Microfluidic Microcirculation Mimetic for Exploring Biophysical Mechanisms of Chemotherapy-Induced Metastasis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microfluidics Microcirculation Mimetic, MMM
2.2. Cell Culture
2.3. Drug Treatments
2.4. Fluorometric Morphometry
2.5. Statistical Analysis
3. Results
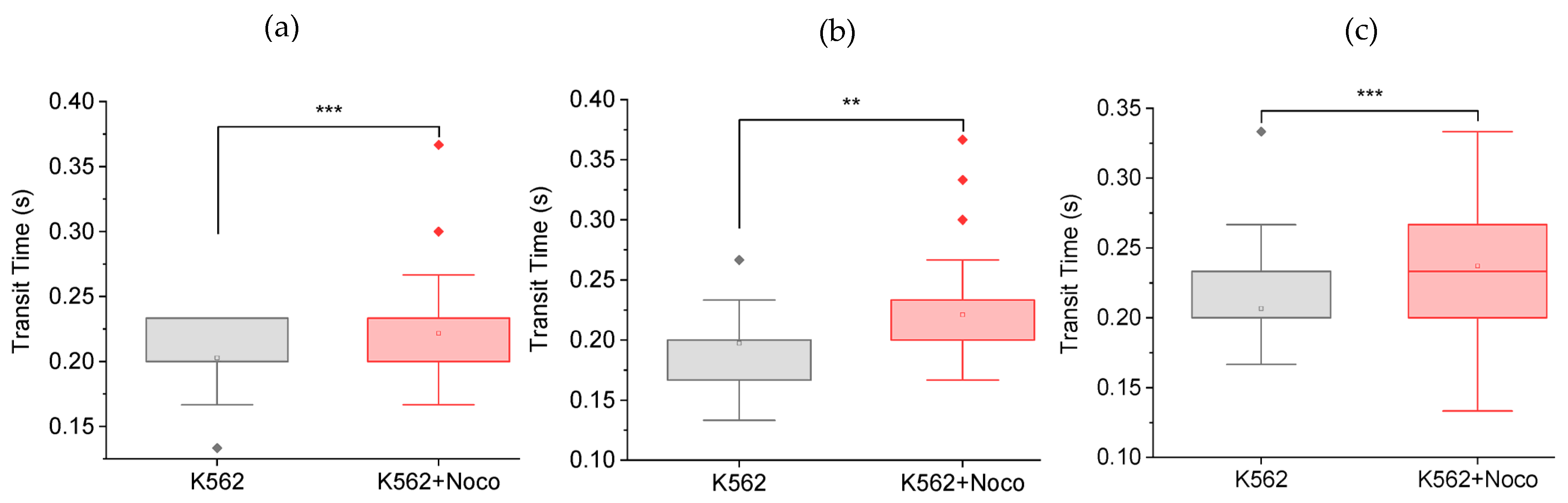
3.1. Nocodazole Increases Cell Transit Time
3.2. Feasibility of Exploring Biophyical Mechanisms
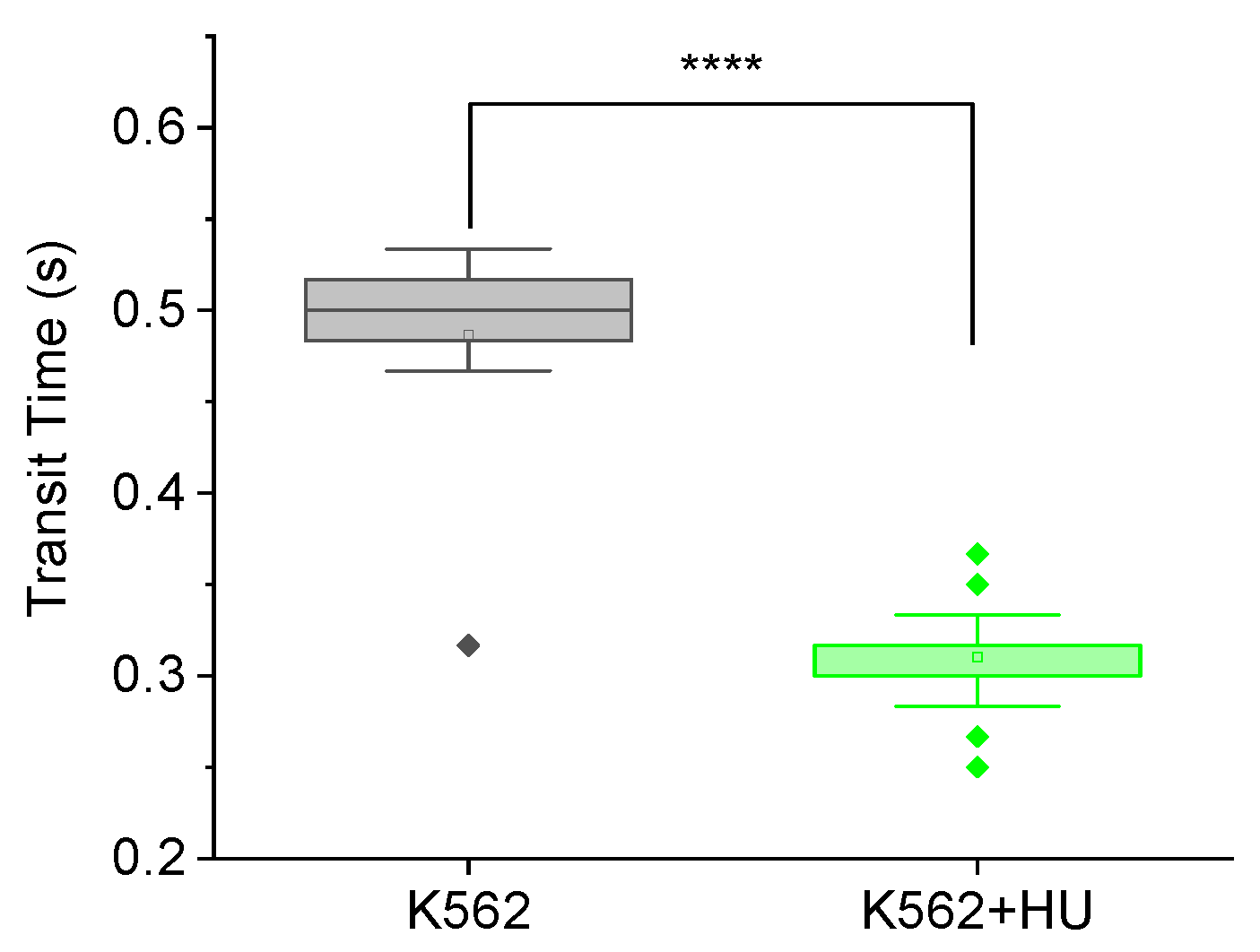
3.2.1. Feasibility Testing Using Hydroxyurea
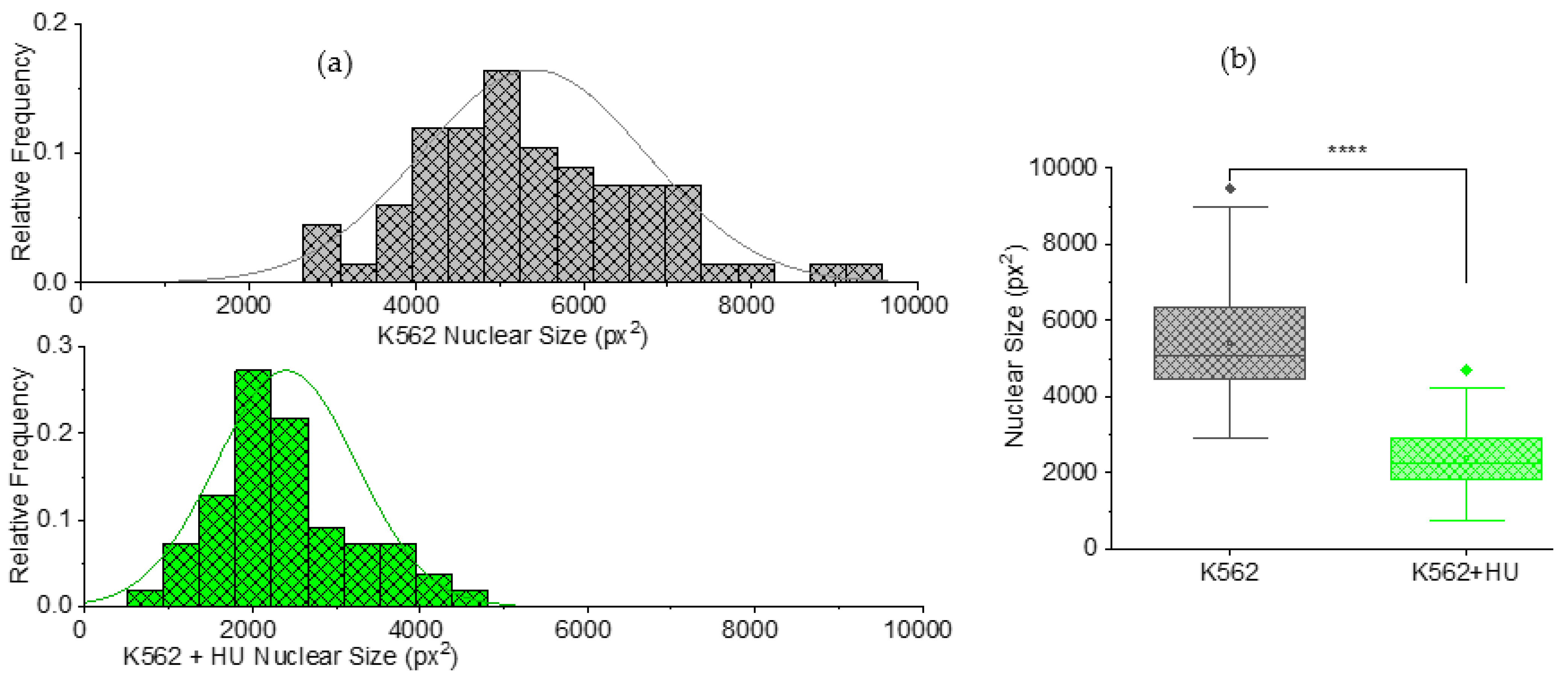
3.2.2. Size Effects: Independent Morphometry Tests
3.3. Important Caveat: Unstable Framerates Can Lead to Inconsistent Results
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Mehlen, P.; Puisieux, A. Metastasis: A question of life or death. Nat. Rev. Cancer 2006, 6, 449–458. [Google Scholar] [CrossRef]
- Seyfried, T.N.; Huysentruyt, L.C. On the origin of cancer metastasis. Crit. Rev. Oncog. 2013, 18, 43–73. [Google Scholar] [CrossRef]
- Steeg, P.S. Tumor metastasis: Mechanistic insights and clinical challenges. Nat. Med. 2006, 12, 895–904. [Google Scholar] [CrossRef]
- Fouad, Y.A.; Aanei, C. Revisiting the hallmarks of cancer. Am. J. Cancer Res. 2017, 7, 1016–1036. [Google Scholar]
- Ran, S. The role of TLR4 in chemotherapy-driven metastasis. Cancer Res. 2015, 75, 2405–2410. [Google Scholar] [CrossRef]
- Weber, G.F. Why does cancer therapy lack effective anti-metastasis drugs? Cancer Lett. 2013, 328, 207–211. [Google Scholar] [CrossRef]
- Volk-Draper, L.; Hall, K.; Griggs, C.; Rajput, S.; Kohio, P.; DeNardo, D.; Ran, S. Paclitaxel therapy promotes breast cancer metastasis in a TLR4-dependent manner. Cancer Res. 2014, 74, 5421–5434. [Google Scholar] [CrossRef]
- Geldof, A.A.; Rao, B.R. Doxorubicin treatment increases metastasis of prostate tumor (R3327-MatLyLu). Anticancer Res. 1988, 8, 1335–1339. [Google Scholar]
- Suresh, S. Biomechanics and biophysics of cancer cells. Acta Biomater. 2007, 3, 413–438. [Google Scholar] [CrossRef]
- Moore, N.M.; Nagahara, L.A. Physical biology in cancer. 1. Cellular physics of cancer metastasis. Am. J. Physiol. Cell Physiol. 2014, 306, C78–C79. [Google Scholar] [CrossRef]
- Man, S.M.; Ekpenyong, A.; Tourlomousis, P.; Achouri, S.; Cammarota, E.; Hughes, K.; Rizzo, A.; Ng, G.; Wright, J.A.; Cicuta, P.; et al. Actin polymerization as a key innate immune effector mechanism to control Salmonella infection. Proc. Natl. Acad. Sci. USA 2014, 111, 17588–17593. [Google Scholar] [CrossRef] [PubMed]
- Prathivadhi-Bhayankaram, S.V.; Ning, J.; Mimlitz, M.; Taylor, C.; Gross, E.; Nichols, M.; Guck, J.; Ekpenyong, A.E. Chemotherapy impedes in vitro microcirculation and promotes migration of leukemic cells with impact on metastasis. Biochem. Biophys. Res. Commun. 2016, 479, 841–846. [Google Scholar] [CrossRef] [PubMed]
- Lam, W.A.; Rosenbluth, M.J.; Fletcher, D.A. Chemotherapy exposure increases leukemia cell stiffness. Blood 2007, 109, 3505–3508. [Google Scholar] [CrossRef] [PubMed]
- Karagiannis, G.S.; Pastoriza, J.M.; Wang, Y.; Harney, A.S.; Entenberg, D.; Pignatelli, J.; Sharma, V.P.; Xue, E.A.; Cheng, E.; D’Alfonso, T.M.; et al. Neoadjuvant chemotherapy induces breast cancer metastasis through a TMEM-mediated mechanism. Sci. Transl. Med. 2017, 9, eaan0026. [Google Scholar] [CrossRef] [PubMed]
- D’Alterio, C.; Scala, S.; Sozzi, G.; Roz, L.; Bertolini, G. Paradoxical effects of chemotherapy on tumor relapse and metastasis promotion. Semin. Cancer Biol. 2020, 60, 351–361. [Google Scholar] [CrossRef] [PubMed]
- Karagiannis, G.S.; Condeelis, J.S.; Oktay, M.H. Chemotherapy-Induced Metastasis: Molecular Mechanisms, Clinical Manifestations, Therapeutic Interventions. Cancer Res. 2019, 79, 4567–4576. [Google Scholar] [CrossRef]
- Ortiz-Otero, N.; Marshall, J.R.; Lash, B.; King, M.R. Chemotherapy-induced release of circulating-tumor cells into the bloodstream in collective migration units with cancer-associated fibroblasts in metastatic cancer patients. BMC Cancer 2020, 20, 873. [Google Scholar] [CrossRef]
- Bellomo, G.; Rainer, C.; Quaranta, V.; Astuti, Y.; Raymant, M.; Boyd, E.; Stafferton, R.; Campbell, F.; Ghaneh, P.; Halloran, C.M.; et al. Chemotherapy-induced infiltration of neutrophils promotes pancreatic cancer metastasis via Gas6/AXL signalling axis. Gut 2022, 71, 2284–2299. [Google Scholar] [CrossRef]
- Mansouri, N.; Keklikoglou, I.; Nassiri, S.; Torchia, B.; Guichard, A.; De Palma, M. Role of extracellular vesicles in chemotherapy-induced lung metastasis. Eur. Respir. J. 2020, 56, 3944. [Google Scholar]
- Zarfati, A.; Martucci, C.; Crocoli, A.; Serra, A.; Persano, G.; Inserra, A. Chemotherapy-induced cavitating Wilms’ tumor pulmonary metastasis: Active disease or scarring? A case report and literature review. Front. Pediatr. 2023, 11, 1083168. [Google Scholar] [CrossRef]
- Monteran, L.; Ershaid, N.; Doron, H.; Zait, Y.; Scharff, Y.; Ben-Yosef, S.; Avivi, C.; Barshack, I.; Sonnenblick, A.; Erez, N. Chemotherapy-induced complement signaling modulates immunosuppression and metastatic relapse in breast cancer. Nat. Commun. 2022, 13, 5797. [Google Scholar] [CrossRef] [PubMed]
- Su, J.X.; Li, S.J.; Zhou, X.F.; Zhang, Z.J.; Yan, Y.; Liu, S.L.; Qi, Q. Chemotherapy-induced metastasis: Molecular mechanisms and clinical therapies. Acta Pharmacol. Sin. 2023, 1–12. [Google Scholar] [CrossRef]
- Li, Q.; Zhao, Y.H.; Xu, C.; Liang, Y.L.; Zhao, Y.; He, Q.M.; Li, J.Y.; Chen, K.L.; Qiao, H.; Liu, N.; et al. Chemotherapy-Induced Senescence Reprogramming Promotes Nasopharyngeal Carcinoma Metastasis by circRNA-Mediated PKR Activation. Adv. Sci. 2023, 10, 2205668. [Google Scholar] [CrossRef]
- Zhu, C.; Liu, P.; Li, C.Y.; Zhang, Y.; Yin, J.; Hou, L.; Zheng, G.; Liu, X. Near-Death Cells Cause Chemotherapy-Induced Metastasis via ATF4-Mediated NF-κB Signaling Activation. Adv. Sci. 2023, 10, 2205835. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Akinade, T.; Zhou, J.; Wang, H.; Tong, Q.; He, S.; Rinebold, E.; Salazar, L.E.V.; Bhansali, D.; Zhong, Y.; et al. Therapeutic Nanocarriers Inhibit Chemotherapy-Induced Breast Cancer Metastasis. Adv. Sci. 2022, 9, 2203949. [Google Scholar] [CrossRef]
- Wirtz, D.; Konstantopoulos, K.; Searson, P.C. The physics of cancer: The role of physical interactions and mechanical forces in metastasis. Nat. Rev. Cancer 2011, 11, 512–522. [Google Scholar] [CrossRef]
- Gensbittel, V.; Kräter, M.; Harlepp, S.; Busnelli, I.; Guck, J.; Goetz, J.G. Mechanical Adaptability of Tumor Cells in Metastasis. Dev. Cell 2021, 56, 164–179. [Google Scholar] [CrossRef]
- Mierke, C.T. Mechanical Cues Affect Migration and Invasion of Cells from Three Different Directions. Front. Cell Dev. Biol. 2020, 8, 946. [Google Scholar]
- Vasquez, R.J.; Howell, B.; Yvon, A.M.C.; Wadsworth, P.; Cassimeris, L. Nanomolar concentrations of nocodazole alter microtubule dynamic instability in vivo and in vitro. Mol. Biol. Cell 1997, 8, 973–985. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef]
- Liu, Y.-N.; Kang, B.-B.; Chen, J.H. Transcriptional regulation of human osteopontin promoter by C/EBPalpha and AML-1 in metastatic cancer cells. Oncogene 2004, 23, 278–288. [Google Scholar] [CrossRef] [PubMed]
- Wu, P.-H.; Aroush, D.R.-B.; Asnacios, A.; Chen, W.-C.; Dokukin, M.E.; Doss, B.L.; Durand-Smet, P.; Ekpenyong, A.; Guck, J.; Guz, N.V.; et al. A comparison of methods to assess cell mechanical properties. Nat. Methods 2018, 15, 491–498. [Google Scholar] [CrossRef] [PubMed]
- Otto, O.; Rosendahl, P.; Mietke, A.; Golfier, S.; Herold, C.; Klaue, D.; Girardo, S.; Pagliara, S.; Ekpenyong, A.; Jacobi, A.; et al. Real-time deformability cytometry: On-the-fly cell mechanical phenotyping. Nat. Methods 2015, 12, 199–202. [Google Scholar] [CrossRef]
- Tse, H.T.K.; Gossett, D.R.; Moon, Y.S.; Masaeli, M.; Sohsman, M.; Ying, Y.; Mislick, K.; Adams, R.P.; Rao, J.; Di Carlo, D. Quantitative diagnosis of malignant pleural effusions by single-cell mechanophenotyping. Sci. Transl. Med. 2013, 5, 212ra163. [Google Scholar] [CrossRef] [PubMed]
- Xue, C.; Wang, J.; Zhao, Y.; Chen, D.; Yue, W.; Chen, J. Constriction Channel Based Single-Cell Mechanical Property Characterization. Micromachines 2015, 6, 1794–1804. [Google Scholar] [CrossRef]
- Bento, D.; Rodrigues, R.O.; Faustino, V.; Pinho, D.; Fernandes, C.S.; Pereira, A.I.; Garcia, V.; Miranda, J.M.; Lima, R. Deformation of Red Blood Cells, Air Bubbles, and Droplets in Microfluidic Devices: Flow Visualizations and Measurements. Micromachines 2018, 9, 151. [Google Scholar] [CrossRef]
- Pinho, D.; Carvalho, V.; Gonçalves, I.M.; Teixeira, S.; Lima, R. Visualization and Measurements of Blood Cells Flowing in Microfluidic Systems and Blood Rheology: A Personalized Medicine Perspective. J. Pers. Med. 2020, 10, 249. [Google Scholar] [CrossRef]
- Miranda, I.; Souza, A.; Sousa, P.; Ribeiro, J.; Castanheira, E.M.S.; Lima, R.; Minas, G. Properties and Applications of PDMS for Biomedical Engineering: A Review. J. Funct. Biomater. 2022, 13, 2. [Google Scholar] [CrossRef]
- Boas, L.V.; Faustino, V.; Lima, R.; Miranda, J.M.; Minas, G.; Fernandes, C.S.V.; Catarino, S.O. Assessment of the Deformability and Velocity of Healthy and Artificially Impaired Red Blood Cells in Narrow Polydimethylsiloxane (PDMS) Microchannels. Micromachines 2018, 9, 384. [Google Scholar] [CrossRef]
- Faustino, V.; Rodrigues, R.O.; Pinho, D.; Costa, E.; Santos-Silva, A.; Miranda, V.; Amaral, J.S.; Lima, R. A Microfluidic Deformability Assessment of Pathological Red Blood Cells Flowing in a Hyperbolic Converging Microchannel. Micromachines 2019, 10, 645. [Google Scholar] [CrossRef]
- Ekpenyong, A.E.; Toepfner, N.; Fiddler, C.; Herbig, M.; Li, W.; Cojoc, G.; Summers, C.; Guck, J.; Chilvers, E.R. Mechanical deformation induces depolarization of neutrophils. Sci. Adv. 2017, 3, e1602536. [Google Scholar] [CrossRef] [PubMed]
- Tietze, S.; Kräter, M.; Jacobi, A.; Taubenberger, A.; Herbig, M.; Wehner, R.; Schmitz, M.; Otto, O.; List, C.; Kaya, B.; et al. Spheroid Culture of Mesenchymal Stromal Cells Results in Morphorheological Properties Appropriate for Improved Microcirculation. Adv. Sci. 2019, 6, 1802104. [Google Scholar] [CrossRef] [PubMed]
- Chan, C.J.; Ekpenyong, A.E.; Golfier, S.; Li, W.; Chalut, K.J.; Otto, O.; Elgeti, J.; Guck, J.; Lautenschläger, F. Myosin II Activity Softens Cells in Suspension. Biophys. J. 2015, 108, 1856–1869. [Google Scholar] [CrossRef] [PubMed]
- Asuquo, M.I.; Effa, E.; Gbotosho, O.; Otu, A.; Toepfner, N.; Ameh, S.; Bhayankaram, S.-P.; Zetocha, N.; Nwakama, C.; Egbe, W.; et al. Microfluidic Microcirculation Mimetic as a Tool for the Study of Rheological Characteristics of Red Blood Cells in Patients with Sickle Cell Anemia. Appl. Sci. 2022, 12, 4394. [Google Scholar] [CrossRef]
- Ekpenyong, A.E.; Toepfner, N.; Chilvers, E.R.; Guck, J. Mechanotransduction in neutrophil activation and deactivation. Biochim. Biophys. Acta—Mol. Cell Res. 2015, 1853, 3105–3116. [Google Scholar]
- Guck, J.; Ananthakrishnan, R.; Mahmood, H.; Moon, T.J.; Cunningham, C.C.; Käs, J. The optical stretcher: A novel laser tool to micromanipulate cells. Biophys. J. 2001, 81, 767–784. [Google Scholar] [CrossRef]
- Ekpenyong, A.E.; Whyte, G.; Chalut, K.; Pagliara, S.; Lautenschläger, F.; Fiddler, C.; Paschke, S.; Keyser, U.F.; Chilvers, E.R.; Guck, J. Viscoelastic Properties of Differentiating Blood Cells Are Fate- and Function-Dependent. PLoS ONE 2012, 7, e45237. [Google Scholar] [CrossRef]
- Rowat, A.C.; Jaalouk, D.E.; Zwerger, M.; Ung, W.L.; Eydelnant, I.A.; Olins, D.E.; Olins, A.L.; Herrmann, H.; Weitz, D.A.; Lammerding, J. Nuclear envelope composition determines the ability of neutrophil-type cells to passage through micron-scale constrictions. J. Biol. Chem. 2013, 288, 8610–8618. [Google Scholar] [CrossRef]
- Prasanth, D.; Suresh, S.; Prathivadhi-Bhayankaram, S.; Mimlitz, M.; Zetocha, N.; Lee, B.; Ekpenyong, A. Microgravity Modulates Effects of Chemotherapeutic Drugs on Cancer Cell Migration. Life 2020, 10, 162. [Google Scholar] [CrossRef]
- Chaffer, C.; Weinberg, R. A Perspective on Cancer Cell Metastasis. Science 2011, 331, 1559–1565. [Google Scholar] [CrossRef]
- Koumoutsakos, P.; Pivkin, I.; Milde, F. The Fluid Mechanics of Cancer and Its Therapy. Annu. Rev. Fluid Mech. 2013, 45, 325–355. [Google Scholar] [CrossRef]
- Luk, V.N.; Mo, G.C.; Wheeler, A.R. Pluronic additives: A solution to sticky problems in digital microfluidics. Langmuir 2008, 24, 6382–6389. [Google Scholar] [CrossRef] [PubMed]
- Madaan, K.; Kaushik, D.; Verma, T. Hydroxyurea: A key player in cancer chemotherapy. Expert Rev. Anticancer Ther. 2012, 12, 19–29. [Google Scholar] [CrossRef] [PubMed]
- Douedi, S.; Carson, M.P. Anthracycline Medications (Doxorubicin); StatPearls Publishing: Orlando, FL, USA, 2020. [Google Scholar]
- Hamm-Alvarez, S.F.; Sonee, M.; Loran-Goss, K.; Shen, W.C. Paclitaxel and nocodazole differentially alter endocytosis in cultured cells. Pharm. Res. 1996, 13, 1647–1656. [Google Scholar] [CrossRef] [PubMed]
- Wojcik, T.; Buczek, E.; Majzner, K.; Kolodziejczyk, A.; Miszczyk, J.; Kaczara, P.; Kwiatek, W.; Baranska, M.; Szymonski, M.; Chlopicki, S. Comparative endothelial profiling of doxorubicin and daunorubicin in cultured endothelial cells. Toxicol. In Vitro 2015, 29, 512–521. [Google Scholar] [CrossRef]
- Walter, Y.; Hubbard, A.; Benoit, A.; Jank, E.; Salas, O.; Jordan, D.; Ekpenyong, A. Development of In Vitro Assays for Advancing Radioimmunotherapy against Brain Tumors. Biomedicines 2022, 10, 1796. [Google Scholar] [CrossRef]
- McKinley, S.; Taylor, A.; Peeples, C.; Jacob, M.; Khaparde, G.; Walter, Y.; Ekpenyong, A. Microgravity-Induced Changes to Drug Response in Cancer Cells Quantified Using Fluorescence Morphometry. Life 2023, 13, 1683. [Google Scholar] [CrossRef]
- Larson, M.G. Analysis of variance. Circulation 2008, 117, 115–121. [Google Scholar] [CrossRef]
- Apraiz, A.; Mitxelena, J.; Zubiaga, A. Studying cell cycle-regulated gene expression by two complementary cell synchronization protocols. J. Vis. Exp. 2017, 2017, 55745. [Google Scholar]
- Grady, M.E.; Composto, R.J.; Eckmann, D.M. Cell elasticity with altered cytoskeletal architectures across multiple cell types. J. Mech. Behav. Biomed. Mater. 2016, 61, 197–207. [Google Scholar] [CrossRef]
- Ito, S.; Okuda, S.; Abe, M.; Fujimoto, M.; Onuki, T.; Nishimura, T.; Takeichi, M. Induced cortical tension restores functional junctions in adhesion-defective carcinoma cells. Nat. Commun. 2017, 8, 1834. [Google Scholar] [CrossRef] [PubMed]
- Ly, C.; Ogana, H.; Kim, H.N.; Hurwitz, S.; Deeds, E.J.; Kim, Y.M.; Rowat, A.C. Altered physical phenotypes of leukemia cells that survive chemotherapy treatment. Integr. Biol. 2023, 15, zyad006. [Google Scholar] [CrossRef] [PubMed]
- Munbodh, R.; Jackson, A. Quantifying cell migration distance as a contributing factor to the development of rectal toxicity after prostate radiotherapy. Med. Phys. 2014, 41, 021724. [Google Scholar] [CrossRef] [PubMed]
- Fritsch, A.; Höckel, M.; Kiessling, T.; Nnetu, K.D.; Wetzel, F.; Zink, M.; Käs, J.A. Are biomechanical changes necessary for tumour progression? Nat. Phys. 2010, 6, 730–732. [Google Scholar] [CrossRef]
- Swaminathan, V.; Mythreye, K.; O’Brien, E.T.; Berchuck, A.; Blobe, G.C.; Superfine, R. Mechanical stiffness grades metastatic potential in patient tumor cells and in cancer cell lines. Cancer Res. 2011, 71, 5075–5080. [Google Scholar] [CrossRef]
- Guck, J.; Schinkinger, S.; Lincoln, B.; Wottawah, F.; Ebert, S.; Romeyke, M.; Lenz, D.; Erickson, H.M.; Ananthakrishnan, R.; Mitchell, D.; et al. Optical deformability as an inherent cell marker for testing malignant transformation and metastatic competence. Biophys. J. 2005, 88, 3689–3698. [Google Scholar] [CrossRef]
- Remmerbach, T.W.; Wottawah, F.; Dietrich, J.; Lincoln, B.; Wittekind, C.; Guck, J. Oral cancer diagnosis by mechanical phenotyping. Cancer Res. 2009, 69, 1728–1732. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abraham, A.; Virdi, S.; Herrero, N.; Bryant, I.; Nwakama, C.; Jacob, M.; Khaparde, G.; Jordan, D.; McCuddin, M.; McKinley, S.; et al. Microfluidic Microcirculation Mimetic for Exploring Biophysical Mechanisms of Chemotherapy-Induced Metastasis. Micromachines 2023, 14, 1653. https://doi.org/10.3390/mi14091653
Abraham A, Virdi S, Herrero N, Bryant I, Nwakama C, Jacob M, Khaparde G, Jordan D, McCuddin M, McKinley S, et al. Microfluidic Microcirculation Mimetic for Exploring Biophysical Mechanisms of Chemotherapy-Induced Metastasis. Micromachines. 2023; 14(9):1653. https://doi.org/10.3390/mi14091653
Chicago/Turabian StyleAbraham, Ashley, Sukhman Virdi, Nick Herrero, Israel Bryant, Chisom Nwakama, Megha Jacob, Gargee Khaparde, Destiny Jordan, Mackenzie McCuddin, Spencer McKinley, and et al. 2023. "Microfluidic Microcirculation Mimetic for Exploring Biophysical Mechanisms of Chemotherapy-Induced Metastasis" Micromachines 14, no. 9: 1653. https://doi.org/10.3390/mi14091653





