Venom of Parasitoid Pteromalus puparum Impairs Host Humoral Antimicrobial Activity by Decreasing Host Cecropin and Lysozyme Gene Expression
Abstract
:1. Introduction
2. Results
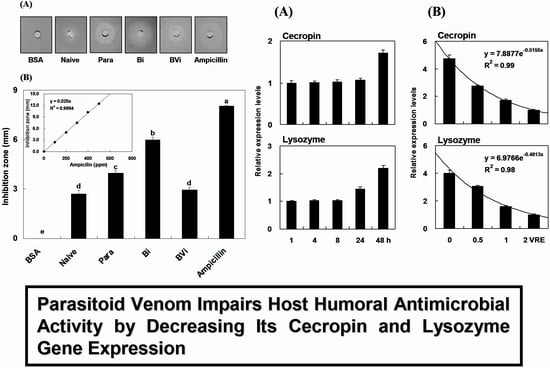
2.1. Induction and Inhibition of Antimicrobial Activity of Host Hemolymph
2.2. Molecular Cloning and Sequence Analyses of The Pr-cec and Pr-lys cDNAs
2.3. Effect of Immune Induction and Inhibition on Pr-cec and Pr-lys Gene Expression
2.4. Time Course of Immune Induction on Pr-cec and Pr-lys Gene Expression
2.5. Tissue Distribution of Pr-cec and Pr-lys Gene Expression
2.6. Reduction of Antimicrobial Activity in the Host Hemolymph by the RNA Interference of Pr-cec and Pr-lys
2.7. Time Course and Dose Effect of Venom Inhibition on Pr-cec and Pr-lys Gene Expression
2.8. Influence of Synthesized Pr-cec and Recombinant Pr-lys on the P. puparum Emergence Rate from the Parasitized Pupae
3. Discussion
4. Experimental Section
4.1. Insect Rearing
4.2. Crude Venom Preparation
4.3. Hemolymph Collection
4.4. In Vitro Assay of Inhibition Zone
4.5. Obtained Full-length cDNA and Sequence Analysis
4.6. Gene Expression Profile Analysis
4.7. RNA Interference
4.8. Production of Synthesized Pr-cec and Recombinant Pr-lys
4.9. Emergence Rate Assay
4.10. Statistical Analysis
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Cherry, S.; Silverman, N. Host-pathogen interactions in Drosophila: New tricks from an old friend. Nat. Immunol. 2006, 7, 911–917. [Google Scholar] [CrossRef] [PubMed]
- Tunaz, H.; Stanley, D. An immunological axis of biocontrol: Infections in field-trapped insects. Naturwissenschaften 2009, 96, 96–115. [Google Scholar] [CrossRef] [PubMed]
- Kanost, M.R.; Jiang, H.; Yu, X.Q. Innate immune responses of a lepidopteran insect, Manduca sexta. Immunol. Rev. 2004, 198, 97–105. [Google Scholar] [CrossRef] [PubMed]
- Beckage, N.E. Insect Immunology, 1st ed.; Academic Press: San Diego, CA, USA, 2008; pp. 13–54. [Google Scholar]
- Hoffmann, J.A. The immune response of Drosophila. Nature 2003, 426, 33–38. [Google Scholar] [CrossRef] [PubMed]
- Lavine, M.D.; Beckage, N.E. Polydnaviruses: Potent mediators of host insect immune dysfunction. Parasitol. Today 1995, 11, 368–378. [Google Scholar] [CrossRef]
- Strand, M.R.; Beck, M.H.; Lavine, M.D.; Clark, K.D. Microplitis demolitor bracovirus inhibits phagocytosis by hemocytes from Pseudoplusia includens. Arch. Insect Biochem. Physiol. 2006, 61, 134–145. [Google Scholar] [CrossRef] [PubMed]
- Ao, J.; Ling, E.; Yu, X.Q. Drosophila C-type lectins enhance cellular encapsulation. Mol. Immunol. 2007, 44, 2541–2548. [Google Scholar] [CrossRef] [PubMed]
- Janeway, C.A. The immune system evolved to discriminate infectious nonself from noninfectious self. Immunol. Today 1992, 13, 11–16. [Google Scholar] [CrossRef]
- Janeway, C.A.; Medzhitov, R. Innate immune recognition. Annu. Rev. Immunol. 2002, 20, 197–216. [Google Scholar] [CrossRef] [PubMed]
- Lavine, M.D.; Strand, M.R. Insect hemocytes and their role in immunity. Insect Biochem. Mol. Biol. 2002, 32, 1295–1309. [Google Scholar] [CrossRef]
- Stanley, D.; Miller, J.; Tunaz, H. Eicosanoid actions in insect immunity. J. Innate Immun. 2009, 1, 282–290. [Google Scholar] [CrossRef] [PubMed]
- Vizioli, J.; Salzet, M. Antimicrobial peptides versus parasitic infections? Trends Parasitol. 2002, 18, 475–476. [Google Scholar] [CrossRef]
- Hetru, C.; Troxler, L.; Hoffmann, J.A. Drosophila melanogaster antimicrobial defense. J. Infect. Dis. 2003, 187, S327–S334. [Google Scholar] [CrossRef] [PubMed]
- Strand, M.R. The insect cellular immune response. Insect Sci. 2008, 15, 1–14. [Google Scholar] [CrossRef]
- Martinson, E.O.; Wheeler, D.; Wright, J.; Mrinalini; Siebert, A.L.; Werren, J.H. Nasonia vitripennis venom causes targeted gene expression changes in its fly host. Mol. Ecol. 2014, 23, 5918–5930. [Google Scholar] [CrossRef] [PubMed]
- Asgari, S.; Rivers, D.B. Venom proteins from endoparasitoid wasps and their role in host-parasite interactions. Annu. Rev. Entomol. 2011, 56, 313–335. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, A.M.; Kim, Y. Parasitism by Cotesia plutellae alters the hemocyte population and immunological function of the diamondback moth, Plutella xylostella. J. Insect Physiol. 2006, 52, 943–950. [Google Scholar] [CrossRef] [PubMed]
- Pennacchio, F.; Strand, M.R. Evolution of developmental strategies in parasitic hymenoptera. Annu. Rev. Entomol. 2006, 51, 233–258. [Google Scholar] [CrossRef] [PubMed]
- Glatz, R.V.; Asgari, S.; Schmidt, O. Evolution of polydnaviruses as insect immune suppressors. Trends Microbiol. 2004, 12, 545–554. [Google Scholar] [CrossRef] [PubMed]
- Barat-Houari, M.; Hilliou, F.; Jousset, F.X.; Sofer, L.; Deleury, E.; Rocher, J.; Ravallec, M.; Galibert, L.; Delobel, P.; Feyereisen, R.; et al. Gene expression profiling of Spodoptera frugiperda hemocytes and fat body using cDNA microarray reveals polydnavirus-associated variations in lepidopteran host genes transcript levels. BMC Genomics 2006, 7, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Volkoff, A.N.; Beliveau, C.; Rocher, J.; Hilgarth, R.; Levasseur, A.; Duonor-Cerutti, M.; Cusson, M.; Webb, B.A. Evidence for a conserved polydnavirus gene family: Ichnovirus homologs of the CsIV repeat element genes. Virology 2002, 300, 316–331. [Google Scholar] [CrossRef] [PubMed]
- Amaya, K.E.; Asgari, S.; Jung, R.; Hongskula, M.; Beckage, N.E. Parasitization of Manduca sexta larvae by the parasitoid wasp Cotesia congregata induces an impaired host immune response. J. Insect Physiol. 2005, 51, 505–512. [Google Scholar] [CrossRef] [PubMed]
- Asgari, S. Venom proteins from polydnavirus-producing endoparasitoids: Their role in host-parasite interactions. Arch. Insect Biochem. Physiol. 2006, 61, 146–156. [Google Scholar] [CrossRef] [PubMed]
- Bae, S.; Kim, Y. Host physiological changes due to parasitism of a braconid wasp, Cotesia plutellae, on diamondback moth, Plutella xylostella. Comp. Biochem. Physiol. A 2004, 138, 39–44. [Google Scholar] [CrossRef] [PubMed]
- Labrosse, C.; Carton, Y.; Dubuffet, A.; Drezen, J.M.; Poirie, M. Active suppression of D. melanogaster immune response by long gland products of the parasitic wasp Leptopilina boulardi. J. Insect Physiol. 2003, 49, 513–522. [Google Scholar] [CrossRef]
- Labrosse, C.; Stasiak, K.; Lesobre, J.; Grangeia, A.; Huguet, E.; Drezen, J.M.; Poirie, M. A RhoGAP protein as a main immune suppressive factor in the Leptopilina boulardi (Hymenoptera, Figitidae)-Drosophila melanogaster interaction. Insect Biochem. Mol. Biol. 2005, 35, 93–103. [Google Scholar] [CrossRef] [PubMed]
- Colinet, D.; Dubuffet, A.; Cazes, D.; Moreau, S.; Drezen, J.M.; Poirié, M. A serpin from the parasitoid wasp Leptopilina boulardi targets the Drosophila phenoloxidase cascade. Dev. Comp. Immunol. 2009, 33, 681–689. [Google Scholar] [CrossRef] [PubMed]
- Colinet, D.; Cazes, D.; Belghazi, M.; Gatti, J.L.; Poirié, M. Extracellular superoxide dismutase in insects: Characterization, function, and interspecific variation in parasitoid wasp venom. J. Biol. Chem. 2011, 286, 40110–40121. [Google Scholar] [CrossRef] [PubMed]
- Mortimer, N.T.; Goecks, J.; Kacsoh, B.Z.; Mobley, J.A.; Bowersock, G.J.; Taylor, J.; Schlenke, T.A. Parasitoid wasp venom SERCA regulates Drosophila calcium levels and inhibits cellular immunity. Proc. Natl. Acad. Sci. USA 2013, 110, 9427–9432. [Google Scholar] [CrossRef] [PubMed]
- Cai, J.; Ye, G.Y.; Hu, C. Parasitism of Pieris rapae (Lepidoptera: Pieridae) by a pupal endoparasitoid, Pteromalus puparum (Hymenoptera: Pteromalidae): Effects of parasitization and venom on host hemocytes. J. Insect Physiol. 2004, 50, 315–322. [Google Scholar] [CrossRef] [PubMed]
- Ye, G.Y.; Zhu, J.Y.; Zhang, Z.; Fang, Q.; Cai, J.; Hu, C. Venom from the endoparasitoid Pteromalus puparum (Hymenoptera: Pteromalidae) adversely affects host hemocytes: Differential toxicity and microstructural and ultrastructural changes in plasmatocytes and granular cells. In Recent Advances in the Biochemistry, Toxicity, and Mode of Action of Parasitic Wasp Venoms, 1st ed.; Rivers, D.B., Yolder, J.A., Eds.; Research Signpost: Kerala, India, 2007; pp. 115–127. [Google Scholar]
- Zhang, Z.; Ye, G.Y.; Cai, J.; Hu, C. Comparative venom toxicity between Pteromalus puparum and Nasonia vitripennis (Hymenoptera: Pteromalidae) toward the hemocytes of their natural hosts, non-target insects and cultured insect cells. Toxicon 2005, 46, 337–349. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.L.; Ye, G.Y.; Zhu, J.Y.; Chen, X.X.; Hu, C. Isolation and characterization of an immunosuppressive protein from venom of the pupa-specific endoparasitoid Pteromalus puparum. J. Invertebr. Pathol. 2008, 99, 186–191. [Google Scholar] [CrossRef] [PubMed]
- Fang, Q.; Wang, L.; Zhu, Y.K.; Stanley, D.W.; Chen, X.X.; Hu, C.; Ye, G.Y. Pteromalus puparum venom impairs host cellular immune responses by decreasing expression of its scavenger receptor gene. Insect Biochem. Mol. Biol. 2011, 41, 852–862. [Google Scholar] [CrossRef] [PubMed]
- Fang, Q.; Wang, F.; Gatehouse, J.A.; Gatehouse, A.M.; Chen, X.X.; Hu, C.; Ye, G.Y. Venom of parasitoid, Pteromalus puparum, suppresses host, Pieris rapae, immune promotion by decreasing host C-type lectin gene expression. PLoS ONE 2011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, L.; Fang, Q.; Qian, C.; Wang, F.; Yu, X.Q.; Ye, G. Inhibition of host cell encapsulation through inhibiting immune gene expression by the parasitic wasp venom calreticulin. Insect Biochem. Mol. Biol. 2013, 43, 936–946. [Google Scholar] [CrossRef] [PubMed]
- Fang, Q.; Wang, L.; Zhu, J.; Li, Y.; Song, Q.; Stanley, D.W.; Akhtar, Z.R.; Ye, G. Expression of immune-response genes in lepidopteran host is suppressed by venom from an endoparasitoid, Pteromalus puparum. BMC Genomics 2010. [Google Scholar] [CrossRef] [PubMed]
- Yi, H.Y.; Chowdhury, M.; Huang, Y.D.; Yu, X.Q. Insect antimicrobial peptides and their applications. Appl. Microbiol. Biotechnol. 2014, 98, 5807–5822. [Google Scholar] [CrossRef] [PubMed]
- Rao, X.J.; Ling, E.; Yu, X.Q. The role of lysozyme in the prophenoloxidase activation system of Manduca sexta: An in vitro approach. Dev. Comp. Immunol. 2010, 34, 264–271. [Google Scholar] [CrossRef] [PubMed]
- Burke, G.R.; Strand, M.R. Systematic analysis of a wasp parasitism arsenal. Mol. Ecol. 2014, 23, 890–901. [Google Scholar] [CrossRef] [PubMed]
- Pruijssers, A.J.; Strand, M.R. PTP-H2 and PTP-H3 from Microplitis demolitor bracovirus localize to focal adhesions and are antiphagocytic in insect immune cells. J. Virol. 2007, 81, 1209–1219. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Beck, M.H.; Wang, Y.; Jiang, H.; Strand, M.R. The viral protein Egf1.0 is a dual activity inhibitor of prophenoloxidase-activating proteinases 1 and 3 from Manduca sexta. J. Biol. Chem. 2008, 283, 21325–21333. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Beck, M.H.; Strand, M.R. Egf1.5 is a second phenoloxidase cascade inhibitor encoded by Microplitis demolitor bracovirus. Insect Biochem. Mol. Biol. 2010, 40, 497–505. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Nalini, M.; Kim, Y. Viral lectin encoded in Cotesia plutellae bracovirus and its immunosuppressive effect on host hemocytes. Comp. Biochem. Physiol. A 2008, 149, 351–361. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.R.; Kim, Y. Antiviral activity of the inducible humoral immunity and its suppression by eleven BEN family members encoded in Cotesia plutellae bracovirus. Comp. Biochem. Physiol. A 2015, 179, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Colinet, D.; Schmitz, A.; Depoix, D.; Crochard, D.; Poirié, M. Convergent use of RhoGAP toxins by eukaryotic parasites and bacterial pathogens. PLoS Pathog. 2007. [Google Scholar] [CrossRef] [PubMed]
- Labrosse, C.; Eslin, P.; Doury, G.; Drezen, J.M.; Poirié, M. Hemocyte changes in D. melanogaster in response to long gland components of the parasitoid wasp Leptopilina boulardi: A Rho-GAP protein as an important factor. J. Insect Physiol. 2005, 51, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Haine, E.R.; Moret, Y.; Siva-Jothy, M.T.; Rolff, J. Antimicrobial defense and persistent infection in insects. Science 2008, 322, 1257–1259. [Google Scholar] [CrossRef] [PubMed]
- Chevignon, G.; Cambier, S.; Da Silva, C.; Poulain, J.; Drezen, J.M.; Huguet, E.; Moreau, S.J. Transcriptomic response of Manduca sexta immune tissues to parasitization by the bracovirus associated wasp Cotesia congregata. Insect Biochem. Mol. Biol. 2015, 62, 86–99. [Google Scholar] [CrossRef] [PubMed]
- Beckage, N.E.; Gelman, D.B. Wasp parasitoid disruption of host development: Implications for new biologically based strategies for insect control. Annu. Rev. Entomol. 2004, 49, 299–330. [Google Scholar] [CrossRef] [PubMed]
- Barandoc, K.P.; Kim, J.; Kim, Y. Cotesia plutellae bracovirus suppresses expression of an antimicrobial peptide, cecropin, in the diamondback moth, Plutella xylostella, challenged by bacteria. J. Microbiol. 2010, 48, 117–123. [Google Scholar] [CrossRef] [PubMed]
- Wertheim, B.; Kraaijeveld, A.R.; Schuster, E.; Blanc, E.; Hopkins, M.; Pletcher, S.D.; Strand, M.R.; Partridge, L.; Godfray, H.C. Genome-wide gene expression in response to parasitoid attack in Drosophila. Genome Biol. 2005, 6, R94. [Google Scholar] [CrossRef] [PubMed]
- Bitra, K.; Suderman, R.J.; Strand, M.R. Polydnavirus Ank proteins bind NF-κB homodimers and inhibit processing of Relish. PLoS Pathog. 2012, 8. [Google Scholar] [CrossRef] [PubMed]
- Kylsten, P.; Samakovlis, C.; Hultmark, D. The cecropin locus in Drosophila: A compact gene cluster involved in the response to infection. EMBO J. 1990, 9, 217–224. [Google Scholar] [PubMed]
- Zhao, C.; Liaw, L.; Lee, I.H.; Lehrer, R.I. cDNA cloning of three cecropin-like antimicrobial peptides (Styelins) from the tunicate, Styela clava. FEBS Lett. 1997, 412, 144–148. [Google Scholar] [CrossRef]
- Andersson, M.; Boman, A.; Boman, H.G. Ascaris nematodes from pig and human make three antibacterial peptides: Isolation of cecropin P1 and two ASABF peptides. Cell. Mol. Life Sci. 2003, 60, 599–606. [Google Scholar] [CrossRef] [PubMed]
- Gudmundsson, G.H.; Lidholm, D.A.; Åsling, B.; Gan, R.; Boman, H.G. The cecropin locus, cloning and expression of a gene cluster encoding three antibacterial peptides in Hyalophora cecropia. J. Biol. Chem. 1991, 266, 11510–11517. [Google Scholar] [PubMed]
- Hara, S.; Taniai, K.; Kato, Y.; Yamakawa, M. Isolation and amidation of the non-amidated form of cecropin D from larvae of Bombyx mori. Comp. Biochem. Physiol. 1994, 108, 303–308. [Google Scholar] [CrossRef]
- Zasloff, M. Antimicrobial peptides of multicellular organisms. Nature 2002, 415, 389–395. [Google Scholar] [CrossRef] [PubMed]
- Plunkett, R.M.; Murray, S.I.; Lowenberger, C.A. Generation and characterization of the antibacterial activity of a novel hybrid antimicrobial peptide comprising functional domains from different insect cecropins. Can. J. Microbiol. 2009, 55, 520–528. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Zhou, F.; Chu, Y.; Zhao, Z.; An, C. Identification and expression profile analysis of antimicrobial peptide/protein in Asian corn borer, Ostrinia furnacalis (Guenée). Int. J. Biol. Sci. 2013, 9, 1004–1012. [Google Scholar] [CrossRef] [PubMed]
- Fujimoto, S.; Toshimori-Tsuda, I.; Kishimoto, K.; Yamano, Y.; Morishima, I. Protein purification, cDNA cloning and gene expression of lysozyme from eri-silkworm, Samia cynthia ricini. Comp. Biochem. Physiol. B 2001, 128, 709–718. [Google Scholar] [CrossRef]
- Lavine, M.D.; Chen, G.; Strand, M.R. Immune challenge differentially affects transcript abundance of three antimicrobial peptides in hemocytes from the moth Pseudoplusia includens. Insect Biochem. Mol. Biol. 2005, 35, 1335–1346. [Google Scholar] [CrossRef] [PubMed]
- Téllez, G.A.; Castaño-Osorio, J.C. Expression and purification of an active cecropin-like recombinant protein against multidrug resistance Escherichia coli. Protein Expr. Purif. 2014, 100, 48–53. [Google Scholar] [CrossRef] [PubMed]
- Valachova, I.; Takac, P.; Majtan, J. Midgut lysozymes of Lucilia sericata—New antimicrobials involved in maggot debridement therapy. Insect Mol. Biol. 2014, 23, 779–787. [Google Scholar] [CrossRef] [PubMed]
- Roos, E.; Bjorklund, G.; Engstrom, Y. In vivo regulation of tissue-specific and LPS-inducible expression of the Drosophila Cecropin genes. Insect Mol. Biol. 1998, 7, 51–62. [Google Scholar] [CrossRef] [PubMed]
- Petersen, U.M.; Kadalayil, L.; Rehorn, K.P.; Hoshizaki, D.K.; Reuter, R.; Engström, Y. Serpent regulates Drosophila immunity genes in the larval fat body through an essential GATA motif. EMBO J. 1999, 18, 4013–4022. [Google Scholar] [CrossRef] [PubMed]
- Hong, S.M.; Kusakabe, T.; Lee, J.M.; Tatsuke, T.; Kawaguchi, Y.; Kang, M.W.; Kang, S.W.; Kim, K.A.; Nho, S.K. Structure and expression analysis of the Cecropin-E gene from the silkworm, Bombyx mori. Biosci. Biotechnol. Biochem. 2008, 72, 1992–1998. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.C.; Asling, B.; Faye, I. Organization and expression of the immunoresponsive lysozyme gene in the giant silk moth, Hyalophora cecropia. J. Biol. Chem. 1991, 266, 6644–6649. [Google Scholar] [PubMed]
- Kim, Y.; Basio, N.A.; Ibrahim, A.M.; Bae, S. Gene structure of Cotesia plutellae bracovirus (CpBV)-IkB and its expression pattern in diamondback moth, Plutella xylostella, parasitized by Cotesia plutellae. Korean J. Appl. Entomol. 2006, 45, 1–10. [Google Scholar]
- Thoetkiattikul, H.; Beck, M.H.; Strand, M.R. Inhibitor κB-like proteins from a polydnavirus inhibit NF-κB activation and suppress the insect immune response. Proc. Natl. Acad. Sci. USA 2005, 102, 11426–11431. [Google Scholar] [CrossRef] [PubMed]
- Bae, S.; Kim, Y. IkB genes encoded in Cotesia plutellae bracovirus suppress an antiviral response and enhance baculovirus pathogenicity against the diamondback moth, Plutella xylostella. J. Invertebr. Pathol. 2009, 102, 79–87. [Google Scholar] [CrossRef] [PubMed]
- Magkrioti, C.; Iatrou, K.; Labropoulou, V. Differential inhibition of BmRelish1-dependent transcription in lepidopteran cells by bracovirus ankyrin-repeat proteins. Insect Biochem. Mol. Biol. 2011, 41, 993–1002. [Google Scholar] [CrossRef] [PubMed]
- Herzner, G.; Schlecht, A.; Dollhofer, V.; Parzefall, C.; Harrar, K.; Kreuzer, A.; Pilsl, L.; Ruther, J. Larvae of the parasitoid wasp Ampulex compressa sanitize their host, the American cockroach, with a blend of antimicrobials. Proc. Natl. Acad. Sci. USA 2013, 110, 1369–1374. [Google Scholar] [CrossRef] [PubMed]
- Zambon, R.A.; Nandakumar, M.; Vakharia, V.N.; Wu, L.P. The Toll pathway is important for an antiviral response in Drosophila. Proc. Natl. Acad. Sci. USA 2005, 102, 7257–7262. [Google Scholar] [CrossRef] [PubMed]
- Kang, C.I.; Kim, S.H.; Kim, H.B.; Park, S.W.; Choe, Y.J.; Oh, M.D.; Kim, E.C.; Choe, K.W. Pseudomonas aeruginosa bacteremia: Risk factors for mortality and influence of delayed receipt of effective antimicrobial therapy on clinical outcome. Clin. Infect. Dis. 2003, 37, 745–751. [Google Scholar] [CrossRef] [PubMed]
- SignalP 4.1 Server. Available online: http://www.cbs.dtu.dk/services/SignalP/ (accessed on 13 November 2015).
- Tanura, K.; Dudley, J.; Nei, M.; Kumar, S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef] [PubMed]
- Clustal Omega. Available online: http://www.ebi.ac.uk/Tools/msa/clustalo/ (accessed on 13 November 2015).
- Primer3. Available online: http://www.simgene.com/Primer3 (accessed on 13 November 2015).
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and 2-∆∆ CT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Wiesner, A.; Götz, P. Silica beads induce cellular and humoral immune responses in Galleria mellonella larvae and in isolated plasmatocytes, obtained by a newly adapted nylon wool separation method. J. Insect Physiol. 1993, 39, 865–876. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Tang, Q.Y.; Zhang, C.X. Data Processing System (DPS) software with experimental design, statistical analysis and data mining developed for use in entomological research. Insect Sci. 2013, 20, 254–260. [Google Scholar] [CrossRef] [PubMed]






| Microorganism | Treatments a | Inhibition Zone (mm) b |
|---|---|---|
| Escherichia coli (K12) | Control | 7.8 ± 0.2 a |
| PBS | 8.2 ± 0.8 a | |
| GFPi | 8.7 ± 0.3 a | |
| CECi | 5.9 ± 0.3 bc | |
| LYSi | 7.3 ± 0.2 ab | |
| CEC-LYSi | 4.2 ± 0.2 c | |
| Micrococcus luteus | Control | 13.5 ± 0.7 a |
| PBS | 13.8 ± 0.6 a | |
| GFPi | 13.3 ± 0.8 a | |
| CECi | 10.5 ± 0.6 b | |
| LYSi | 7.8 ± 0.5 c | |
| CEC-LYSi | 7.1 ± 0.2 c |
| Gene Names | Primer Sequences (from 5' to 3') a | Functions | |
|---|---|---|---|
| Pr-cec | SP | TTTCGCAACCACCTACAT | qPCR |
| AP | TTCCAGCATTTCCATCAG | qPCR | |
| Pr-lys | SP | TTGGGTATGTCTCGTTGAA | qPCR |
| AP | TTGTGATGTCGTCCGTTGT | qPCR | |
| 18S rRNA | SP | TTTGCCTTATCAACTTTCG | qPCR |
| AP | TGTGGTAGCCGTTTCTCA | qPCR | |
| Pr-lys | Sub-SP | TAGAGCTCATGAAGTTAGCAGTATTCATTTTTG b | expression |
| Sub-AP | ATGTCGAC TTAACAAGAACTTATGTCAGGGAG b | expression | |
| Primer Directions | Primer Names | Primer Sequences (from 5' to 3') a |
|---|---|---|
| Forward | T7PrCEC-F | GGATCGTAATACGACTCACTATAGGATGAATTTCGGAAAATTGTTTTTG |
| PrCEC-F | ATGAATTTCGGAAAATTGTTTTTG | |
| T7PrLYS-F | GGATCGTAATACGACTCACTATAGGATGAAGTTAGCAGTATTCATTTTTG | |
| PrLYS-F | ATGAAGTTAGCAGTATTCATTTTTG | |
| T7GFP-F | GGATCGTAATACGACTCACTATAGGAAGGGCGAGGAGCTGTTCACCG | |
| GFP-F | AAGGGCGAGGAGCTGTTCACCG | |
| Reverse | T7PrCEC-R | GGATCGTAATACGACTCACTATAGGCTATTTTCCTTTATAGATGGTGGCA |
| PrCEC-R | CTATTTTCCTTTATAGATGGTGGCA | |
| T7PrLYS-R | GGATCGTAATACGACTCACTATAGGTTAACAAGAACTTATGTCAGGGAG | |
| PrLYS-R | TTAACAAGAACTTATGTCAGGGAG | |
| T7GFP-R | GGATCGTAATACGACTCACTATAGGCAGCAGGACCATGTGATCGCGC | |
| GFP-R | CAGCAGGACCATGTGATCGCGC |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fang, Q.; Wang, B.-B.; Ye, X.-H.; Wang, F.; Ye, G.-Y. Venom of Parasitoid Pteromalus puparum Impairs Host Humoral Antimicrobial Activity by Decreasing Host Cecropin and Lysozyme Gene Expression. Toxins 2016, 8, 52. https://doi.org/10.3390/toxins8020052
Fang Q, Wang B-B, Ye X-H, Wang F, Ye G-Y. Venom of Parasitoid Pteromalus puparum Impairs Host Humoral Antimicrobial Activity by Decreasing Host Cecropin and Lysozyme Gene Expression. Toxins. 2016; 8(2):52. https://doi.org/10.3390/toxins8020052
Chicago/Turabian StyleFang, Qi, Bei-Bei Wang, Xin-Hai Ye, Fei Wang, and Gong-Yin Ye. 2016. "Venom of Parasitoid Pteromalus puparum Impairs Host Humoral Antimicrobial Activity by Decreasing Host Cecropin and Lysozyme Gene Expression" Toxins 8, no. 2: 52. https://doi.org/10.3390/toxins8020052
APA StyleFang, Q., Wang, B.-B., Ye, X.-H., Wang, F., & Ye, G.-Y. (2016). Venom of Parasitoid Pteromalus puparum Impairs Host Humoral Antimicrobial Activity by Decreasing Host Cecropin and Lysozyme Gene Expression. Toxins, 8(2), 52. https://doi.org/10.3390/toxins8020052