Combining Electro-Osmotic Flow and FTA® Paper for DNA Analysis on Microfluidic Devices
Abstract
:1. Introduction
2. Materials and Methods
2.1. Manufacture and Functionalisation of Microfluidic Devices
2.2. Sample Preparation
2.3. DNA Purification Procedure
2.3.1. Conventional Method
2.3.2. Microfluidic Method
2.4. DNA Quantification
2.5. DNA Amplification Procedure
3. Results and Discussion
3.1. Optimisation of Electro-Osmotic Movement
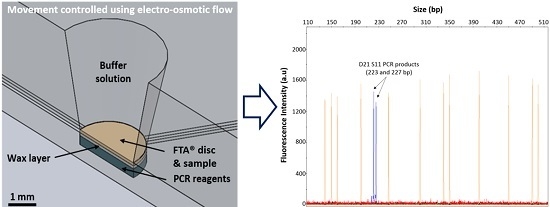
3.2. Integrated DNA Purification and Amplification
3.3. Analysis of Different Sample Types
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| DNA | Deoxyribonucleic acid |
| DTT | Dithiothreitol |
| EDTA | Ethylenediaminetetraacetic acid |
| EOF | Electro-osmotic flow |
| PCR | Polymerase chain reaction |
| qPCR | Real-time quantitative PCR |
| TE | Tris/EDTA buffer |
References
- Park, B.H.; Kim, Y.T.; Jung, J.H.; Seo, T.S. Integration of sample pretreatment, μPCR, and detection for a total genetic analysis microsystem. Microchim. Acta 2014, 181, 1655–1668. [Google Scholar] [CrossRef]
- Njoroge, S.K.; Chen, H.W.; Witek, M.A.; Soper, S.A. Integrated microfluidic systems for DNA analysis. In Microfluidics; Springer: Berlin/Heidelberg, Germany, 2011; pp. 203–260. [Google Scholar]
- Wen, J.; Legendre, L.A.; Bienvenue, J.M.; Landers, J.P. Purification of nucleic acids in microfluidic devices. Anal. Chem. 2008, 80, 6472–6479. [Google Scholar] [CrossRef] [PubMed]
- Amasia, M.; Madou, M. Large-volume centrifugal microfluidic device for blood plasma separation. Bioanalysis 2010, 2, 1701–1710. [Google Scholar] [CrossRef] [PubMed]
- Jebrail, M.J.; Sinha, A.; Vellucci, S.; Renzi, R.F.; Ambriz, C.; Gondhalekar, C.; Schoeniger, J.S.; Patel, K.D.; Branda, S.S. World-to-digital-microfluidic interface enabling extraction and purification of RNA from human whole blood. Anal. Chem. 2014, 86, 3856–3862. [Google Scholar] [CrossRef] [PubMed]
- Ferrance, J.P.; Wu, Q.R.; Giordano, B.; Hernandez, C.; Kwok, Y.; Snow, K.; Thibodeau, S.; Landers, J.P. Developments toward a complete micro-total analysis system for Duchenne muscular dystrophy diagnosis. Anal. Chim. Acta 2003, 500, 223–236. [Google Scholar] [CrossRef]
- Chia, B.T.; Yang, X.-Y.; Cheng, M.-Y.; Lin, C.-W.; Yang, Y.-J. An electromagnetically-driven microfluidic platform with indirect-heating thermo-pneumatic valves. Biochip J. 2011, 5, 97–105. [Google Scholar] [CrossRef]
- Ferguson, B.S.; Buchsbaum, S.F.; Wu, T.-T.; Hsieh, K.; Xiao, Y.; Sun, R.; Soh, H.T. Genetic Analysis of H1N1 Influenza Virus from Throat Swab Samples in a Microfluidic System for Point-of-Care Diagnostics. J. Am. Chem. Soc. 2011, 133, 9129–9135. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.G.; Cheong, K.H.; Huh, N.; Kim, S.; Choi, J.W.; Ko, C. Microchip-based one step DNA extraction and real-time PCR in one chamber for rapid pathogen identification. Lab Chip 2006, 6, 886–895. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.; Mauk, M.; Chen, D.; Qiu, X.; Kim, J.; Gale, B.; Bau, H.H. A PCR reactor with an integrated alumina membrane for nucleic acid isolation. Analyst 2010, 135, 2408–2414. [Google Scholar] [CrossRef] [PubMed]
- Hagan, K.A.; Reedy, C.R.; Bienvenue, J.M.; Dewald, A.H.; Landers, J.P. A valveless microfluidic device for integrated solid phase extraction and polymerase chain reaction for short tandem repeat (STR) analysis. Analyst 2011, 136, 1928–1937. [Google Scholar] [CrossRef] [PubMed]
- Shaw, K.J.; Joyce, D.A.; Docker, P.T.; Dyer, C.E.; Greenway, G.M.; Greenman, J.; Haswell, S.J. Development of a real-world direct interface for integrated DNA extraction and amplification in a microfluidic device. Lab Chip 2011, 11, 443–448. [Google Scholar] [CrossRef] [PubMed]
- GE Healthcare. Sample collection kits and cards, punching technologies, and rapid STR instrumentation handbook. 2015, pp. 8–10. Available online: http://www.gelifesciences.com/gehcls_images/GELS/Related%20Content/Files/1456105290232/litdoc29005583_20160222024111.pdf (accessed on 1 June 2016).
- GE Healthcare. FTA cards manual (Data file 28-9843-54 AA). 2011, pp. 1–3. Available online: https://www.gelifesciences.com/gehcls_images/GELS/Related%20Content/Files/1357903115683/litdoc28984354_20130114101035.pdf (accessed on 1 May 2016).
- Liu, C.; Geva, E.; Mauk, M.; Qiu, X.; Abrams, W.R.; Malamud, D.; Curtis, K.; Owen, S.M.; Bau, H.H. An isothermal amplification reactor with an integrated isolation membrane for point-of-care detection of infectious diseases. Analyst 2011, 136, 2069–2076. [Google Scholar] [CrossRef] [PubMed]
- Gan, W.; Zhuang, B.; Zhang, P.; Han, J.; Li, C.X.; Liu, P. A filter paper-based microdevice for low-cost, rapid, and automated DNA extraction and amplification from diverse sample types. Lab Chip Miniatur. Chem. Biol. 2014, 14, 3719–3728. [Google Scholar] [CrossRef] [PubMed]
- Le Roux, D.; Root, B.E.; Hickey, J.A.; Scott, O.N.; Tsuei, A.; Li, J.; Saul, D.J.; Chassagne, L.; Landers, J.P.; de Mazancourt, P. An integrated sample-in-answer-out microfluidic chip for rapid human identification by STR analysis. Lab Chip Miniatur. Chem. Biol. 2014, 14, 4415–4425. [Google Scholar] [CrossRef] [PubMed]
- Czurratis, D.; Beyl, Y.; Zinober, S.; Lärmer, F.; Zengerle, R. A novel concept for long-term pre-storage and release of liquids for pressure-driven lab-on-a-chip devices. J. Micromech. Microeng. 2015, 25. [Google Scholar] [CrossRef]
- Selvakumar, S.; Linares, R.; Oppenheimer, A.; Anthony, B. Variation analysis of flow rate delivered using a blister pump. Proc. SPIE 2012, 8251. [Google Scholar] [CrossRef]
- Dai, M.; Jin, S.; Nugen, S.R. Water-soluble electrospun nanofibers as a method for on-chip reagent storage. Biosensors 2012, 2, 388–395. [Google Scholar] [CrossRef] [PubMed]
- Ahlford, A.; Kjeldsen, B.; Reimers, J.; Lundmark, A.; Romani, M.; Wolff, A.; Syvanen, A.-C.; Brivio, M. Dried reagents for multiplex genotyping by tag-array minisequencing to be used in microfluidic devices. Analyst 2010, 135, 2377–2385. [Google Scholar] [CrossRef] [PubMed]
- McCreedy, T. Fabrication techniques and materials commonly used for the production of microreactors and micro total analytical systems. Trac-Trends Anal. Chem. 2000, 19, 396–401. [Google Scholar] [CrossRef]
- Erill, I.; Campoy, S.; Erill, N.; Barbe, J.; Aguilo, J. Biochemical analysis and optimization of inhibition and adsorption phenomena in glass-silicon PCR-chips. Sens. Actuators B Chem. 2003, 96, 685–692. [Google Scholar] [CrossRef]
- Hartshorne, H.; Backhouse, C.J.; Lee, W.E. Ferrofluid-based microchip pump and valve. Sens. Actuators B Chem. 2004, 99, 592–600. [Google Scholar] [CrossRef]
- Krenke, B.E.; Tereba, A.; Anderson, S.J.; Buel, E.; Culhane, S.; Finis, C.J.; Tomsey, C.S.; Zachetti, J.M.; Masibay, A.; Rabbach, D.R.; et al. Validation of a 16-locus fluorescent multiplex system. J. Forensic Sci. 2002, 47, 773–785. [Google Scholar] [CrossRef] [PubMed]
- Butler, J.M. Genetics and Genomics of Core Short Tandem Repeat Loci Used in Human Identity Testing. J. Forensic Sci. 2006, 51, 253–265. [Google Scholar] [CrossRef] [PubMed]
- Bienvenue, J.M.; Duncalf, N.; Marchiarullo, D.; Ferrance, J.P.; Landers, J.P. Microchip-based cell lysis and DNA extraction from sperm cells for application to forensic analysis. J. Forensic Sci. 2006, 51, 266–273. [Google Scholar] [CrossRef] [PubMed]




| Applied Voltage (Vcm−1) | 50 | 75 | 100 | 125 | 150 |
|---|---|---|---|---|---|
| FTA® paper | 77.8% | 83.8% | 87.3% | 42.9% | 29.8% |
| Anode | - | - | - | 17.3% | 26.8% |
| Cathode | - | - | - | - | - |
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wimbles, R.; Melling, L.M.; Shaw, K.J. Combining Electro-Osmotic Flow and FTA® Paper for DNA Analysis on Microfluidic Devices. Micromachines 2016, 7, 119. https://doi.org/10.3390/mi7070119
Wimbles R, Melling LM, Shaw KJ. Combining Electro-Osmotic Flow and FTA® Paper for DNA Analysis on Microfluidic Devices. Micromachines. 2016; 7(7):119. https://doi.org/10.3390/mi7070119
Chicago/Turabian StyleWimbles, Ryan, Louise M. Melling, and Kirsty J. Shaw. 2016. "Combining Electro-Osmotic Flow and FTA® Paper for DNA Analysis on Microfluidic Devices" Micromachines 7, no. 7: 119. https://doi.org/10.3390/mi7070119