Nucleotide Identification in DNA Using Dielectrophoresis Spectroscopy
Abstract
:1. Introduction
2. Theory
3. Materials and Methods
3.1. Sample Preparation
- 5′-(biotin) TGTTGTGCGA-3′
- 5′-(biotin) TGTTGTGCGT-3′
- 5′-(biotin) TGTTGTGCGG-3′
- 5′-(biotin) TGTTGTGCGC-3′
- 5′-(biotin) TGTTGTGCAC-3′
- 5′-(biotin) TGTTGTGCTC-3′
- 5′-(biotin) TGTTGTGCCC-3′
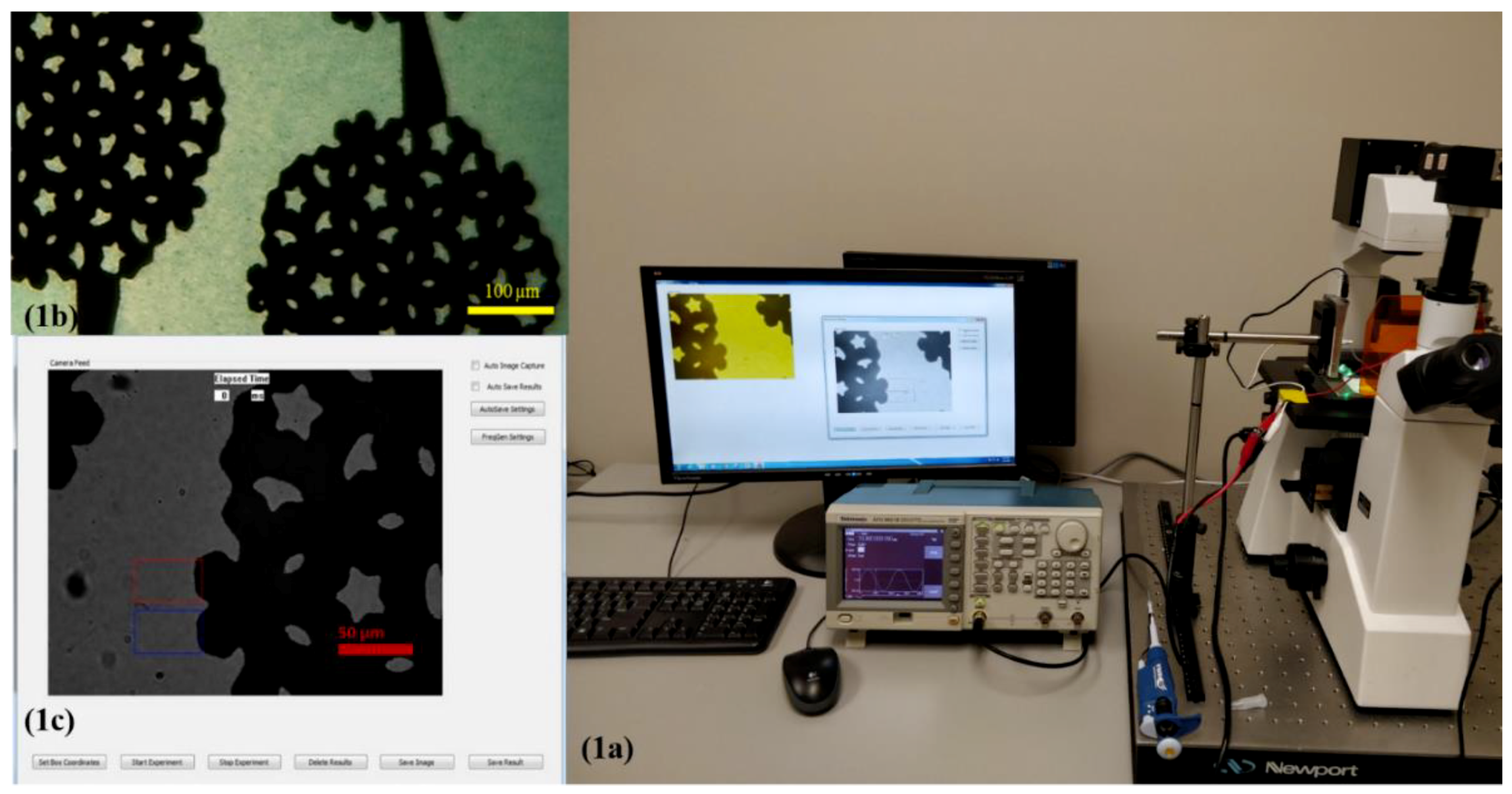
3.2. DEP Spectroscopy: Hardware and Software
3.3. Spectrum Measurement
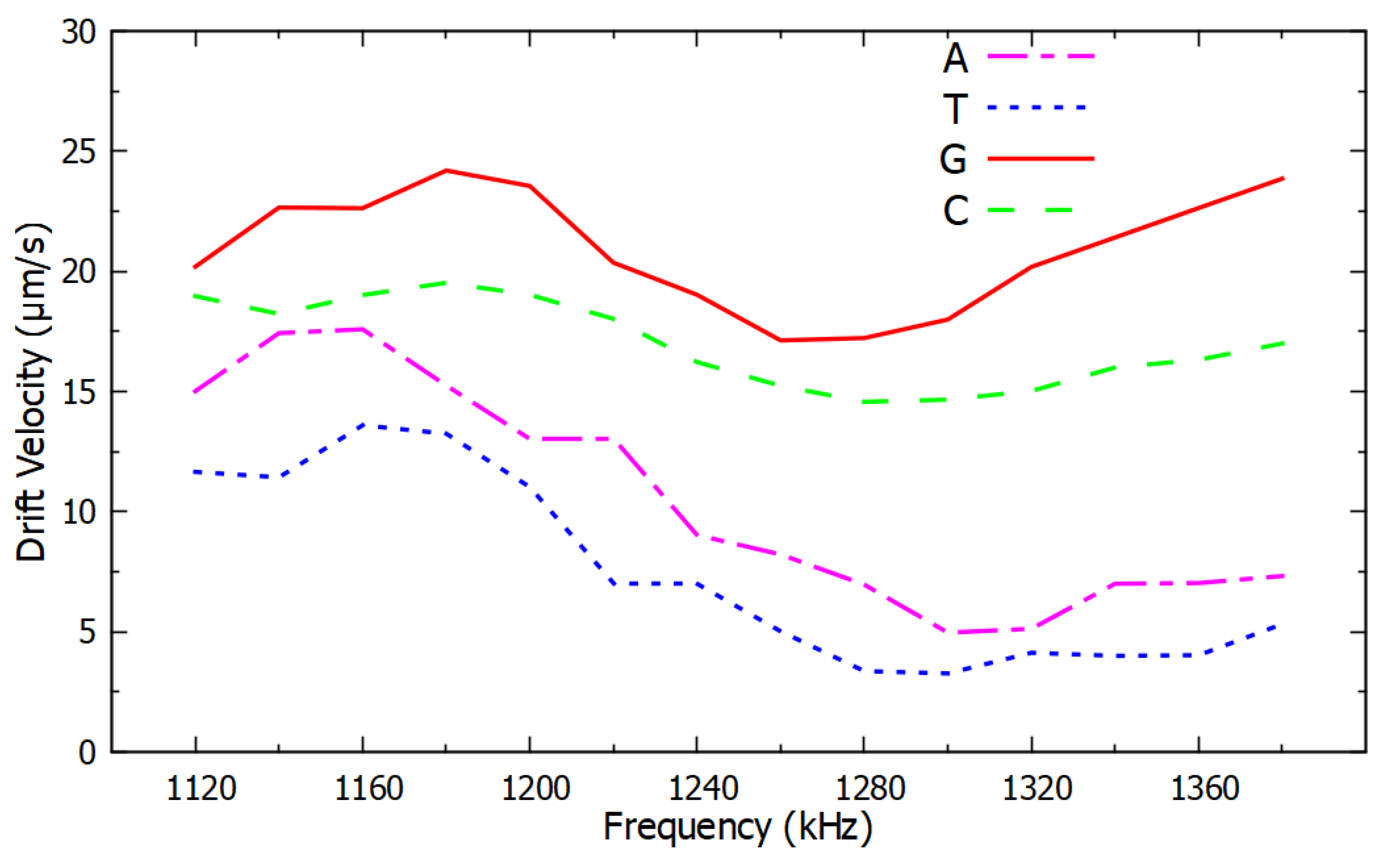
4. Results
5. Conclusions
6. Patent
Author Contributions
Funding
Conflicts of Interest
References
- Chen, C.; Keith, J.; Chen, C.C.; Schwender, H.; Keith, J.; Nunkesser, R.; Mengersen, K.; Macrossan, P. Methods for identifying SNP interactions: A review on variations of logic regression, random forest and bayesian logistic. IEEE/ACM Trans. Comput. Biol. Bioinform. 2011, 8, 1580–1591. [Google Scholar] [CrossRef] [PubMed]
- Bush, W.S.; Moore, J.H. Chapter 11: Genome-wide association studies. PLoS Comput. Biol. 2012, 8, 1–11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, Y.; Wan, Z.; Coarfa, C.; Drabek, R.; Chen, L.; Ostrowski, E.A.; Liu, Y.; Weinstock, G.M.; Wheeler, D.A.; Gibbs, R.A.; et al. A SNP discovery method to assess variant allele probability from next-generation resequencing data. Genome Res. 2010, 20, 273–280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yasukochi, Y.; Sakuma, J.; Takeuchi, I.; Kato, K.; Oguri, M.; Fujimaki, T.; Horibe, H.; Yamada, Y. Two novel susceptibility loci for type 2 diabetes mellitus identified by longitudinal exome-wide association studies in a Japanese population. Genomics 2019, 111, 34–42. [Google Scholar] [CrossRef]
- Roy, J.; Mallick, B. Altered gene expression in late-onset Alzheimer’s disease due to SNPs within 3′UTR microRNA response elements. Genomics 2017, 109, 177–185. [Google Scholar] [CrossRef]
- Sato, M.; Soma, M.; Nakayama, T.; Kanmatsuse, K. Dopamine D1 receptor gene polymorphism is associated with essential hypertension. Hypertension 2000, 36, 183–186. [Google Scholar] [CrossRef] [Green Version]
- Koptides, M.; Mean, R.; Stavrou, C.; Pierides, A.; Demetriou, K.; Nakayama, T.; Hildebrandt, F.; Fuchshuber, A.; Deltas, C.C. Novel NPR1 polymorphic variants and its exclusion as a candidate gene for medullary cystic kidney disease (ADMCKD) type 1. Mol. Cell. Probes 2001, 15, 357–361. [Google Scholar] [CrossRef]
- Nakayama, T.; Asai, S.; Sato, N.; Soma, M. Genotype and haplotype association study of the STRK1 region on 5q12 among Japanese: A case-control study. Stroke 2006, 37, 69–76. [Google Scholar] [CrossRef] [Green Version]
- Brænne, I.; Zeng, L.; Willenborg, C.; Tragante, V.; Kessler, T.; Consortium, C.; Consortium, C.; Willer, C.J.; Laakso, M.; Wallentin, L.; et al. Genomic correlates of glatiramer acetate adverse cardiovascular effects lead to a novel locus mediating coronary risk. PLoS ONE 2017, 12, e0182999. [Google Scholar] [CrossRef] [Green Version]
- Rusu, V. Hoch type 2 diabetes variants disrupt function of SLC16A11 through two distinct mechanisms. Cell 2017, 170, 199–212. [Google Scholar] [CrossRef]
- Maher, B. Personal genomes: The case of the missing heritability. Nature 2008, 456, 18–21. [Google Scholar] [CrossRef] [PubMed]
- Toma, T.T.; Williams, Z.; Dawson, J.; Adjeroh, D. What Can One Chromosome Tell us about human Biogeographical Ancestry? In Proceedings of the 2017 IEEE International Conference on Bioinformatics and Biomedicine (BIBM), Kansas City, MO, USA, 13–16 November 2017; pp. 188–193. [Google Scholar]
- He, J.; Zelikovsky, A. Informative SNP selection methods based on SNP prediction. IEEE Trans. Nanobiosci. 2007, 6, 60–67. [Google Scholar] [CrossRef] [PubMed]
- LaFramboise, T. Single nucleotide polymorphism arrays: A decade of biological, computational and technological advances. Nucleic Acids Res. 2009, 37, 4181–4193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kwok, P.Y.; Chen, X. Detection of single nucleotide polymorphisms. Curr. Issues Mol. Biol. 2003, 5, 43–60. [Google Scholar]
- Talukder, A.K.; Gandham, S.; Prahalad, H.A.; Bhattacharyya, N.P. Cloud-MAQ: The Cloud-enabled Scalable Whole Genome Reference Assembly Application. In Proceedings of the 2010 Seventh International Conference on Wireless and Optical Communications Networks-(WOCN), Colombo, Sri Lanka, 25 May 2010; pp. 0–4. [Google Scholar]
- Li, R.; Li, Y.; Fang, X.; Yang, H.; Wang, J.J.; Kristiansen, K.; Wang, J.J. SNP detection for massively parallel whole-genome resequencing-ppt. Genome Res. 2009, 19, 1124–1132. [Google Scholar] [CrossRef] [Green Version]
- Cheng, Y.H.; Kuo, C.N.; Lai, C.M. Effective natural PCR-RFLP primer design for SNP genotyping using teaching-learning-based optimization with elite strategy. IEEE Trans. Nanobioscience 2016, 15, 657–665. [Google Scholar] [CrossRef]
- Hall, J.G.; Eis, P.S.; Law, S.M.; Reynaldo, L.P.; Prudent, J.R.; Marshall, D.J.; Allawi, H.T.; Mast, A.L.; Dahlberg, J.E.; Kwiatkowski, R.W.; et al. Sensitive detection of DNA polymorphisms by the serial invasive signal amplification reaction. Proc. Natl. Acad. Sci. USA 2000, 97, 8272–8277. [Google Scholar] [CrossRef] [Green Version]
- Schmalzing, D.; Belenky, A.; Novotny, M.A.; Koutny, L.; Salas-Solano, O.; El-Difrawy, S.; Adourian, A.; Matsudaira, P.; Ehrlich, D. Microchip electrophoresis: A method for high-speed SNP detection. Nucleic Acids Res. 2000, 28, E43. [Google Scholar] [CrossRef]
- Ross, P.; Hall, L.; Smirnov, I.; Haff, L. High level multiplex genotyping by MALDI-TOF mass spectrometry. Nat. Biotechnol. 1998, 16, 1347. [Google Scholar] [CrossRef]
- Landegren, U.; Kaiser, R.; Sanders, J.; Hood, L. A ligase-mediated gene detection technique. Science 1988, 241, 1077–1080. [Google Scholar] [CrossRef]
- Wu, D.Y.; Wallace, R.B. The ligation amplification reaction (LAR)—Amplification of specific DNA sequences using sequential rounds of template-dependent ligation. Genomics 1989, 4, 560–569. [Google Scholar] [CrossRef]
- Tobe, V.O.; Taylor, S.L.; Nickerson, D.A. Single-well genotyping of diallelic sequence variations by a two-color ELISA-based oligonucleotide ligation assay. Nucleic Acids Res. 1996, 24, 3728–3732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huh, Y.S.; Lowe, A.J.; Strickland, A.D.; Batt, C.A.; Erickson, D. Surface-enhanced Raman scattering based ligase detection reaction. J. Am. Chem. Soc. 2009, 131, 2208–2213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Y.; Karalkar, N.B.; Kool, E.T. Nonenzymatic autoligation in direct three-color detection of RNA and DNA point mutations. Nat. Biotechnol. 2001, 19, 148–152. [Google Scholar] [CrossRef] [PubMed]
- Sando, S.; Abe, H.; Kool, E.T. Quenched auto-ligating dnas: Multicolor identification of nucleic acids at single nucleotide resolution. J. Am. Chem. Soc. 2004, 126, 1081–1087. [Google Scholar] [CrossRef]
- Wallace, R.B.; Shaffer, J.; Murphy, R.F.; Bonner, J.; Hirose, T.; Itakura, K. Hybridization of synthetic oligodeoxyribonucleotides to phi chi 174 DNA: The effect of single base pair mismatch. Nucleic Acids Res. 1979, 6, 3543–3557. [Google Scholar] [CrossRef]
- Liu, G.; Wan, Y.; Gau, V.; Zhang, J.; Wang, L.; Song, S.; Fan, C. An enzyme-based E-DNA sensor for sequence-specific detection of femtomolar DNA targets. J. Am. Chem. Soc. 2008, 130, 6820–6825. [Google Scholar] [CrossRef]
- Huang, Y.; Zhang, Y.-L.; Xu, X.; Jiang, J.-H.; Shen, G.-L.; Yu, R.-Q. Highly specific and sensitive electrochemical genotyping via gap ligation reaction and surface hybridization detection. J. Am. Chem. Soc. 2009, 131, 2478–2480. [Google Scholar] [CrossRef]
- Swami, N.; Chou, C.F.; Ramamurthy, V.; Chaurey, V. Enhancing DNA hybridization kinetics through constriction-based dielectrophoresis. Lab Chip 2009, 9, 3212–3220. [Google Scholar] [CrossRef]
- Kirmani, S.A.M.; Gudagunti, F.D.; Velmanickam, L.; Nawarathna, D.; Lima, I.T. Negative dielectrophoresis spectroscopy for rare analyte quantification in biological samples. J. Biomed. Opt. 2017, 22, 037006. [Google Scholar] [CrossRef] [Green Version]
- Gudagunti, F.D.; Velmanickam, L.; Nawarathna, D.; Lima, I.T. Label-free biosensing method for the detection of a pancreatic cancer biomarker based on dielectrophoresis spectroscopy. Chemosensors 2018, 6, 33. [Google Scholar] [CrossRef] [Green Version]
- Yahya, N.N.; Aziz, N.A.; Buyong, M.R.; Majlis, B.Y. Size-based particles separation utilizing dielectrophoresis technique. In Proceedings of the 2017 IEEE Regional Symposium on Micro and Nanoelectronics (RSM), Penang, Malaysia, 23–25 August 2017; pp. 10–13. [Google Scholar]
- Pethig, R. Dielectrophoresis: Status of the theory, technology, and applications. Biomicrofluidics 2010, 4, 1–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Viefhues, M.; Eichhorn, R. DNA dielectrophoresis: Theory and applications a review. Electrophoresis 2017, 38, 1483–1506. [Google Scholar] [CrossRef] [PubMed]
- Khoshmanesh, K.; Nahavandi, S.; Baratchi, S.; Mitchell, A.; Kalantar-zadeh, K. Dielectrophoretic platforms for bio-microfluidic systems. Biosens. Bioelectron. 2011, 26, 1800–1814. [Google Scholar] [CrossRef]
- Negr, F.; Cavallini, A. Effect of dielectrophoretic forces on nanoparticles. IEEE Trans. Dielectr. Electr. Insul. 2017, 24, 1708–1717. [Google Scholar] [CrossRef]
- Altinagac, E.; Ozcan, S.S.; Genc, Y.; Kizil, H.; Trabzon, L. Biological particle manipulation: An example of Jurkat enrichment. In Proceedings of the 2015 IEEE 10th International Conference NanoMicro Engineered and Molecular System NEMS 2015, Hong Kong, 7 November 2015; Volume 10, pp. 25–27. [Google Scholar]
- Camacho-alanis, F.; Ros, A. HHS public access. Bioanalysis 2015, 7, 353–371. [Google Scholar] [CrossRef] [Green Version]
- Yafouz, B.; Kadri, N.A.; Ibrahim, F. Dielectrophoretic manipulation and separation of microparticles using microarray dot electrodes. Sensors 2014, 14, 6356–6369. [Google Scholar] [CrossRef]
- Cui, L.; Holmes, D.; Morgan, H. The dielectrophoretic levitation and separation of latex beads in microchips. Electrophoresis 2001, 22, 3893–3901. [Google Scholar] [CrossRef]
- Green, N.G.; Morgan, H. Dielectrophoresis of submicrometer latex spheres. 1. experimental results. J. Phys. Chem. B 1999, 103, 41–50. [Google Scholar] [CrossRef] [Green Version]
- Weng, P.Y.; Chen, I.A.; Yeh, C.K.; Chen, P.Y.; Juang, J.Y. Size-dependent dielectrophoretic crossover frequency of spherical particles. Biomicrofluidics 2016, 10, 011909. [Google Scholar] [CrossRef] [Green Version]
- Chu, C.-H.; Sarangadharan, I.; Regmi, A.; Chen, Y.-W.; Hsu, C.-P.; Chang, W.-H.; Lee, G.-Y.; Chyi, J.-I.; Chen, C.-C.; Shiesh, S.-C.; et al. Beyond the Debye length in high ionic strength solution: Direct protein detection with field-effect transistors (FETs) in human serum. Sci. Rep. 2017, 7, 5256. [Google Scholar] [CrossRef] [PubMed]
- Green, N.M. Avidin. 3. The nature of the biotin-binding site. Biochem. J. 1963, 89, 599–609. [Google Scholar] [CrossRef] [PubMed]
- Rohani, A.; Moore, J.H.; Kashatus, J.A.; Sesaki, H.; Kashatus, D.F.; Swami, N.S. Label-free quantification of intracellular mitochondrial dynamics using dielectrophoresis. Anal. Chem. 2017, 89, 5757–5764. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Camacho-Alanis, F.; Ros, A. Protein dielectrophoresis and the link to dielectric properties. Bioanalysis 2015, 7, 353–371. [Google Scholar] [CrossRef] [PubMed] [Green Version]





© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gudagunti, F.D.; Velmanickam, L.; Nawarathna, D.; Lima, I.T., Jr. Nucleotide Identification in DNA Using Dielectrophoresis Spectroscopy. Micromachines 2020, 11, 39. https://doi.org/10.3390/mi11010039
Gudagunti FD, Velmanickam L, Nawarathna D, Lima IT Jr. Nucleotide Identification in DNA Using Dielectrophoresis Spectroscopy. Micromachines. 2020; 11(1):39. https://doi.org/10.3390/mi11010039
Chicago/Turabian StyleGudagunti, Fleming Dackson, Logeeshan Velmanickam, Dharmakeerthi Nawarathna, and Ivan T. Lima, Jr. 2020. "Nucleotide Identification in DNA Using Dielectrophoresis Spectroscopy" Micromachines 11, no. 1: 39. https://doi.org/10.3390/mi11010039





