Rational PCR Reactor Design in Microfluidics
Abstract
:1. Introduction
2. Design Parameters for POC RT-qPCR
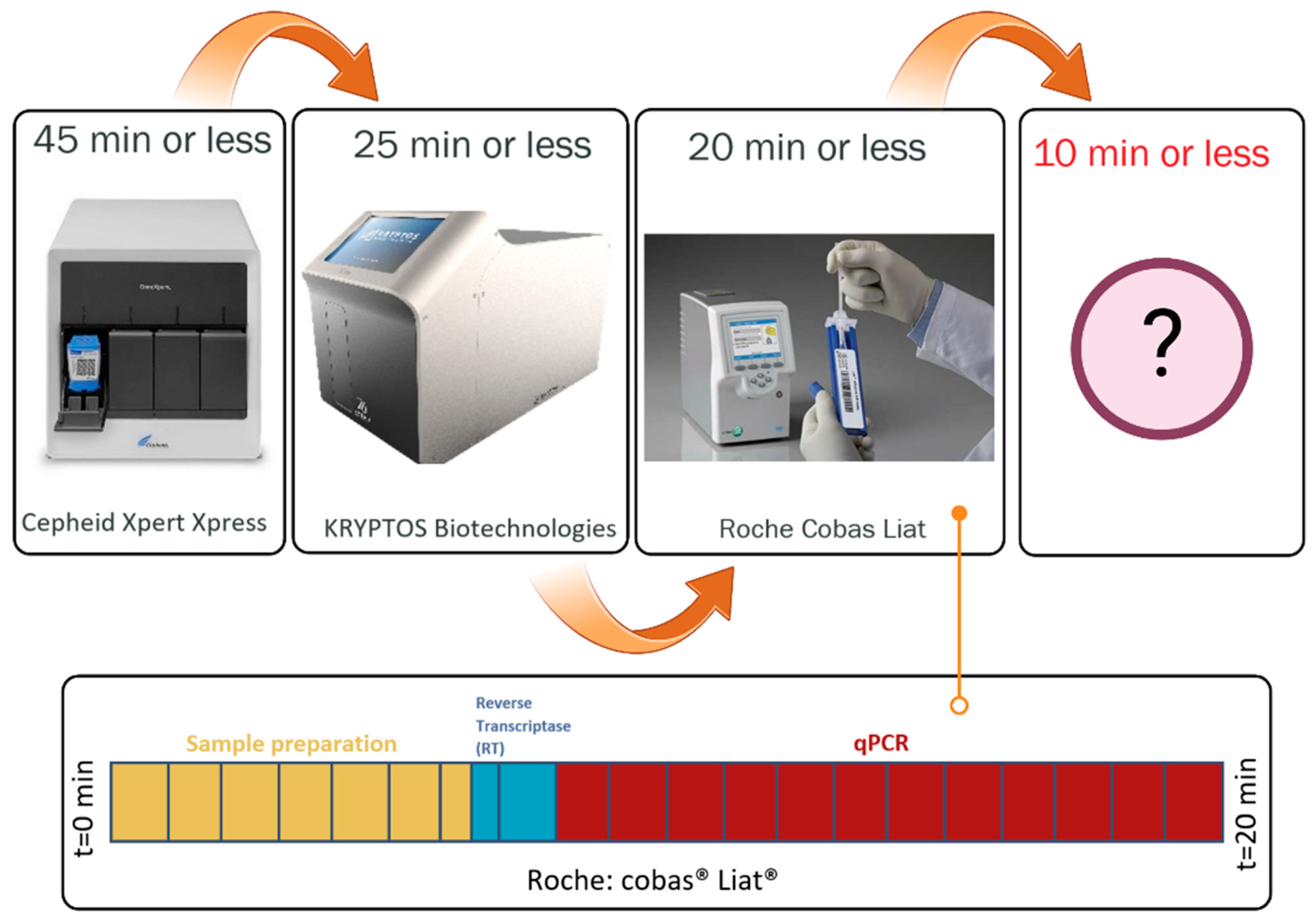
2.1. Sample to Answer Time (t)
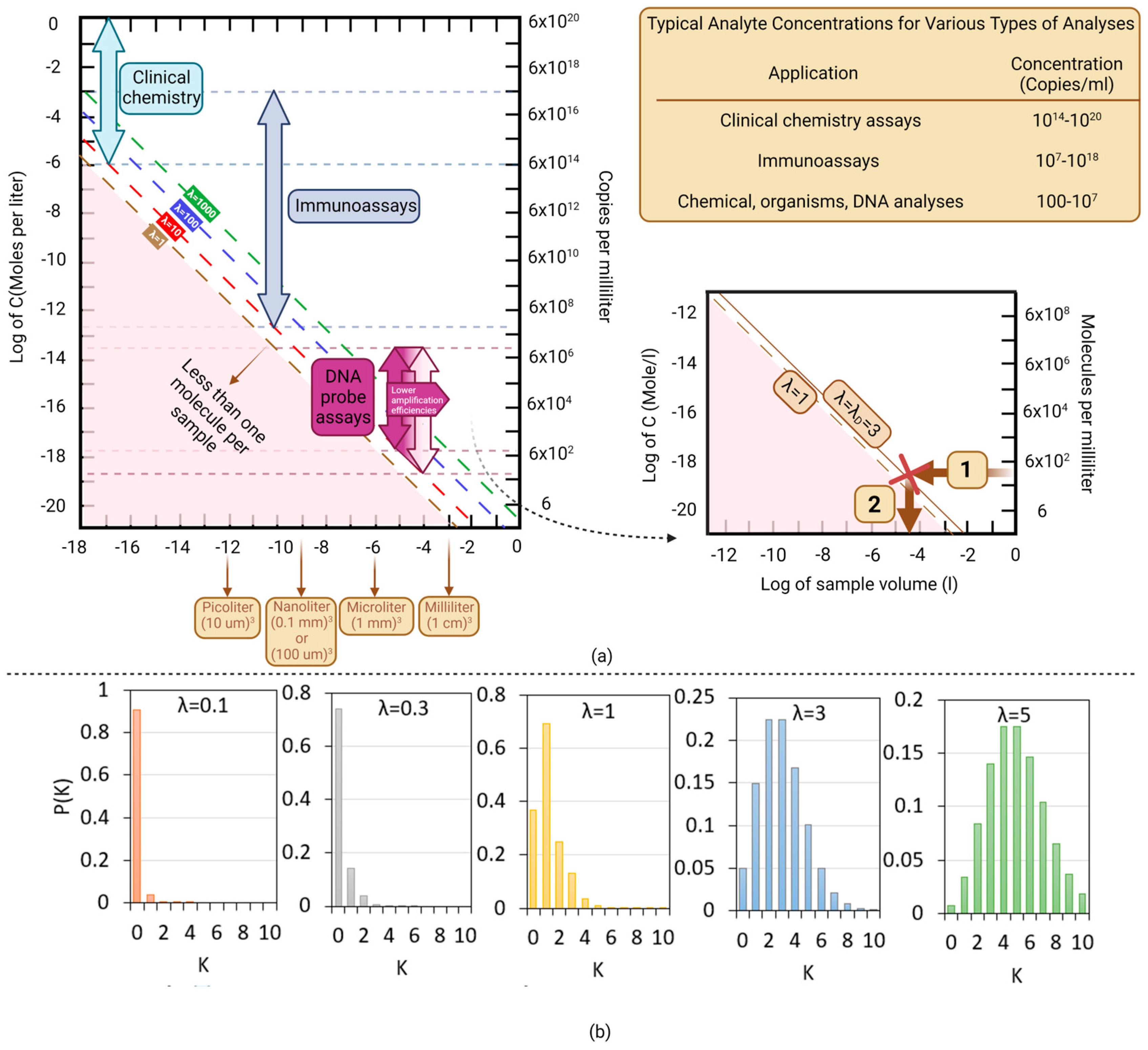
2.2. Limit of Detection–Minimal Number of Copies per Volume (λ)
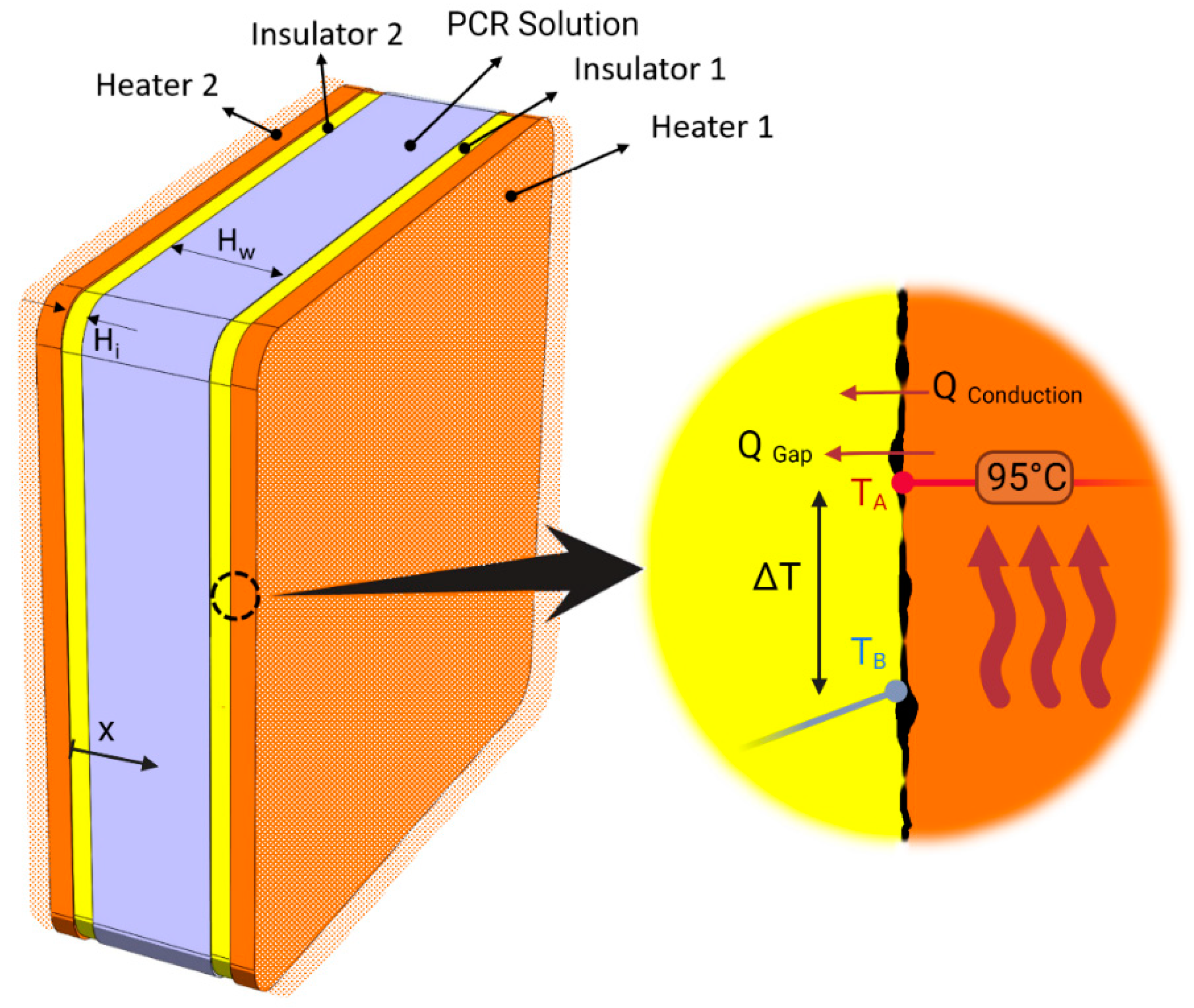
2.3. System Thermal Efficiency γ
3. Numerical Simulation
3.1. Governing Equations and Boundary Conditions
3.2. Geometry and Dimensions
3.3. Solving Method
3.4. Results and Discussions
4. Candidate PCR Reactors in the Market or under Development
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Müller, K.; Daßen, S.; Holowachuk, S.; Zwirglmaier, K.; Stehr, J.; Buersgens, F.; Ullerich, L.; Stoecker, K. Pulse-Controlled Amplification–a new powerful tool for front-line diagnostics. medRxiv 2020. [Google Scholar] [CrossRef]
- Available online: https://lexdiagnostics.com/ (accessed on 1 July 2023).
- Available online: https://www.ttp.com/case-studies/amplifying-dna-as-fast-as-the-biology-permits/ (accessed on 1 July 2023).
- Rychlik, W.; Spencer, W.J.; Rhoads, R.E. Optimization of the annealing temperature for DNA amplification in vitro. Nucleic Acids Res. 1990, 18, 6409–6412. [Google Scholar] [CrossRef] [Green Version]
- Najafov, A.; Hoxhaj, G. PCR Guru: An Ultimate Benchtop Reference for Molecular Biologists; Academic Press: Cambridge, MA, USA, 2016; ISBN 012804246X. [Google Scholar]
- Ferré, F. Gene Quantification; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2012; ISBN 1461241642. [Google Scholar]
- Cha, R.S.; Thilly, W.G. Specificity, efficiency, and fidelity of PCR. Genome Res. 1993, 3, S18–S29. [Google Scholar] [CrossRef]
- Hwu, A.T.; Madadelahi, M.; Nakajima, R.; Shamloo, E.; Perebikovsky, A.; Kido, H.; Jain, A.; Jasinskas, A.; Prange, S.; Felgner, P. Centrifugal disc liquid reciprocation flow considerations for antibody binding to COVID antigen array during microfluidic integration. Lab Chip 2022, 22, 2695–2706. [Google Scholar] [CrossRef]
- Kordzadeh-Kermani, V.; Dartoomi, H.; Azizi, M.; Ashrafizadeh, S.N.; Madadelahi, M. Investigating the Performance of the Multi-Lobed Leaf-Shaped Oscillatory Obstacles in Micromixers Using Bulk Acoustic Waves (BAW): Mixing and Chemical Reaction. Micromachines 2023, 14, 795. [Google Scholar] [CrossRef]
- Khandurina, J.; McKnight, T.E.; Jacobson, S.C.; Waters, L.C.; Foote, R.S.; Ramsey, J.M. Integrated system for rapid PCR-based DNA analysis in microfluidic devices. Anal. Chem. 2000, 72, 2995–3000. [Google Scholar] [CrossRef]
- Kordzadeh-Kermani, V.; Madadelahi, M.; Ashrafizadeh, S.N.; Kulinsky, L.; Martinez-Chapa, S.O.; Madou, M.J. Electrified lab on disc systems: A comprehensive review on electrokinetic applications. Biosens. Bioelectron. 2022, 114381. [Google Scholar] [CrossRef]
- Neuzil, P.; Zhang, C.; Pipper, J.; Oh, S.; Zhuo, L. Ultra fast miniaturized real-time PCR: 40 cycles in less than six minutes. Nucleic Acids Res. 2006, 34, e77. [Google Scholar] [CrossRef]
- Yin, H.; Wu, Z.; Shi, N.; Qi, Y.; Jian, X.; Zhou, L.; Tong, Y.; Cheng, Z.; Zhao, J.; Mao, H. Ultrafast multiplexed detection of SARS-CoV-2 RNA using a rapid droplet digital PCR system. Biosens. Bioelectron. 2021, 188, 113282. [Google Scholar] [CrossRef]
- Available online: https://www.businessinsider.com/ (accessed on 1 July 2023).
- Available online: https://www.staffcare.com/ (accessed on 1 July 2023).
- Pardee, K.; Green, A.A.; Takahashi, M.K.; Braff, D.; Lambert, G.; Lee, J.W.; Ferrante, T.; Ma, D.; Donghia, N.; Fan, M. Rapid, low-cost detection of Zika virus using programmable biomolecular components. Cell 2016, 165, 1255–1266. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.-N.; Ko, Y.J.; Seong, M.-W.; Kim, J.-S.; Shin, B.-M.; Sung, H. Analytical and clinical validation of six commercial Middle East Respiratory Syndrome coronavirus RNA detection kits based on real-time reverse-transcription PCR. Ann. Lab. Med. 2016, 36, 450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Draz, M.S.; Shafiee, H. Applications of gold nanoparticles in virus detection. Theranostics 2018, 8, 1985. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.; Duong Bang, D.; Wolff, A. 2019 novel coronavirus disease (COVID-19): Paving the road for rapid detection and point-of-care diagnostics. Micromachines 2020, 11, 306. [Google Scholar] [CrossRef] [Green Version]
- Lu, X.; Whitaker, B.; Sakthivel, S.K.K.; Kamili, S.; Rose, L.E.; Lowe, L.; Mohareb, E.; Elassal, E.M.; Al-sanouri, T.; Haddadin, A. Real-time reverse transcription-PCR assay panel for Middle East respiratory syndrome coronavirus. J. Clin. Microbiol. 2014, 52, 67–75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, C.; Cui, J.; Huang, L.; Du, B.; Chen, L.; Xue, G.; Li, S.; Zhang, W.; Zhao, L.; Sun, Y. Rapid and visual detection of 2019 novel coronavirus (SARS-CoV-2) by a reverse transcription loop-mediated isothermal amplification assay. Clin. Microbiol. Infect. 2020, 26, 773–779. [Google Scholar] [CrossRef]
- Available online: https://diagnostics.roche.com/ (accessed on 1 July 2023).
- Arnaout, R.; Lee, R.A.; Lee, G.R.; Callahan, C.; Yen, C.F.; Smith, K.P.; Arora, R.; Kirby, J.E. SARS-CoV2 testing: The limit of detection matters. bioRxiv 2020. [Google Scholar] [CrossRef]
- Tan, L.L.; Loganathan, N.; Agarwalla, S.; Yang, C.; Yuan, W.; Zeng, J.; Wu, R.; Wang, W.; Duraiswamy, S. Current commercial dPCR platforms: Technology and market review. Crit. Rev. Biotechnol. 2023, 43, 433–464. [Google Scholar] [CrossRef]
- Goldberg, S. Probability: An Introduction; Courier Corporation: North Chelmsford, MA, USA, 1986; ISBN 0486652521. [Google Scholar]
- Allen, J.J. Micro Electro Mechanical System Design; CRC Press: Boca Raton, FL, USA, 2005; ISBN 0429116861. [Google Scholar]
- Millington, A.L.; Houskeeper, J.A.; Quackenbush, J.F.; Trauba, J.M.; Wittwer, C.T. The kinetic requirements of extreme qPCR. Biomol. Detect. Quantif. 2019, 17, 100081. [Google Scholar] [CrossRef]
- Yin, I.X.; Zhang, J.; Zhao, I.S.; Mei, M.L.; Li, Q.; Chu, C.H. The antibacterial mechanism of silver nanoparticles and its application in dentistry. Int. J. Nanomed. 2020, 15, 2555–2562. [Google Scholar] [CrossRef] [Green Version]
- Innis, M.A.; Myambo, K.B.; Gelfand, D.H.; Brow, M.A. DNA sequencing with Thermus aquaticus DNA polymerase and direct sequencing of polymerase chain reaction-amplified DNA. Proc. Natl. Acad. Sci. USA 1988, 85, 9436–9440. [Google Scholar] [CrossRef]
- Farrar, J.S.; Wittwer, C.T. Extreme PCR: Efficient and specific DNA amplification in 15–60 seconds. Clin. Chem. 2015, 61, 145–153. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Liu, L.; Tu, Y.; Zhang, J.; Miao, G.; Zhang, L.; Ge, S.; Xia, N.; Yu, D.; Qiu, X. Rapid PCR powered by microfluidics: A quick review under the background of COVID-19 pandemic. TrAC Trends Anal. Chem. 2021, 143, 116377. [Google Scholar] [CrossRef] [PubMed]
- Shi, L.; Chew, M.Y.L. Fire behaviors of polymers under autoignition conditions in a cone calorimeter. Fire Saf. J. 2013, 61, 243–253. [Google Scholar] [CrossRef]
- Brandrup, J.; Immergut, E.H.; Grulke, E.A.; Abe, A.; Bloch, D.R. (Eds.) Polymer Data Handbook; Wiley: New York, NY, USA, 1999. [Google Scholar]
- Mallegni, N.; Milazzo, M.; Cristallini, C.; Barbani, N.; Fredi, G.; Dorigato, A.; Cinelli, P.; Danti, S. Characterization of Cyclic Olefin Copolymers for Insulin Reservoir in an Artificial Pancreas. J. Funct. Biomater. 2023, 14, 145. [Google Scholar] [CrossRef] [PubMed]
- Weidenfeller, B.; Höfer, M.; Schilling, F.R. Thermal conductivity, thermal diffusivity, and specific heat capacity of particle filled polypropylene. Compos. Part A Appl. Sci. Manuf. 2004, 35, 423–429. [Google Scholar] [CrossRef]
- Bergman, T.L.; Bergman, T.L.; Incropera, F.P.; Dewitt, D.P.; Lavine, A.S. Fundamentals of Heat and Mass Transfer; John Wiley & Sons: Hoboken, NJ, USA, 2011; ISBN 0470501979. [Google Scholar]
- Northrup, M.A.; Benett, B.; Hadley, D.; Landre, P.; Lehew, S.; Richards, J.; Stratton, P. A miniature analytical instrument for nucleic acids based on micromachined silicon reaction chambers. Anal. Chem. 1998, 70, 918–922. [Google Scholar] [CrossRef]
- Gibbins, J. Thermal Contact Resistance of Polymer Interfaces. Master’s Thesis, University of Waterloo, Waterloo, ON, Canada, 2006. [Google Scholar]
- Saad, Y. Iterative Methods for Sparse Linear Systems; SIAM: Philadelphia, PA, USA, 2003; ISBN 0898715342. [Google Scholar]
- Madadelahi, M.; Azimi-Boulali, J.; Madou, M.; Martinez-Chapa, S. Characterization of Fluidic-Barrier-Based Particle Generation in Centrifugal Microfluidics. Micromachines 2022, 13, 881. [Google Scholar] [CrossRef]
- Madadelahi, M.; Madou, M.J.; Nokoorani, Y.D.; Shamloo, A.; Martinez-Chapa, S.O. Fluidic barriers in droplet-based centrifugal microfluidics: Generation of multiple emulsions and microspheres. Sens. Actuators B Chem. 2020, 311, 127833. [Google Scholar] [CrossRef]
- Available online: https://www.nextgenpcr.com/ (accessed on 1 July 2023).
- Available online: https://www.selectscience.net/products/xxpress-thermal-cycler/?prodID=113721#tab-2 (accessed on 1 July 2023).
- Houssin, T.; Cramer, J.; Grojsman, R.; Bellahsene, L.; Colas, G.; Moulet, H.; Minnella, W.; Pannetier, C.; Leberre, M.; Plecis, A. Ultrafast, sensitive and large-volume on-chip real-time PCR for the molecular diagnosis of bacterial and viral infections. Lab Chip 2016, 16, 1401–1411. [Google Scholar] [CrossRef]
- An, Y.-Q.; Huang, S.-L.; Xi, B.-C.; Gong, X.-L.; Ji, J.-H.; Hu, Y.; Ding, Y.-J.; Zhang, D.-X.; Ge, S.-X.; Zhang, J. Ultrafast Microfluidic PCR Thermocycler for Nucleic Acid Amplification. Micromachines 2023, 14, 658. [Google Scholar] [CrossRef]




| Material | Thermal Conductivity (W/m·K) | Density (kg/m3) | Specific Thermal Capacity (kJ/kg K) | Thermal Diffusivity (m2/s) | Ref. | |
|---|---|---|---|---|---|---|
| Solids | PMMA | 0.21 | 1202.9 | 1.46 | 1.19 × 10−7 | [32] |
| PDMS | 0.15 | 970 | 1.46 | 1.06 × 10−7 | [33] | |
| PC | 0.2 | 1226.7 | 1.22 | 1.34 × 10−7 | [32] | |
| COC 8007 | Between 0.12 and 0.15 | 1020 | 1.2 | 1.17 × 10−7 | [34] | |
| PP | 0.22 | 900 | 1.330 to 2.400 | 0.961 × 10−7 | [35] | |
| Silicon | 148 (3.2 to 150) | 2330 | 0.712 | 9 × 10−5 | [36,37] | |
| Teflon | 0.35 | 2200 | 0.97 | 1.64 × 10−7 | [36] | |
| Glass | 1.4 | 2500 | 0.75 | 7.46 × 10−7 | [36] | |
| Liquid | Water | 0.613 | 997 | 4.17 | 1.47 × 10−7 | [36] |
| Oil | 0.131 | 837 | 1.67 | 7.85 × 10−8 | [36] |
| Type of System | Device/Company | Time to Result | Characteristics | Thermal Contact Resistance | γ = Time/Volume |
|---|---|---|---|---|---|
| Plate system, not an integrated system, up to 96 wells |  NextGenPCR® MBS Molecular Biology Systems (Goes, The Netherlands) | 2 min without sample preparation |
| Low (squeezing pressure) | 2 min/20 µL = 0.1 |
| Integrated POC system |  Lex Diagnostics (Melbourn, UK) | Sample to answer of 5 min and thermal cycling time of less than 3 min |
| Very low (engineered permanent contact) | 3 min/5 µL = 0.6 With heater on both sides: 3 min/10 µL = 0.3 |
| Plate system, not an integrated system, up to 96 wells |  XXPress BJS Technologies (Duluth, GA, USA) | 10 min without sample preparation |
| Lowest (engineered permanent contact) | 10 min/40 µL = 0.4 |
| Integrated POC system |  Roche’s Cobas Liat (Basel, Switzerland) | 20 min with sample preparation, 12 min for PCR |
| Low (squeezing pressure) | 12 min/80 µL = 0.15 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Madadelahi, M.; Madou, M.J. Rational PCR Reactor Design in Microfluidics. Micromachines 2023, 14, 1533. https://doi.org/10.3390/mi14081533
Madadelahi M, Madou MJ. Rational PCR Reactor Design in Microfluidics. Micromachines. 2023; 14(8):1533. https://doi.org/10.3390/mi14081533
Chicago/Turabian StyleMadadelahi, Masoud, and Marc J. Madou. 2023. "Rational PCR Reactor Design in Microfluidics" Micromachines 14, no. 8: 1533. https://doi.org/10.3390/mi14081533





