FISHprep: A Novel Integrated Device for Metaphase FISH Sample Preparation
Abstract
: We present a novel integrated device for preparing metaphase chromosomes spread slides (FISHprep). The quality of cytogenetic analysis from patient samples greatly relies on the efficiency of sample pre-treatment and/or slide preparation. In cytogenetic slide preparation, cell cultures are routinely used to process samples (for culture, arrest and fixation of cells) and/or to expand limited amount of samples (in case of prenatal diagnostics). Arguably, this expansion and other sample pretreatments form the longest part of the entire diagnostic protocols spanning over 3–4 days. We present here a novel device with an integrated expansion chamber to culture, arrest and fix metaphase cells followed by a subsequent splashing protocol leading to ample metaphase chromosome spreads on a glass slide for metaphase FISH analysis. The device provides an easy, disposable, low cost, integrated solution with minimal handling for metaphase FISH slide preparation.1. Introduction
Fluorescence In-Situ Hybridization (FISH) is an indispensable molecular cytogenetic technique for diagnosis of both inherited and acquired chromosomal abnormalities at a much higher resolution than conventional karyotyping [1-10]. Novel FISH based techniques are commonly used in diagnosis of various diseases [4,8,11-18]. Recently, interphase FISH has gained much popularity, and as a result metaphase FISH has received lesser attention, which has significantly derailed the progress made in progressing metaphase FISH analysis technologies [19-21]. Interphase FISH offers numerous advantages compared to metaphase FISH, such as better resolution (down to 1–15 kB) [22-25], wide range of commercial probes [26-30], shorter analysis time (for prenatal studies, dysmorphology, tumor-specific markers) [5,22,31-36] and finally advantages of commercial imaging systems reducing data analysis time [37]. However, interphase FISH has limitations with respect to identification of unknown chromosome abnormalities and rearrangements like translocations. This is because interphase FISH relies on the availability of probes, which limits its applications only for the identification of known translocations [38-40]. As a result, metaphase FISH, still continues to be widely used for diagnosis of chromosomal aberrations in case of unknown translocations [41].
Most of these FISH-based techniques have very similar sample preparation protocols and despite such widespread use of FISH analysis, the sample preparation continues to be a manual, cumbersome and lengthy process leading to significant delays in the diagnosis as well as subsequent treatment of patients. Slide quality is one of the most important factors affecting the efficiency of FISH probe hybridization, and also the intensity and clarity of the FISH signals. Also, it is widely known that in all conventional metaphase preparations there are a large number of nuclei present suitable for interphase FISH studies [7]. Hence an automated system for FISH sample preparation can prove beneficial in multiple venues [6]. Considering the need for culturing various other cell types for FISH sample preparation (lymphocytes, amniocytes, chorionic villi and solid tumors), it would be highly beneficial to integrate the culturing protocol in such a system. This is particularly true in case of prenatal diagnosis, where the starting sample volume is not large enough to run all the necessary diagnosis tests.
Carefully evaluating the sample preparation protocol, it becomes evident that the most time consuming step is the expansion or culture of the lymphocytes (or other cell types like amniocytes, solid tumors, etc.) often taking from 72 h up to two weeks. Due to our interest, we only focus on metaphase FISH analysis and will not detail the steps of immobilization and preparation of interphase cells. However, every metaphase preparation includes cells in the interphase phase as well. For more information kindly refer to novel techniques for interphase FISH and related protocol for cells immobilization [19-21].
Molecular Cytogeneticists are extremely aware of the importance of preparing good metaphase chromosome spreads for getting a reliable FISH analysis [42-44]. The conventional short term protocol for preparing metaphase chromosome spreads from lymphocytes includes a 72 h cell expansion (with mitogenic stimulation) step, followed by arrest of cells in metaphase and later fixation in Carnoy's fixative (1:3 vol:vol—acetic acid and methanol). Once fixed, the cells are splashed on a glass slide with a thin film of water on it, in order to form the chromosome spreads. While it sounds trivial and continues to be a very common protocol in cytogenetic labs, there are numerous variants of slightly modified protocols prevailing among the community with differences in time of culture, fixation, volume of stimulant, mitotic arrest and a vast number of different methods to prepare the chromosome spreads on the slides [42-44].
While many devices for controlled spreading of chromosomes exist, none of these devices have included integrated cell expansion and fixation chamber into the spreading device [44-46]. As a result, these steps (expansion, arrest and fixation) still need to be performed in traditional culture flasks. Also, the size of these existing spreading devices tend to be much larger compared to the proposed FISHprep device. This has led to reduced applicability or usage of these chromosome spreader or dropper tools, as they can be readily replaced by a pipette in the hands of a skilled technician. We have recently presented an integrated device to perform Metaphase FISH on a chip and included a splashing protocol for preparation of these metaphase spreads [38]. But we feel that lack of a complete sample pre-processing device (i.e., Culture, Arrest and Fixation of cells) coupled with a mechanism to prepare chromosome slides has been the missing link for designing a fully automated sample-to-FISH analysis device. Hence, we have developed FISHprep—a novel splashing device integrated with a microfluidic cell culture chamber capable of cell expansion, arrest, fixation and finally splashing of fixed cells on a glass slide to provide metaphase chromosomes spreads for further FISH analysis. This device provides an easy to handle, low cost, disposable, integrated solution for the entire metaphase FISH slide preparation protocol.
2. Experimental Section
2.1. Materials and Chemicals
Polycarbonate (PC) sheets procured from Nordplast (Denmark) were used to fabricate the FISHprep device. Glass slides (SuperFrost), syringes, 3 port valves, paper clips, silicone tubings, Teflon tubings and a 5 μm pore sized PC membrane (Whatman 7060-4713) were ordered from VWR Denmark. Chemical reagents such as Phosphate Buffered Saline (PBS), phytohemagglutinin (PHA), Roswell Park Memorial Institute (RPMI) medium, Fetal bovine serum (FBS) were ordered from Sigma-Aldrich. Carboxyfluorescein succinimidyl ester (CFSE) stain used for viability studies was ordered from Invitrogen Germany. DAPI (4′,6-diamidino-2-phenylindole) (Invitrogen) was used as a counter stain for coloring the chromosomes. Experiments were conducted using blood samples from unknown donors received from the Blood Bank of the Rigshospitalet in Denmark.
2.2. Apparatus
A micromilling machine (Folken Industries, Glendale, AZ, USA) was used for milling the FISHprep devices parts and a UV light source (DYMAX EC5000) was used for treating the surfaces before bonding them under pressure in a bonding press (P/O/Weber, Remshalden, Germany). Finally, a Zeiss Axio Observer Z1 Fluorescent microscope was used for analysis of the spreads and a flow cytometer (Accuri c6) was used for analysis of cell proliferation.
2.3. Fabrication
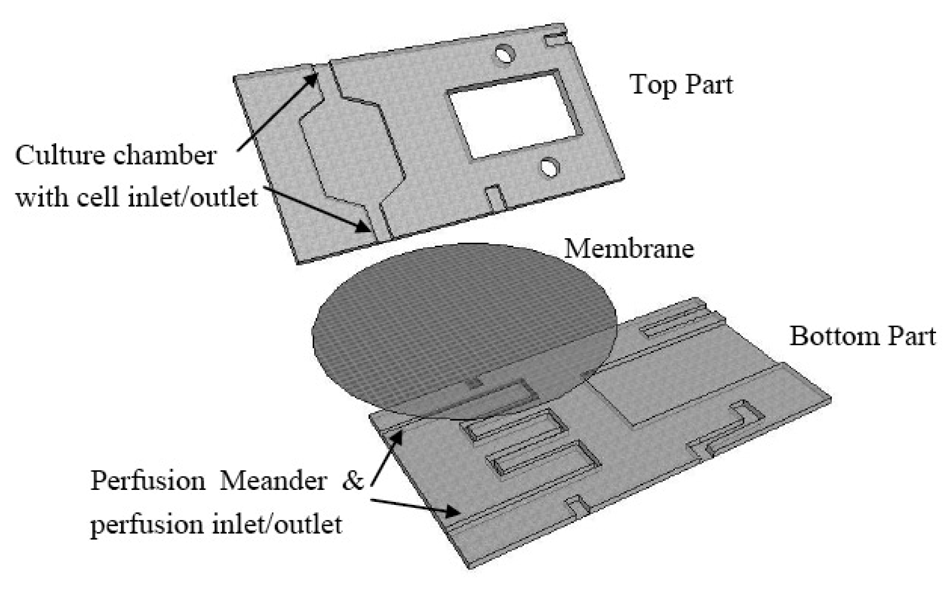
A schematic of the FISHprep device is shown in Figure 1.
The device is fabricated by micromilling in Polycarbonate Sheets. The bottom part contains a meander channel (1 mm wide, 300 μm deep) for perfusion and a slot for sliding in glass slides (56 × 26.2 mm). The top part contains a cell culture chamber (250 μm deep) with a cell inlet channel and a cell outlet channel connected to the splashing chamber's fixed cells inlet. It also includes another channel to act as an inlet for splashing water on the glass slide. A polycarbonate membrane (5 μm pore size) is sandwiched between the perfusion channel and the cell culture chamber during the bonding process of the two parts. The two parts are bonded together using UV activated bonding [47]. First the two parts are wiped clean with IPA followed by rigorous wash with a detergent soap and water. Subsequently the two parts are thoroughly air dried and exposed to UV for 45 s. Before putting the two parts together a PC membrane and interconnection plugs (Silicon tubing: Outer Diameter (OD) 3 mm, Inner Diameter (ID) 1 mm) are placed on the lower part. A Silicon tubing U-plug is placed at the cell outlet of the culture chamber and connected to the fixed cells inlet of the splashing chamber. Finally, the top part is placed on the lower part and put in a P/O/Weber bonding press at 130 °C for 30 min.
2.4. Paper Clip Valve: Leakage Test
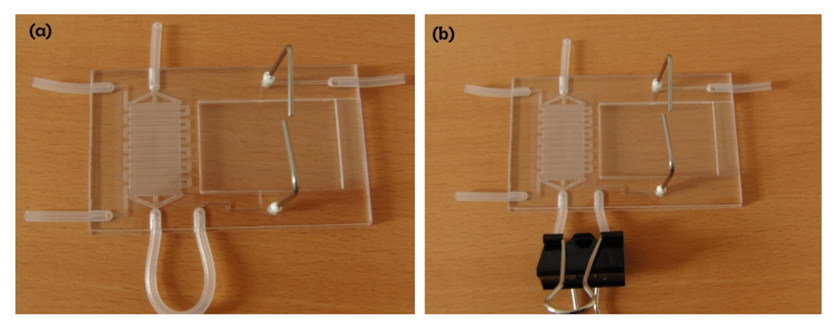
In order to connect the culture chamber and splashing device, we have devised a simple and easy paper clip based valving strategy. This strategy was adopted in order to utilize tools commonly available in the cell labs and also to keep the protocol simple. A 3 mm OD silicone tubing is bonded from the cells outlet to the cells inlet of the splashing device (Figure 2). The tubing forms an external U-section out of the device providing enough room for putting on a paper clip to stop the flow. The paper clip valve was tested at increasing flow rates to identify the maximum permissible flow rate before leakage occurs and to validate the applicability of these valves for the cell culture protocol. The flow rate was increased gradually using a syringe pump and the flow rate at which the device leaked was recorded.
2.5. Culturing Protocol
The device is cleaned and primed by flushing 10% ethanol solution for 5 min and later washed with PBS for 10 min to remove any traces of ethanol from the culture chamber. This priming and sterilization helps to remove any trapped air in the culture and perfusion chamber. Before cells are seeded in the culture chamber, the device is perfused with RPMI 1640 + 10% FBS for 1 h at the flow rate of 37.5 μL/h (the device is kept at 37 °C in a CO2 incubator). A 500 μL buffy coat with a cell count of 2 × 106 cells/mL was seeded into the culture chamber through the cell inlet by opening only the perfusion outlet connected to the waste collector. This protocol for seeding ensures that all the cells get trapped onto the membrane while the suspension media is filtered out from the perfusion outlet. Finally, the cells inlet is closed and a perfusion of fresh media is started through the perfusion inlet at 75 μL/h with RPMI 1640 + 10% FBS containing 10 μL/mL PHA. After 72 h of mitogenic stimulation, the perfusion is stopped and after wait time of 5 min, (which ensures that the flow has completely stopped) the paper clip valve is opened and the spreading protocol is initiated. Cells seeded in a well plate served as a control for the expansion experiments.
2.6. CFSE Staining Protocol
In order to confirm the expansion of lymphocytes on the culture chamber, the cells were stained with CellTrace™ CFSE fluorescence stain (Invitrogen, Germany), at a concentration of 0.7 μM in phosphate buffer saline (PBS) and incubated at 37 °C for 15 min. The cells were later re-pelleted and resuspended in pre-warmed cell culture medium and incubated for 30 min. The cells were then centrifuged and resuspended in pre-warmed fresh medium and subjected to culturing in the FISHprep culture chamber. The CFSE stained cells were stimulated with PHA and cultured for 72 h. Finally, the cells were collected through the cell outlet. In order to collect the cells after culture to analyze their expansion, the U-plug was sliced via a scalpel and disconnected from the splashing chamber. Later, the cells were extracted from the cell outlet by flushing culture media from the cell inlet. To conclude on the expansion, the collected cells were analyzed for fluorescence intensity using a flow cytometer.
2.7. Spreading Protocol
In these experiments, the aim was to perform the FISH sample pretreatment. Therefore, after the culture protocol we replaced the media with 75 mM KCL solution at 0.4 chamber volume/min (150 μL/h) for 25 min. This hypotonic treatment induced swelling of the cells. Finally, in order to fix the cells, the chamber is perfused with freshly prepared fixative (acetic acid:methanol-3:1) at 0.4 chamber volume/min (150 μL/h) for 30 min. Between each change of perfusion, by use of 3 port stopcock, it was ensured that the bubbles were removed before starting the perfusion. For control, simultaneous cultures were conducted in culture flasks followed by the traditional FISH slide preparation protocol [7].
On completion of the sample preparation protocol, the paper clip valve was opened, which connected the spreading chamber to the cells culture chamber outlet. This allows the flushing of fixed cells from the culture chamber on the glass slide. A glass slide treated with corona for 1 min is inserted into the splashing chamber slide slot. The corona treatment helps to activate the surface and improves the wetting behavior of the slide leading to better spreads [48]. Finally, in order to create metaphase spreads on the slide, a drop of cold water is dropped on to the glass slide via the separate inlet for water. This is quickly followed by a drop of fixed cells suspension from the cell chamber by opening the paper-clip valve. The slide is allowed to dry for 2 min before removing it from the FISHprep device. This is currently done manually by use of syringe pumps, which opens the possibility of automation in future. The process is repeated onto three to four slides in order to have ample slides to have quantified FISH analysis (it is a routine process in standard FISH analysis to prepare at least three slides). For the control cultures, the traditional metaphase FISH sample preparation protocol is followed and later, the chromosome spreads are prepared using the dropping technique by an experienced technician [7].
2.7. Analysis
The slides with chromosome spreads are stained with DAPI and sealed with a coverslip. Later they are analyzed using an AxioVision Z1 Observer microscope to analyze the metaphase spreads and thereby validate the applicability of the FISHprep device for metaphase FISH sample preparation. As a final step, the slides prepared with FISHprep device were analyzed using the traditional metaphase FISH protocol [7]. A centromeric probe targeting X-chromosome (Kreatech, NL) was used for the FISH analysis.
3. Results and Discussion
3.1. Fabrication
The fabricated FISHprep device is shown in Figure 2. The interconnections to the syringe pumps are made by inserting a Teflon tubing (OD 1.2 mm) in the silicon plugs (ID 1 mm) bonded between the PC sheets. At every interconnection before the syringe, a 3 way valve is attached to the Teflon tubing to allow for easy changing of reagents (sterilizing compounds, media, fixative, etc.) and removal of bubbles for bubble-free operation device operation.
3.2. Paper Clip Valve: Leakage Test
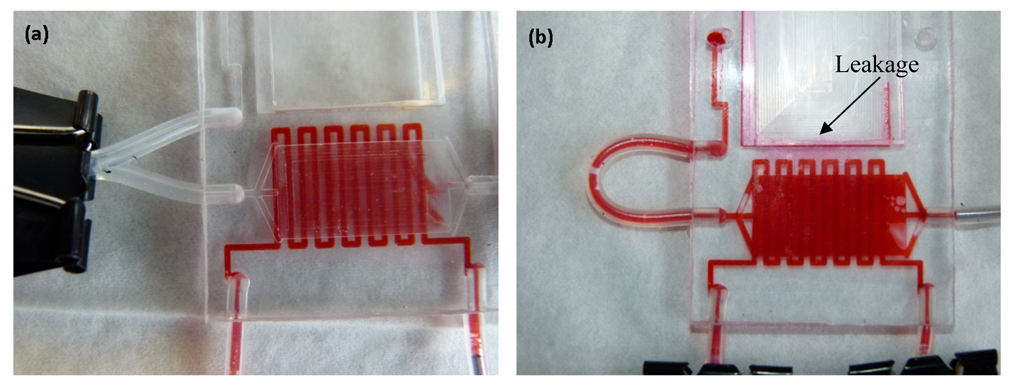
The paper clip based valving technique was tested for isolation between the culture chamber and the splashing chamber until the cells had been fixed in the culture chamber for 72 h. The FISHprep devices were tested for increasing flow rates starting from 150 μL/h up to 750 μL/h. The flow rate of 150 μL/h represents the maximum perfusion rate for the fixation protocol used for sample preparation in the culture and fixation chamber (Figure 3(a)). The first signs of leakage in the device were visible only after 500 μL/min flow rate due to lateral flow in the PC membrane at the bonding interface (Figure 3(b)). Even at such high flow rates, there was no flow through the paper clip valve; the leakage only occurred through the short bonding edge in the FISHprep device. Also, it was noticed that the device was still functioning well without any leakage, when the flow rate was again reduced to 150 μL/h (Figure 3(b)).
3.3. Expansion and Spreading Protocol
Figure 4(a) shows the seeded lymphocytes on top of the membrane in the culture chamber. The pores in the PC membrane can be seen in the background. After 72 h of mitogenic stimulation with PHA, we could see a significant increase in the size of the cell cytoplasm (Figure 4(b)).
3.4. CFSE Proliferation Assay
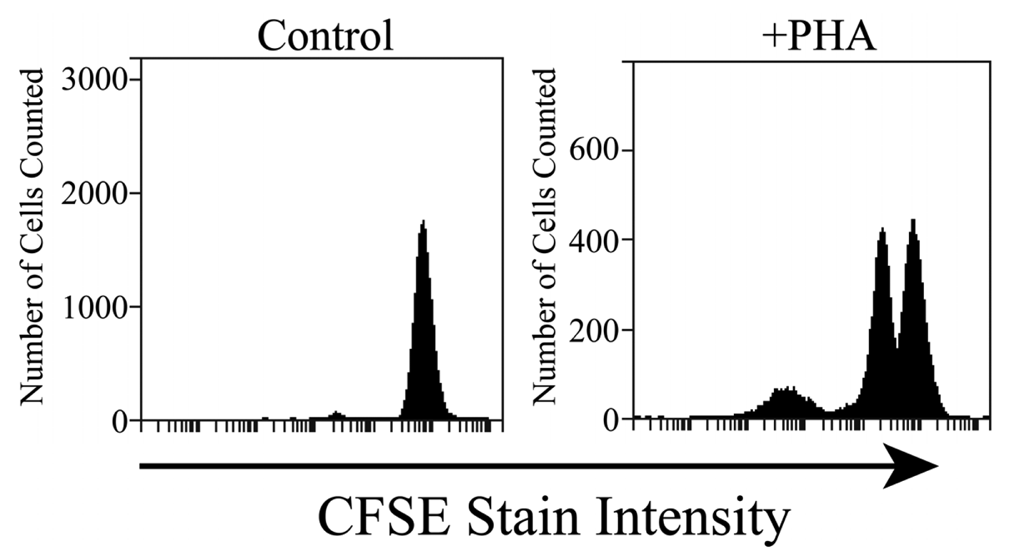
The cells collected from the FISHprep culture chamber were analyzed in a flow cytometer by using CFSE staining of the cells. CFSE stain is retained within the cells throughout development and mitosis. On proliferation, the label is inherited by daughter cells but with a lower florescence intensity which can be monitored on a flow cytometer. Figure 5 shows results of fluorescence analysis of 20 μL of FISHprep culture samples. The cells show signature peaks of proliferation as depicted by the decrease in fluorescence intensity. In contrast, the negative cultures of lymphocytes (without PHA) on the FISHprep device, hardly show any growth which confirms that the FISHprep device does not induce any activation or expansion of the lymphocytes due to microfluidic handling. As our interests were related to procuring ample FISH spreads to conduct a FISH analysis, we didn't culture the cells for longer time or quantify the culture dynamics in the FISHprep culture chamber. It might be of interest at a later point to culture cells for longer durations for characterization and comparison of the growth pattern of cells on the FISHprep device with traditional cultures. There is a possibility that cells in FISHprep culture device might follow a different expansion cycle compared to the culture flask [49].
3.5. Analysis of the Spreads
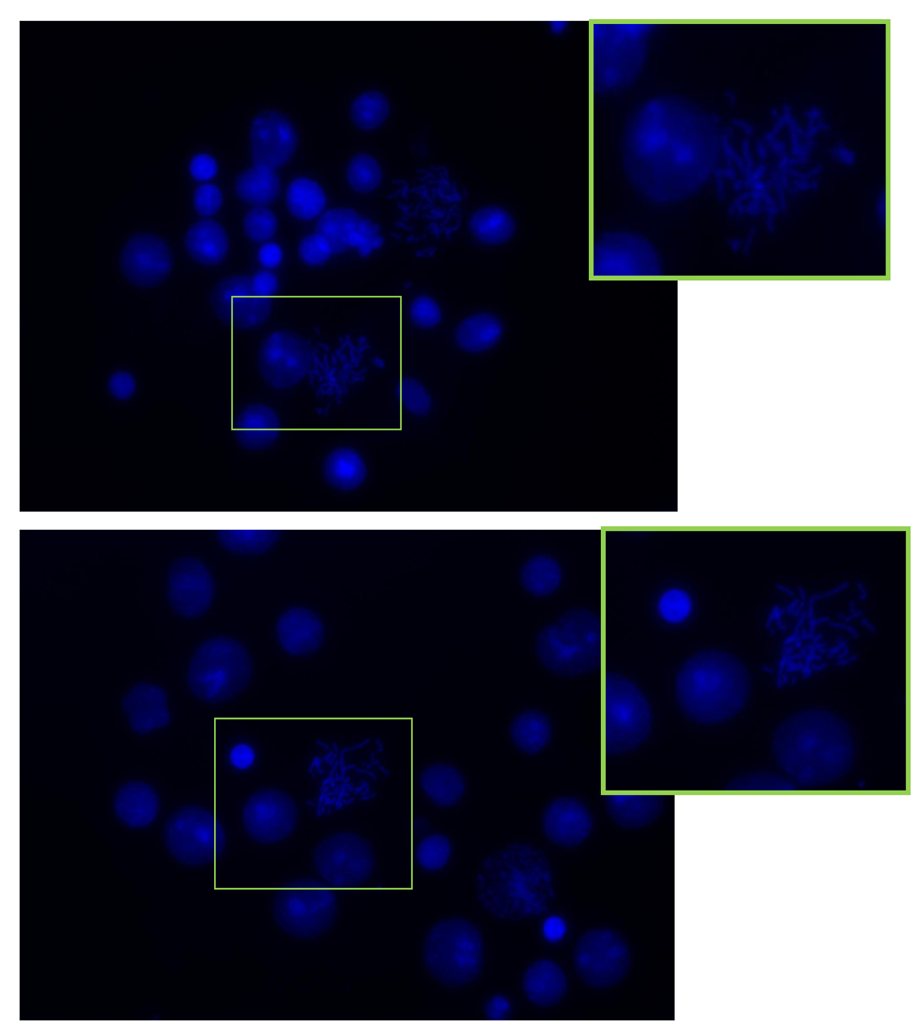
The spreads obtained from the integrated device and the control flask cultures were stained with DAPI and analyzed using an Axio Observer Z1 microscope. Figure 6(a,b) presents the spreads achieved from the FISHprep device and control flask samples respectively. The mitotic index in both the slides was found to be above 75%. The spreads on the FISHprep device were comparable to the spreads received with control samples.
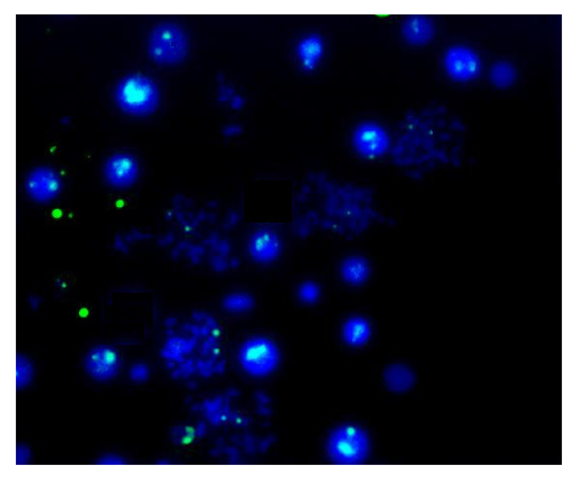
Finally, Figure 7 shows the results of the traditional metaphase FISH protocol applied to the samples prepared using the FISHprep device. The FISH signals acquired for the X-chromosomes in the spreads validate the compatibility of this proposed sample preparation technique for cytogenetic analysis. Hence, we concluded the promising applicability of FISHprep device for metaphase FISH sample preparation.
4. Conclusions
A novel, simple, minimal handling, integrated device (FISHprep) was presented for processing samples and preparing slides for FISH analysis. The presented device provides a simple, low cost, disposable alternative to traditional sample preparation technique including culture, arrest, fixation and subsequent preparation of glass slides with metaphase chromosome spreads. The inclusion of a membrane based culture chamber into the FISHprep device opens possibilities for expanding the applicability of the device to other cell types like amniocytes or chorionic villus, where controlled long term expansion of cells is of great interest due to small amounts of available sample volume. In the near future, we aim to integrate the Metaphase FISH on chip protocol into the FISHprep protocol to create a chromosome total analysis system.

Acknowledgments
The authors would like to thank the Danish Research Council for Technology and Production and the Lundbeck Foundation for their financial support to make this research possible. Finally, Peder Skafte-Pedersen for assistance with microscope for image acquisition. The Wilhelm Johannsen Centre for Functional Genome Research is established by The Danish National Research Foundation.
References
- Liehr, T. Fluorescence In-Situ Hybridization (FISH): Application Guide, 1st ed.; Springer-Verlag: Berlin/Heidelberg, Germany, 2009; p. 451. [Google Scholar]
- Ward, B.E.; Gersen, S.L.; Carelli, M.P.; McGuire, N.M.; Dackowski, W.R.; Weinstein, M.; Sandlin, C.; Warren, R.; Klinger, K.W. Rapid prenatal diagnosis of chromosomal aneuploidies by fluorescence in situ hybridization: Clinical experience with 4,500 specimens. Am. J. Hum. Genet. 1993, 52, 854–865. [Google Scholar]
- Sasai, K.; Tanaka, R.; Kawamura, M.; Honjo, K.; Matsunaga, N.; Nakada, T.; Homma, K.; Fujimura, H. Chromosome spreading techniques for primary gastrointestinal tumors. J. Gastroenterol. 1996, 31, 505–511. [Google Scholar]
- Nath, J.; Johnson, K.L. A review of fluorescence in situ hybridization (FISH): Current status and future prospects. Biotech. Histochem. 2000, 75, 54–78. [Google Scholar]
- Tepperberg, J.; Pettenati, M.J.; Rao, P.N.; Lese, C.M.; Rita, D.; Wyandt, H.; Gersen, S.; White, B.; Schoonmaker, M.M. Prenatal diagnosis using interphase fluorescence in situ hybridization (FISH): 2-year multi-center retrospective study and review of the literature. Prenatal Diag. 2001, 21, 293–301. [Google Scholar]
- Jiang, F.; Katz, R.L. Use of interphase fluorescence in situ hybridization as a powerful diagnostic tool in cytology. Diagn. Molecul. Pathol. 2002, 11, 47–57. [Google Scholar]
- Squire, J.A.; Marrano, P.; Kolomietz, E. FISH in Clinical Cytogenetics, 1st ed.; Oxford Publisher: Oxford, New York, NY, USA, 2002. [Google Scholar]
- Liehr, T.; Starke, H.; Heller, A.; Kosyakova, N.; Mrasek, K.; Gross, M.; Karst, C.; Steinhaeuser, U.; Hunstig, F.; Fickelscher, I.; Kuechler, A.; Trifonov, V.; Romanenko, S.A.; Weise, A. Multicolor fluorescence in situ hybridization (FISH) applied to FISH-banding. Cytogenet. Genome Res. 2006, 114, 240–244. [Google Scholar]
- Weise, A.; Liehr, T. Fluorescence in situ hybridization for prenatal screening of chromosomal aneuploidies. Expert Rev. Mol. Diagn. 2008, 8, 355–357. [Google Scholar]
- Tanas, M.R.; Goldblum, J.R. Fluorescence In Situ Hybridization in the diagnosis of soft tissue neoplasms: A review. Adv. Anat. Pathol. 2009, 16, 383–391. [Google Scholar]
- Braselmann, H.; Kulka, U.; Baumgartner, A.; Eder, C.; Muller, I.; Figel, M.; Zitzelsberger, H. SKY and FISH analysis of radiation-induced chromosome aberrations: A comparison of whole and partial genome analysis. Mutat. Res-Fundam. Mol. Mech. Mut. 2005, 578, 124–133. [Google Scholar]
- Ried, T. Multiparametric fluorescence-in-situ-hybridization (fish) and spectral karyotyping (sky) in diagnosis of human malignancies. Cytometry Part A 2007, 71A, 22. [Google Scholar]
- Takubo, K.; Fujita, M.; Izumiyama, N.; Nakamura, K.; Ishikawa, N.; Poon, S.S.; Fujiwara, M.; Sawabe, M.; Matsuura, M.; Grabsch, H.; Arai, T.O.; Aida, J. Q-FISH analysis of telomere and chromosome instability in the oesophagus with and without squamous cell carcinoma in situ. J. Pathol. 2010, 221, 201–209. [Google Scholar]
- Bystricka, D.; Zemanova, Z.; Brezinova, J.; Gancarcikova, M.; Grosova, L.; Sarova, I.; Izakova, S.; Berkova, A.; Michalova, K. The assessment of array comparative genomic hybridization in complex karyotype analyses. Folia Biol. 2010, 56, 223–230. [Google Scholar]
- Gue, M.; Messaoudi, C.; Sun, J.S.; Boudier, T. Smart 3D-FISH: Automation of distance analysis in nuclei of interphase cells by image processing. Cytometry Part A 2005, 67A, 18–26. [Google Scholar]
- Raap, T. Colour barcoding genes by Fiber-FISH. Eur. J. Human Genet. 1998, 6. PL05. [Google Scholar]
- van de Rijke, F.M.; Florijn, R.J.; Tanke, H.J.; Raap, A.K. DNA fiber-FISH staining mechanism. J. Histochem. Cytochem. 2000, 48, 743–745. [Google Scholar]
- Volpi, E.V.; Bridger, J.M. FISH glossary: An overview of the fluorescence in situ hybridization technique. Biotechniques 2008, 45, 385. [Google Scholar]
- Sieben, V.J.; Marun, C.S.D.; Pilarski, P.M.; Kaigala, G.V.; Pilarski, L.M.; Backhouse, C.J. FISH and chips: Chromosomal analysis on microfluidic platforms. IEEE Nanobiotechnol. 2007, 1, 27–35. [Google Scholar]
- Sieben, V.J.; Debes-Marun, C.S.; Pilarski, L.M.; Backhouse, C.J. An integrated microfluidic chip for chromosome enumeration using fluorescence in situ hybridization. Lab Chip 2008, 8, 2151–2156. [Google Scholar]
- Zanardi, A.; Bandiera, D.; Bertolini, F.; Corsini, C.A.; Gregato, G.; Milani, P.; Barborini, E.; Carbone, R. Miniaturized FISH for screening of onco-hematological malignancies. Biotechniques 2010, 49, 497–504. [Google Scholar]
- Jobanputra, V.; Sobrino, A.; Kinney, A.; Kline, J.; Warburton, D. Multiplex interphase FISH as a screen for common aneuploidies in spontaneous abortions. Human Reprod. 2002, 17, 1166–1170. [Google Scholar]
- Pergament, E.; Chen, P.X.; Thangavelu, M.; Fiddler, M. The clinical application of interphase FISH in prenatal diagnosis. Prenatal Diag. 2000, 20, 215–220. [Google Scholar]
- Garson, J.A.; Vandenberghe, J.A.; Kemshead, J.T. High-resolution in situ hybridization technique using biotinylated NMYC oncogene probe reveals periodic structure of HSRs in human neuroblastoma. Cytogenet. Cell Genet. 1987, 45, 10–15. [Google Scholar]
- Garson, J.A.; Vandenberghe, J.A.; Kemshead, J.T. Novel non-isotopic in situ hybridization technique detects small (1 Kb) unique sequences in routinely G-banded human chromosomes: Fine mapping of N-myc and beta-NGF genes. Nucl. Acid. Res. 1987, 15, 4761–4770. [Google Scholar]
- Cremer, T.; Lichter, P.; Borden, J.; Ward, D.C.; Manuelidis, L. Detection of chromosome aberrations in metaphase and interphase tumor cells by in situ hybridization using chromosome-specific library probes. Human Genet. 1988, 80, 235–246. [Google Scholar]
- Julien, C.; Bazin, A.; Guyot, B.; Forestier, F.; Daffos, F. Rapid prenatal diagnosis of Down's syndrome with in-situ hybridisation of fluorescent DNA probes. Lancet 1986, 2, 863–864. [Google Scholar]
- Guyot, B.; Bazin, A.; Sole, Y.; Julien, C.; Daffos, F.; Forestier, F. Prenatal diagnosis with biotinylated chromosome specific probes. Prenatal Diag. 1988, 8, 485–493. [Google Scholar]
- Stumm, M.; Wegner, R.D.; Bloechle, M.; Eckel, H. Interphase M-FISH applications using commercial probes in prenatal and PGD diagnostics. Cytogenet. Genome Res. 2006, 114, 296–301. [Google Scholar]
- Dziubinski, J.M.; Sarosdy, M.F.; Kahn, P.R.; Ziffer, M.D.; Love, W.R.; Zadra, J.A.; Schellhammer, P.F.; Bokinsky, G.B.; Stenzel, T.T. Multiple fluorescence in situ hybridization (FISH) probes increase sensitivity for detection of bladder cancer. J. Urol. 2006, 175, 890. [Google Scholar]
- Brown, J.; Saracoglu, K.; Uhrig, S.; Knight, S.J.L.; Lucas, S.J.A.; Speicher, M.R.; Eils, R.; Kearney, L. Development of a multicolour FISH assay for subtelomeric chromosome rearrangements in leukaemia. Cytogenet. Cell Genet. 1999, 85, 18. [Google Scholar]
- Kearney, L. The impact of the new fish technologies on the cytogenetics of haematological malignancies. Brit. J. Haematol. 1999, 104, 648–658. [Google Scholar]
- Kearney, L. Multiplex-FISH (M-FISH): Technique, developments and applications. Cytogenet. Genome Res. 2006, 114, 189–198. [Google Scholar]
- Insabato, L.; Bernasconi, B.; Glatz, K.; Dirnhofer, S.; Krugman, J.; Fend, F.; Capella, C.; Tibiletti, M.G.; Di Vizio, D.; Pettinato, G.; Sauter, G.; Tornillo, L.; Terracciano, L. High-throughput tissue microarray FISH analysis in 226 surgically resected gastric B-cell lymphomas. Frequencies of trisomy 1,3,12 and gains of X chromosome. Lab. Invest. 2002, 82, 541. [Google Scholar]
- Tibiletti, M.G. Interphase FISH as a new tool in tumor pathology. Cytogenet. Genome Res. 2007, 118, 229–236. [Google Scholar]
- Xu, J.; Chen, Z. Advances in molecular cytogenetics for the evaluation of mental retardation. Am. J. Med. Genet. Part C 2003, 117C, 15–24. [Google Scholar]
- Smith, G.D.; Riding, M.; Oswald, K.; Bentz, J.S. Integrating a FISH imaging system into the cytology laboratory. Cyto J. 2010, 7. [Google Scholar] [CrossRef]
- Vedarethinam, I.; Shah, P.; Dimaki, M.; Tumer, Z.; Tommerup, N.; Svendsen, W.E. Metaphase FISH on a Chip: Miniaturized Microfluidic Device for Fluorescence in situ Hybridization. Sensors 2010, 10, 9831–9846. [Google Scholar]
- American College of Medical Genetics. Prenatal interphase fluorescence in situ hybridization (FISH) policy statement. Am. J. Human Genet. 1993, 53, 526–527. [Google Scholar]
- Schwartz, S. Efficacy and applicability of interphase fluorescence in situ hybridization for prenatal diagnosis. Am. J. Human Genet. 1993, 52, 851–853. [Google Scholar]
- Christel, E.-S.; Stefan, G.; Inga, N.; Andrea, H.; Susanne, B.; Simone, H.; Monika, K.; Christian, T.; Reiner, S.; Almuth, C. Conflicting results of prenatal FISH with different probes for Down's Syndrome critical regions associated with mosaicism for a de novo del(21)(q22) characterised by molecular karyotyping: Case report. Mol. Cytogenet. 2010, 3. [Google Scholar] [CrossRef]
- Claussen, U.; Michel, S.; Muhlig, P.; Westermann, M.; Grummt, U.W.; Kromeyer-Hauschild, K.; Liehr, T. Demystifying chromosome preparation and the implications for the concept of chromosome condensation during mitosis. Cytogenet. Genome Res. 2002, 98, 136–146. [Google Scholar]
- Hliscs, R.; Muhlig, P.; Claussen, U. The spreading of metaphases is a slow process which leads to a stretching of chromosomes. Cytogenet. Cell Genet. 1997, 76, 167–171. [Google Scholar]
- Henegariu, O.; Heerema, N.A.; Wright, L.L.; Bray-Ward, P.; Ward, D.C.; Vance, G.H. Improvements in cytogenetic slide preparation: Controlled chromosome spreading, chemical aging and gradual denaturing. Cytometry 2001, 43, 101–109. [Google Scholar]
- Qu, Y.Y.; Xing, L.Y.; Hughes, E.D.; Saunders, T.L. Chromosome dropper tool: Effect of slide angles on chromosome spread quality for murine embryonic stem cells. J. Histotechnol. 2008, 31, 75–79. [Google Scholar]
- Yamada, K.; Kakinuma, K.; Tateya, H.; Miyasaka, C. Development of an instrument for chromosome slide preparation. J. Radiat. Res. 1992, 33, 242–249. [Google Scholar]
- Sabourin, D.; Dufva, M.; Jensen, T.; Kutter, J.; Snakenborg, D. One-step fabrication of microfluidic chips with in-plane, adhesive-free interconnections. J. Micromechanic. Microengineer 2010, 20, 037001–037008. [Google Scholar]
- Haubert, K.; Drier, T.; Beebe, D. PDMS bonding by means of a portable, low-cost corona system. Lab Chip 2006, 6, 1548–1549. [Google Scholar]
- Yeon, J.H.; Park, J.K. Microfluidic cell culture systems for cellular analysis. Biochip J. 2007, 1, 17–27. [Google Scholar]
© 2011 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Shah, P.; Vedarethinam, I.; Kwasny, D.; Andresen, L.; Skov, S.; Silahtaroglu, A.; Tümer, Z.; Dimaki, M.; Svendsen, W.E. FISHprep: A Novel Integrated Device for Metaphase FISH Sample Preparation. Micromachines 2011, 2, 116-128. https://doi.org/10.3390/mi2020116
Shah P, Vedarethinam I, Kwasny D, Andresen L, Skov S, Silahtaroglu A, Tümer Z, Dimaki M, Svendsen WE. FISHprep: A Novel Integrated Device for Metaphase FISH Sample Preparation. Micromachines. 2011; 2(2):116-128. https://doi.org/10.3390/mi2020116
Chicago/Turabian StyleShah, Pranjul, Indumathi Vedarethinam, Dorota Kwasny, Lars Andresen, Søren Skov, Asli Silahtaroglu, Zeynep Tümer, Maria Dimaki, and Winnie E. Svendsen. 2011. "FISHprep: A Novel Integrated Device for Metaphase FISH Sample Preparation" Micromachines 2, no. 2: 116-128. https://doi.org/10.3390/mi2020116
APA StyleShah, P., Vedarethinam, I., Kwasny, D., Andresen, L., Skov, S., Silahtaroglu, A., Tümer, Z., Dimaki, M., & Svendsen, W. E. (2011). FISHprep: A Novel Integrated Device for Metaphase FISH Sample Preparation. Micromachines, 2(2), 116-128. https://doi.org/10.3390/mi2020116




