash1, Kmt6, Hat1, Gcn5, Hda1, Hda2 | n.p. | ? | ? | [72,73,74] |
| PKSs | ||||||
| PKS 1/NRPS | Equisetin/ trichosetin | Kmt2, Kmt6, Gcn5 | n.p. | ? | ? | [75] |
| PKS 3 | Fusarubins | Kmt2, Kdm5, Ash1, Kmt6, Hda2 | Kmt6, RID/Dim-2 | ? | ? | [51] |
| PKS 4 | Bikaverin | Kmt2, Kmt5, Kdm5, Ash1, Gcn5, Hda1, Hda2 | n.p. | Kmt3, | ? | [76] |
| PKS 6 | Fusaric acid | Kmt2, Kmt3, Ash1, Kmt6, Hat1, Gcn5, Hda1, Hda2 | n.p. | ? | ? | [72,73,74] |
| PKS 10/NRPS | Fusarin C | Kmt2, Kmt2, Kmt3, Kmt5, Kdm5, Ash1, Gcn5, Hda1, Hda2 | Kmt5, Kmt6, Kdm5 | ? | [72] | |
| PKS 11 | Fumonisins | Kmt2, Kmt3, Ash1, Kmt6, Gcn5, Hda1 | n.p. | Kmt1, Kmt2, Kmt3 | n.p. | [72] |
| PKS 13 | Gibepyrones | Kmt3, Ash1, Gcn5, Hda1 | Kmt6 | ? | ? | [55] |
| PKS 19 | Fujikurins | Kmt6 | n.p. | n.p. | n.p. | [77] |
| PKS22 | Zearalenone | n.p. | Kmt5, Kmt6, Gcn5, Sas3 | n.p. | n.p. | [78] |
| PKS23 | Fusaristatins | n.p. | Kmt6 | n.p. | n.p. | [63,64] |
| PKS24 | Fusarielins | n.p. | Kmt6, Kdm5, Sas3 | n.p. | n.p. | [79,80,81,82] |
| PKS25 | Aurofusarin | n.p. | Kmt2, Kmt6, Gcn5, Sas3, Hep1, RID/Dim-2 | n.p. | n.p. | [83] |
| PKS26 | Zearalenone | n.p. | Kmt5, Kmt6, Gcn5, Sas3 | n.p. | n.p. | [78] |
| PKS27 | Orcinol | n.p. | Kmt6, GCN5, Sas3, RID/Dim-2 | n.p. | n.p. | [84] |
| PKS 39 | Depudecin | n.p. | n.p. | ? | n.p. | [85] |
| PKS 40 | Fusapyrone | n.p. | n.p. | ? | Kmt1 | [86,87] |
| DMATSs | ||||||
| DMATS1 | r-N-DMAT | Gcn5 | n.p. | ? | ? | [88] |
| TCs | ||||||
| DTC1-1 | Gibberellins | Kmt2, Ash1, Kmt5, Kmt6, HAT1, Gcn5, Hda1, Hda2 | n.p. | n.p. | ? | [89] |
| TeTC1 | Carotenoids | Kmt2, Kmt3, Ash1, Kmt6, Gcn5 | Kmt6, Gcn5, Sas3, RID/Dim-2 | ? | ? | [90] |
| STC1 | Germacrene | Kmt2, Kmt3, Ash1, Kmt6, Kdm5, Gcn5, Hda1 | ? | ? | ? | [91] |
| STC3 | Eremophilene | Ash1 | n.p. | ? | ? | [91] |
| STC4 | Koraiol | Kmt2, Ash1, Gcn5 | Kmt6 | ? | ? | [92] |
| STC5 | Guaiadiene | Kmt6, GCN5 | Kmt6 | n.p. | ? | [91] |
| STC6 | Acorenol | Kmt6 | Kmt6 | ? | ? | [92] |
| STC9 | Culmorin | Gcn5 | Kmt6 | ? | ? | [93] |
| TRI5 | Deoxynivalenol | n.p. | Kmt2, Kmt5, Kmt6, Kdm5, Gcn5, Sas3, Elp3, Hep1, Hda2, RID/Dim-2 | n.p. | n.p. | [23] |
| FUP1 | Fusaproliferin | n.p. | n.p. | n.p. | ? | [60] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Atanasoff-Kardjalieff, A.K.; Studt, L. Secondary Metabolite Gene Regulation in Mycotoxigenic Fusarium Species: A Focus on Chromatin. Toxins 2022, 14, 96. https://doi.org/10.3390/toxins14020096
Atanasoff-Kardjalieff AK, Studt L. Secondary Metabolite Gene Regulation in Mycotoxigenic Fusarium Species: A Focus on Chromatin. Toxins. 2022; 14(2):96. https://doi.org/10.3390/toxins14020096
Chicago/Turabian StyleAtanasoff-Kardjalieff, Anna Katharina, and Lena Studt. 2022. "Secondary Metabolite Gene Regulation in Mycotoxigenic Fusarium Species: A Focus on Chromatin" Toxins 14, no. 2: 96. https://doi.org/10.3390/toxins14020096






