Genotypes of Staphylococcus aureus Clinical Isolates Are Associated with Phenol-Soluble Modulin (PSM) Production
Abstract
:1. Introduction
2. Results
2.1. Molecular Characteristics of S. aureus Isolates
2.2. In Vitro δ-Toxin Production by S. aureus Clinical Isolates
2.3. Association between In Vitro PSM Production, Methicillin-Resistance, SCCmec Type, and spa Type
3. Discussion
4. Conclusions
5. Materials and Methods
5.1. Bacterial Strains and Growth Conditions
5.2. The spa Typing and SCCmec Typing
5.3. PSM Quantification by LC–MS
5.4. Statistical Analysis
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Lowy, F.D. Staphylococcus aureus Infections. N. Engl. J. Med. 1998, 339, 520–532. [Google Scholar] [CrossRef] [PubMed]
- David, M.Z.; Daum, R.S. Community-associated methicillin-resistant Staphylococcus aureus: Epidemiology and clinical consequences of an emerging epidemic. Clin. Microbiol. Rev. 2010, 23, 616–687. [Google Scholar] [CrossRef] [PubMed]
- Tong, S.Y.C.; Davis, J.S.; Eichenberger, E.; Holland, T.L.; Fowler, V.G. Staphylococcus aureus infections: Epidemiology, pathophysiology, clinical manifestations, and management. Clin. Microbiol. Rev. 2015, 28, 603–661. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanian, D.; Harper, L.; Shopsin, B.; Torres, V.J. Staphylococcus aureus pathogenesis in diverse host environments. Pathog. Dis. 2017, 75, ftx005. [Google Scholar] [CrossRef]
- Tam, K.; Torres, V.J. Staphylococcus aureus Secreted Toxins and Extracellular Enzymes. Microbiol. Spectr. 2019, 7. [Google Scholar] [CrossRef]
- Wang, R.; Braughton, K.R.; Kretschmer, D.; Bach, T.-H.L.; Queck, S.Y.; Li, M.; Kennedy, A.D.; Dorward, D.W.; Klebanoff, S.J.; Peschel, A.; et al. Identification of novel cytolytic peptides as key virulence determinants for community-associated MRSA. Nat. Med. 2007, 13, 1510–1514. [Google Scholar] [CrossRef]
- Kobayashi, S.D.; Malachowa, N.; Whitney, A.R.; Braughton, K.R.; Gardner, D.J.; Long, D.; Wardenburg, J.B.; Schneewind, O.; Otto, M.; DeLeo, F.R. Comparative Analysis of USA300 Virulence Determinants in a Rabbit Model of Skin and Soft Tissue Infection. J. Infect. Dis. 2011, 204, 937–941. [Google Scholar] [CrossRef]
- Yarwood, J.M.; Schlievert, P.M. Quorum sensing in Staphylococcus infections. J. Clin. Investig. 2003, 112, 1620–1625. [Google Scholar] [CrossRef]
- Novick, R.P. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 2003, 48, 1429–1449. [Google Scholar] [CrossRef]
- Bronner, S.; Monteil, H.; Prévost, G. Regulation of virulence determinants in Staphylococcus aureus: Complexity and applications. FEMS Microbiol. Rev. 2004, 28, 183–200. [Google Scholar] [CrossRef]
- Queck, S.Y.; Jameson-Lee, M.; Villaruz, A.E.; Bach, T.H.L.; Khan, B.A.; Sturdevant, D.E.; Ricklefs, S.M.; Li, M.; Otto, M. RNAIII-Independent Target Gene Control by the agr Quorum-Sensing System: Insight into the Evolution of Virulence Regulation in Staphylococcus aureus. Mol. Cell 2008, 32, 150–158. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, S.S.; Otto, M. How can Staphylococcus aureus phenol-soluble modulins be targeted to inhibit infection? Future Microbiol. 2013, 8, 693–696. [Google Scholar] [CrossRef] [PubMed]
- Otto, M. Phenol-soluble modulins. Int. J. Med. Microbiol. 2014, 304, 164–169. [Google Scholar] [CrossRef] [PubMed]
- Peschel, A.; Otto, M. Phenol-soluble modulins and staphylococcal infection. Nat. Rev. Microbiol. 2013, 11, 667–673. [Google Scholar] [CrossRef]
- Nguyen, T.H.; Cheung, G.Y.C.; Rigby, K.M.; Kamenyeva, O.; Kabat, J.; Sturdevant, D.E.; Villaruz, A.E.; Liu, R.; Piewngam, P.; Porter, A.R.; et al. Rapid pathogen-specific recruitment of immune effector cells in the skin by secreted toxins. Nat. Microbiol. 2021, 7, 62–72. [Google Scholar] [CrossRef]
- Fogel, L.A.; Bubeck Wardenburg, J. Staphylococcus aureus PSMs are a double-edged sword. Nat. Microbiol. 2021, 7, 12–13. [Google Scholar] [CrossRef]
- Kretschmer, D.; Gleske, A.-K.; Rautenberg, M.; Wang, R.; Köberle, M.; Bohn, E.; Schöneberg, T.; Rabiet, M.-J.; Boulay, F.; Klebanoff, S.J.; et al. Human Formyl Peptide Receptor 2 Senses Highly Pathogenic Staphylococcus aureus. Cell Host Microbe 2010, 7, 463–473. [Google Scholar] [CrossRef]
- Schwartz, K.; Syed, A.K.; Stephenson, R.E.; Rickard, A.H.; Boles, B.R. Functional Amyloids Composed of Phenol Soluble Modulins Stabilize Staphylococcus aureus Biofilms. PLoS Pathog. 2012, 8, e1002744. [Google Scholar] [CrossRef]
- Periasamy, S.; Joo, H.-S.; Duong, A.C.; Bach, T.-H.L.; Tan, V.Y.; Chatterjee, S.S.; Cheung, G.Y.C.; Otto, M. How Staphylococcus aureus biofilms develop their characteristic structure. Proc. Natl. Acad. Sci. USA 2012, 109, 1281–1286. [Google Scholar] [CrossRef]
- Wang, R.; Khan, B.A.; Cheung, G.Y.C.; Bach, T.-H.L.; Jameson-Lee, M.; Kong, K.-F.; Queck, S.Y.; Otto, M. Staphylococcus epidermidis surfactant peptides promote biofilm maturation and dissemination of biofilm-associated infection in mice. J. Clin. Investig. 2011, 121, 238–248. [Google Scholar] [CrossRef]
- Chatterjee, S.S.; Joo, H.S.; Duong, A.C.; Dieringer, T.D.; Tan, V.Y.; Song, Y.; Fischer, E.R.; Cheung, G.Y.C.; Li, M.; Otto, M. Essential Staphylococcus aureus toxin export system. Nat. Med. 2013, 19, 364–367. [Google Scholar] [CrossRef] [PubMed]
- Gagnaire, J.; Dauwalder, O.; Boisset, S.; Khau, D.; Freydière, A.-M.; Ader, F.; Bes, M.; Lina, G.; Tristan, A.; Reverdy, M.-E.; et al. Detection of Staphylococcus aureus Delta-Toxin Production by Whole-Cell MALDI-TOF Mass Spectrometry. PLoS ONE 2012, 7, e40660. [Google Scholar] [CrossRef] [PubMed]
- Janzon, L.; Löfdahl, S.; Arvidson, S. Identification and nucleotide sequence of the delta-lysin gene, hld, adjacent to the accessory gene regulator (agr) of Staphylococcus aureus. Mol. Gen. Genet. MGG 1989, 219, 480–485. [Google Scholar] [CrossRef] [PubMed]
- Queck, S.Y.; Khan, B.A.; Wang, R.; Bach, T.-H.L.; Kretschmer, D.; Chen, L.; Kreiswirth, B.N.; Peschel, A.; DeLeo, F.R.; Otto, M. Mobile Genetic Element-Encoded Cytolysin Connects Virulence to Methicillin Resistance in MRSA. PLoS Pathog. 2009, 5, e1000533. [Google Scholar] [CrossRef]
- Chatterjee, S.S.; Chen, L.; Joo, H.-S.; Cheung, G.Y.C.; Kreiswirth, B.N.; Otto, M. Distribution and Regulation of the Mobile Genetic Element-Encoded Phenol-Soluble Modulin PSM-mec in Methicillin-Resistant Staphylococcus aureus. PLoS ONE 2011, 6, e28781. [Google Scholar] [CrossRef]
- Li, M.; Diep, B.A.; Villaruz, A.E.; Braughton, K.R.; Jiang, X.; DeLeo, F.R.; Chambers, H.F.; Lu, Y.; Otto, M. Evolution of virulence in epidemic community-associated methicillin-resistant Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 2009, 106, 5883–5888. [Google Scholar] [CrossRef]
- Su, M.; Lyles, J.T.; Petit III, R.A.; Peterson, J.; Hargita, M.; Tang, H.; Solis-Lemus, C.; Quave, C.L.; Read, T.D. Genomic analysis of variability in Delta-toxin levels between Staphylococcus aureus strains. PeerJ 2020, 8, e8717. [Google Scholar] [CrossRef]
- Berlon, N.R.; Qi, R.; Sharma-Kuinkel, B.K.; Joo, H.-S.; Park, L.P.; George, D.; Thaden, J.T.; Messina, J.A.; Maskarinec, S.A.; Mueller-Premru, M.; et al. Clinical MRSA isolates from skin and soft tissue infections show increased in vitro production of phenol soluble modulins. J. Infect. 2015, 71, 447–457. [Google Scholar] [CrossRef]
- Qi, R.; Joo, H.-S.; Sharma-Kuinkel, B.; Berlon, N.R.; Park, L.; Fu, C.; Messina, J.A.; Thaden, J.T.; Yan, Q.; Ruffin, F.; et al. Increased in vitro phenol-soluble modulin production is associated with soft tissue infection source in clinical isolates of methicillin-susceptible Staphylococcus aureus. J. Infect. 2016, 72, 302–308. [Google Scholar] [CrossRef]
- Bae, E.; Kim, C.K.; Jang, J.-H.; Sung, H.; Choi, Y.; Kim, M.-N. Impact of Community-Onset Methicillin-Resistant Staphylococcus aureus on Staphylococcus aureus Bacteremia in a Central Korea Veterans Health Service Hospital. Ann. Lab. Med. 2019, 39, 158–166. [Google Scholar] [CrossRef]
- Wang, B.; Xu, Y.; Zhao, H.; Wang, X.; Rao, L.; Guo, Y.; Yi, X.; Hu, L.; Chen, S.; Han, L.; et al. Methicillin-resistant Staphylococcus aureus in China: A multicenter longitudinal study and whole-genome sequencing. Emerg. Microbes Infect. 2022, 11, 532–542. [Google Scholar] [CrossRef] [PubMed]
- Young, B.C.; Wu, C.H.; Charlesworth, J.; Earle, S.; Price, J.R.; Gordon, N.C.; Cole, K.; Dunn, L.; Liu, E.; Oakley, S.; et al. Antimicrobial resistance determinants are associated with Staphylococcus aureus bacteraemia and adaptation to the healthcare environment: A bacterial genome-wide association study. Microb. Genom. 2021, 7, 700. [Google Scholar] [CrossRef] [PubMed]
- Kaku, N.; Sasaki, D.; Ota, K.; Miyazaki, T.; Yanagihara, K. Changing molecular epidemiology and characteristics of MRSA isolated from bloodstream infections: Nationwide surveillance in Japan in 2019. J. Antimicrob. Chemother. 2022, 77, 2130–2141. [Google Scholar] [CrossRef] [PubMed]
- Choi, S.-H.; Lee, J.; Jung, J.; Kim, E.S.; Kim, M.J.; Chong, Y.P.; Kim, S.-H.; Lee, S.-O.; Choi, S.-H.; Woo, J.H.; et al. A Longitudinal Study of Adult Patients with Staphylococcus aureus Bacteremia over 11 Years in Korea. J. Korean Med. Sci. 2021, 36, e104. [Google Scholar] [CrossRef] [PubMed]
- Kang, G.S.; Jung, Y.H.; Kim, H.S.; Lee, Y.S.; Park, C.; Lee, K.J.; Cha, J.O. Prevalence of Major Methicillin-Resistant Staphylococcus aureus Clones in Korea Between 2001 and 2008. Ann. Lab. Med. 2016, 36, 536–541. [Google Scholar] [CrossRef] [PubMed]
- Traber, K.; Novick, R. A slipped-mispairing mutation in AgrA of laboratory strains and clinical isolates results in delayed activation of agr and failure to translate δ- and α-haemolysins. Mol. Microbiol. 2006, 59, 1519–1530. [Google Scholar] [CrossRef] [PubMed]
- Chong, Y.P.; Kim, E.S.; Park, S.-J.; Park, K.-H.; Kim, T.; Kim, M.-N.; Kim, S.-H.; Lee, S.-O.; Choi, S.-H.; Woo, J.H.; et al. Accessory Gene Regulator ( agr ) Dysfunction in Staphylococcus aureus Bloodstream Isolates from South Korean Patients. Antimicrob. Agents Chemother. 2013, 57, 1509–1512. [Google Scholar] [CrossRef]
- Traber, K.E.; Lee, E.; Benson, S.; Corrigan, R.; Cantera, M.; Shopsin, B.; Novick, R.P. agr function in clinical Staphylococcus aureus isolates. Microbiology 2008, 154, 2265–2274. [Google Scholar] [CrossRef]
- Painter, K.L.; Krishna, A.; Wigneshweraraj, S.; Edwards, A.M. What role does the quorum-sensing accessory gene regulator system play during Staphylococcus aureus bacteremia? Trends Microbiol. 2014, 22, 676–685. [Google Scholar] [CrossRef]
- Cheung, G.Y.C.; Yeh, A.J.; Kretschmer, D.; Duong, A.C.; Tuffuor, K.; Fu, C.L.; Joo, H.S.; Diep, B.A.; Li, M.; Nakamura, Y.; et al. Functional characteristics of the Staphylococcus aureus δ-toxin allelic variant G10S. Sci. Rep. 2015, 5, 18023. [Google Scholar] [CrossRef]
- Verdon, J.; Girardin, N.; Lacombe, C.; Berjeaud, J.-M.; Héchard, Y. δ-hemolysin, an update on a membrane-interacting peptide. Peptides 2009, 30, 817–823. [Google Scholar] [CrossRef] [PubMed]
- Cheung, G.Y.C.; Joo, H.-S.; Chatterjee, S.S.; Otto, M. Phenol-soluble modulins—Critical determinants of staphylococcal virulence. FEMS Microbiol. Rev. 2014, 38, 698–719. [Google Scholar] [CrossRef]
- Kaito, C.; Saito, Y.; Ikuo, M.; Omae, Y.; Mao, H.; Nagano, G.; Fujiyuki, T.; Numata, S.; Han, X.; Obata, K.; et al. Mobile Genetic Element SCCmec-encoded psm-mec RNA Suppresses Translation of agrA and Attenuates MRSA Virulence. PLoS Pathog. 2013, 9, e1003269. [Google Scholar] [CrossRef]
- Kaito, C.; Saito, Y.; Nagano, G.; Ikuo, M.; Omae, Y.; Hanada, Y.; Han, X.; Kuwahara-Arai, K.; Hishinuma, T.; Baba, T.; et al. Transcription and Translation Products of the Cytolysin Gene psm-mec on the Mobile Genetic Element SCCmec Regulate Staphylococcus aureus Virulence. PLoS Pathog. 2011, 7, e1001267. [Google Scholar] [CrossRef]
- Oliveira, D.C.; De Lencastre, H. Multiplex PCR strategy for rapid identification of structural types and variants of the mec element in methicillin-resistant Staphylococcus aureus. Antimicrob. Agents Chemother. 2002, 46, 2155–2161. [Google Scholar] [CrossRef] [PubMed]
- Harmsen, D.; Claus, H.; Witte, W.; Rothgänger, J.; Claus, H.; Turnwald, D.; Vogel, U. Typing of methicillin-resistant Staphylococcus aureus in a university hospital setting by using novel software for spa repeat determination and database management. J. Clin. Microbiol. 2003, 41, 5442–5448. [Google Scholar] [CrossRef] [PubMed]
- Kondo, Y.; Ito, T.; Ma, X.X.; Watanabe, S.; Kreiswirth, B.N.; Etienne, J.; Hiramatsu, K. Combination of multiplex PCRs for staphylococcal cassette chromosome mec type assignment: Rapid identification system for mec, ccr, and major differences in junkyard regions. Antimicrob. Agents Chemother. 2007, 51, 264–274. [Google Scholar] [CrossRef] [PubMed]
- Joo, H.-S.; Otto, M. The Isolation and Analysis of Phenol-Soluble Modulins of Staphylococcus epidermidis. In Staphylococcus Epidermidis. Methods in Molecular Biology (Methods and Protocols); Fey, P.D., Ed.; Humana Press: Totowa, NJ, USA, 2014; pp. 93–100. ISBN 9781627037358. [Google Scholar]

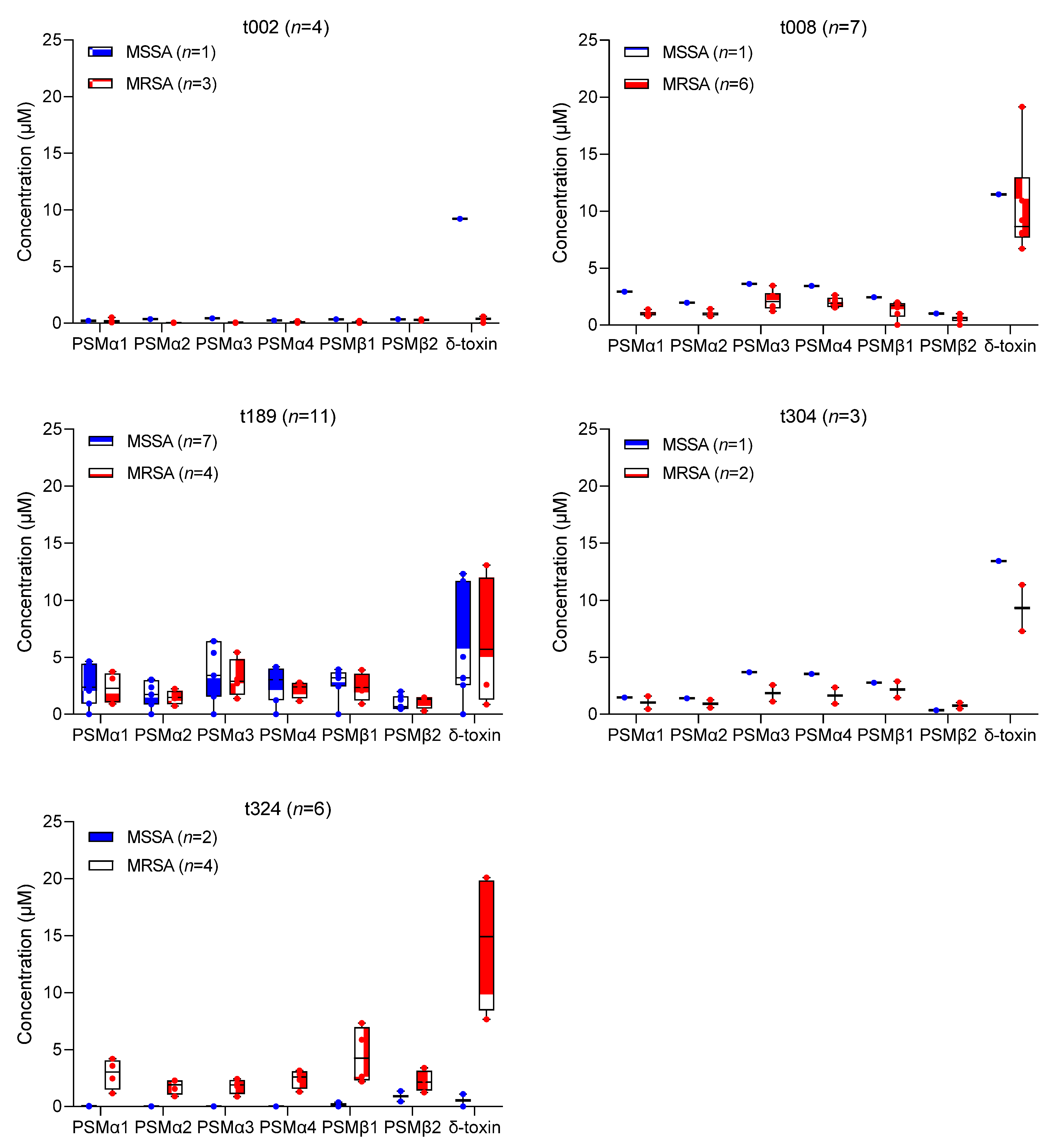
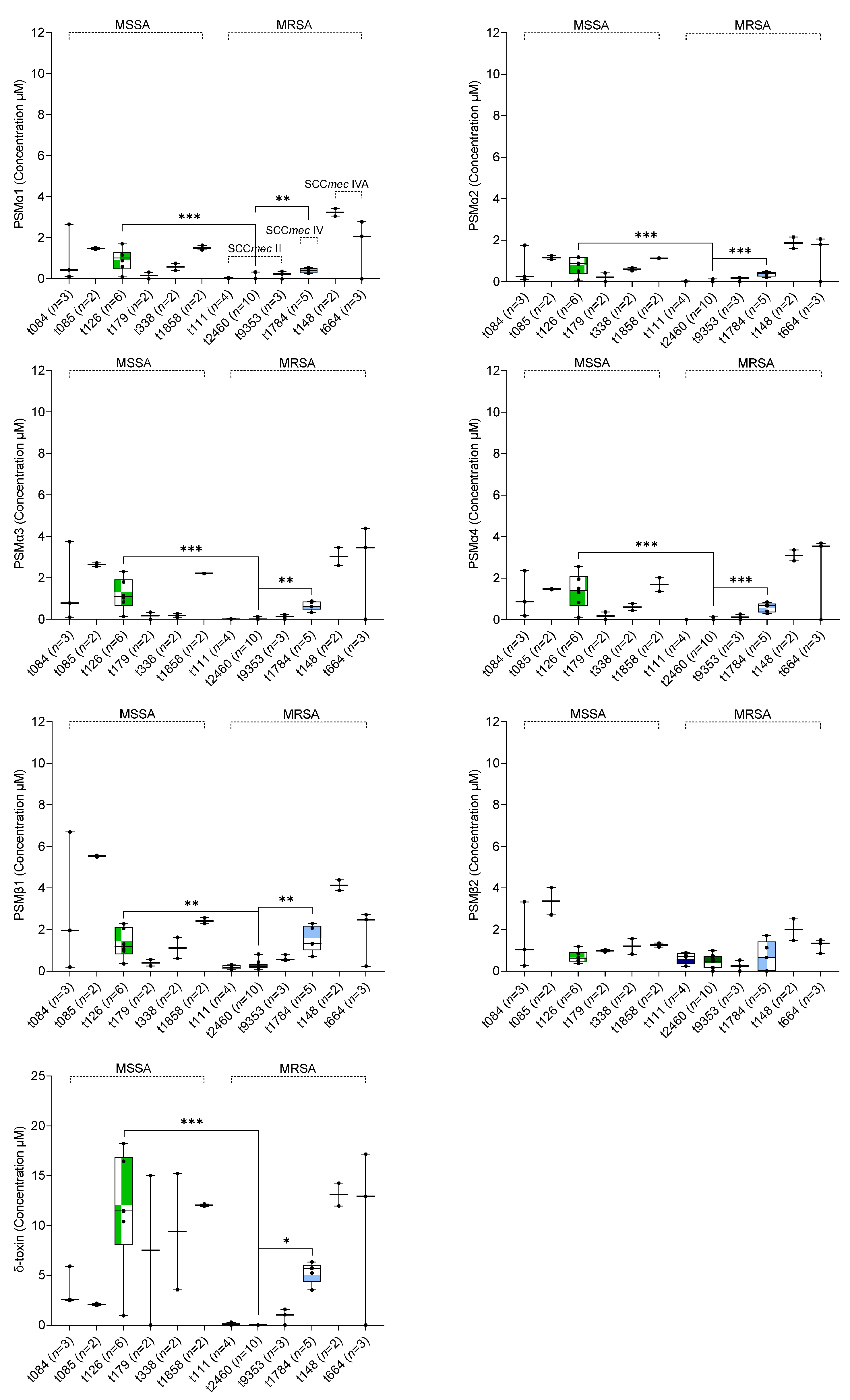
| spa Type | S. aureus Isolates n (%) | Phenotype | SCCmec Type for MRSA | |
|---|---|---|---|---|
| MSSA n (%) | MRSA n (%) | |||
| MSSA and MRSA | ||||
| t189 | 11 (10.4) | 7 (6.6) | 4 (3.8) | IV |
| t008 | 7 (6.6) | 1 (0.9) | 6 (5.7) | IV |
| t324 | 6 (5.7) | 2 (1.9) | 4 (3.8) | IVA |
| t002 | 4 (3.8) | 1 (0.9) | 3 (2.8) | II |
| t304 | 3 (2.8) | 1 (0.9) | 2 (1.9) | IV |
| MSSA | ||||
| t126 | 6 (5.7) | 6 (5.7) | - | O |
| t084 | 3 (2.8) | 3 (2.8) | - | O |
| t085 | 2 (1.9) | 2 (1.9) | - | O |
| t179 | 2 (1.9) | 2 (1.9) | - | O |
| t338 | 2 (1.9) | 2 (1.9) | - | O |
| t1858 | 2 (1.9) | 2 (1.9) | - | O |
| t005 | 1 (0.9) | 1 (0.9) | - | O |
| t019 | 1 (0.9) | 1 (0.9) | - | O |
| t021 | 1 (0.9) | 1 (0.9) | - | O |
| t127 | 1 (0.9) | 1 (0.9) | - | O |
| t177 | 1 (0.9) | 1 (0.9) | - | O |
| t346 | 1 (0.9) | 1 (0.9) | - | O |
| t363 | 1 (0.9) | 1 (0.9) | - | O |
| t386 | 1 (0.9) | 1 (0.9) | - | O |
| t416 | 1 (0.9) | 1 (0.9) | - | O |
| t521 | 1 (0.9) | 1 (0.9) | - | O |
| t571 | 1 (0.9) | 1 (0.9) | - | O |
| t1333 | 1 (0.9) | 1 (0.9) | - | O |
| t1361 | 1 (0.9) | 1 (0.9) | - | O |
| t1767 | 1 (0.9) | 1 (0.9) | - | O |
| t1950 | 1 (0.9) | 1 (0.9) | - | O |
| t4727 | 1 (0.9) | 1 (0.9) | - | O |
| t4956 | 1 (0.9) | 1 (0.9) | - | O |
| t10234 | 1 (0.9) | 1 (0.9) | - | O |
| t10686 | 1 (0.9) | 1 (0.9) | - | O |
| t12605 | 1 (0.9) | 1 (0.9) | - | O |
| undefined | 1 (0.9) | 1 (0.9) | - | O |
| MRSA | ||||
| t2460 | 10 (9.4) | - | 10 (9.4) | II |
| t1784 | 5 (4.7) | - | 5 (4.7) | IV |
| t111 | 4 (3.8) | - | 4 (3.8) | II |
| t664 | 3 (2.8) | - | 3 (2.8) | IVA |
| t9353 | 3 (2.8) | - | 3 (2.8) | II |
| t148 | 2 (1.9) | - | 2 (1.9) | IVA |
| t034 | 1 (0.9) | - | 1 (0.9) | V |
| t062 | 1 (0.9) | - | 1 (0.9) | II |
| t242 | 1 (0.9) | - | 1 (0.9) | IV |
| t264 | 1 (0.9) | - | 1 (0.9) | II |
| t893 | 1 (0.9) | - | 1 (0.9) | II |
| t1081 | 1 (0.9) | - | 1 (0.9) | V |
| t1154 | 1 (0.9) | 1 (0.9) | II | |
| t1560 | 1 (0.9) | - | 1 (0.9) | II |
| t3092 | 1 (0.9) | - | 1 (0.9) | V |
| t4359 | 1 (0.9) | - | 1 (0.9) | IVA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lade, H.; Chung, S.H.; Lee, Y.; Joo, H.-S.; Kim, J.-S. Genotypes of Staphylococcus aureus Clinical Isolates Are Associated with Phenol-Soluble Modulin (PSM) Production. Toxins 2022, 14, 556. https://doi.org/10.3390/toxins14080556
Lade H, Chung SH, Lee Y, Joo H-S, Kim J-S. Genotypes of Staphylococcus aureus Clinical Isolates Are Associated with Phenol-Soluble Modulin (PSM) Production. Toxins. 2022; 14(8):556. https://doi.org/10.3390/toxins14080556
Chicago/Turabian StyleLade, Harshad, Sung Hee Chung, Yeonhee Lee, Hwang-Soo Joo, and Jae-Seok Kim. 2022. "Genotypes of Staphylococcus aureus Clinical Isolates Are Associated with Phenol-Soluble Modulin (PSM) Production" Toxins 14, no. 8: 556. https://doi.org/10.3390/toxins14080556





