On-Chip Isoniazid Exposure of Mycobacterium smegmatis Penicillin-Binding Protein (PBP) Mutant Using Time-Lapse Fluorescent Microscopy
Abstract
:1. Introduction
2. Materials and Methods
2.1. Growth of the Cells
2.2. Antibiotic Responses
2.3. Environmental Stresses
2.4. Minimum Inhibitory Concentration (MIC)
2.5. Transposon (Tn) Mutagenesis and Identification of the msm0031::Tn Mutant
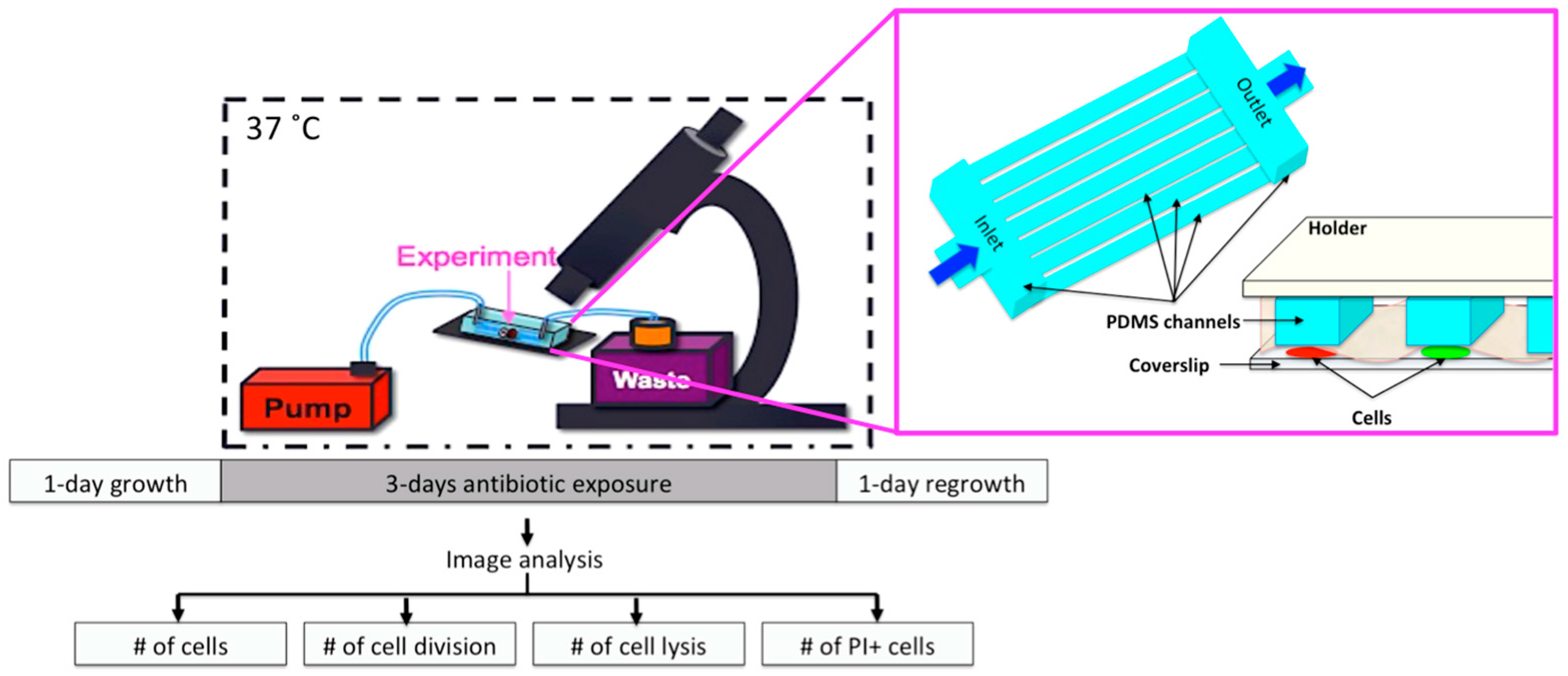
2.6. Microfluidic Chip and Live-Cell Imaging
2.7. Image Analysis
2.8. Statistical Analysis
3. Results
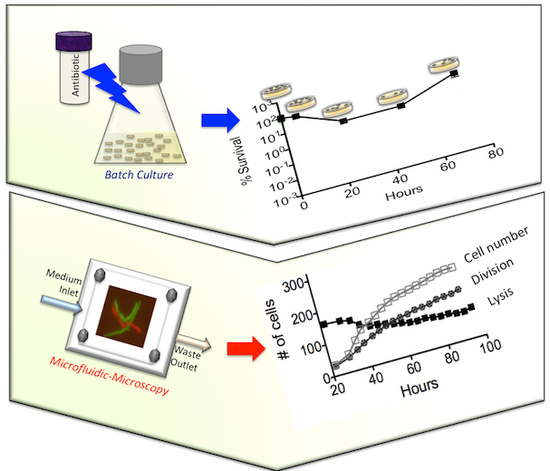
3.1. Batch-Cultue Behavior of the msm0031 Transposon Mutant and Wild-Type M. smegmatis Cells
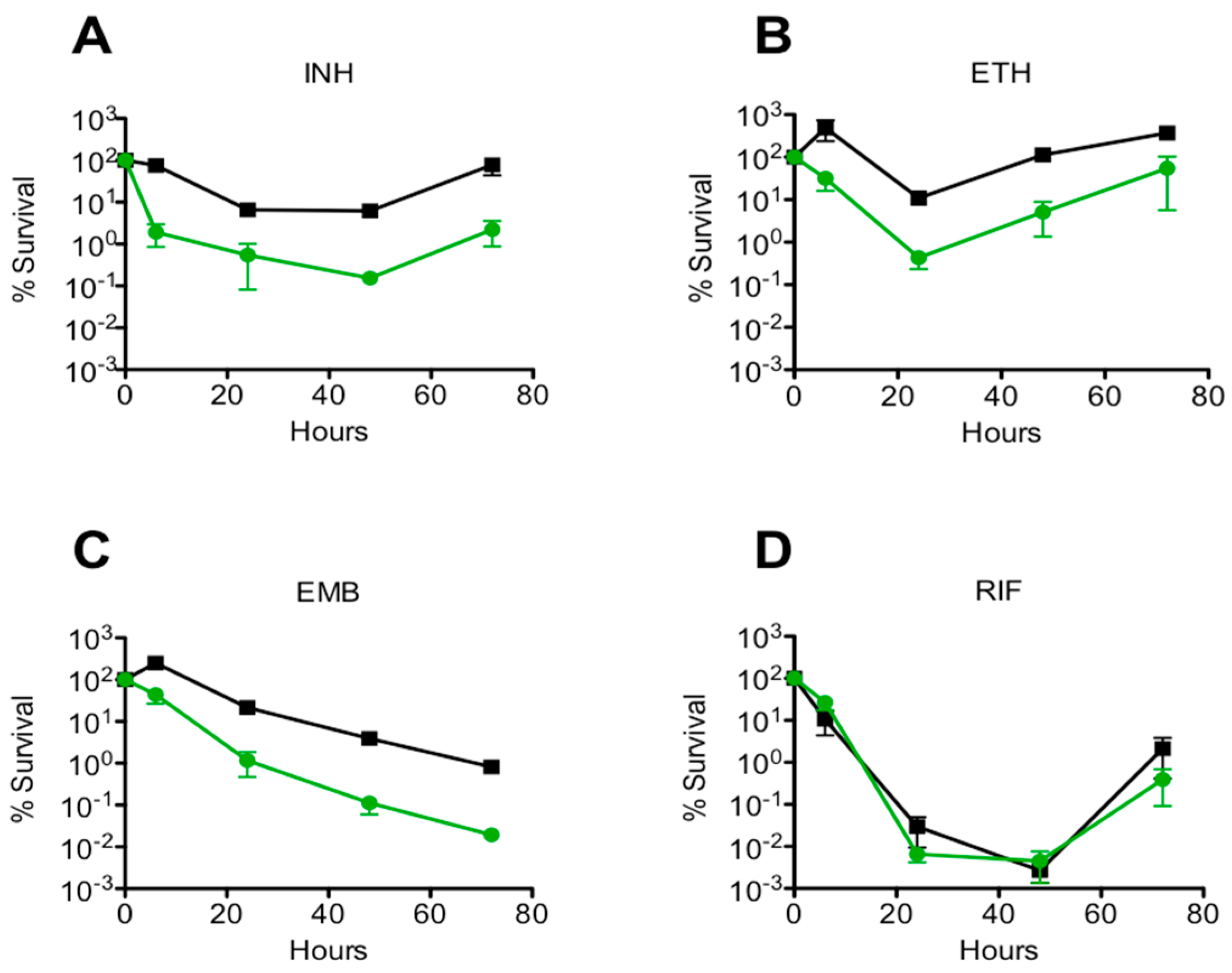
3.2. Drug Specificity
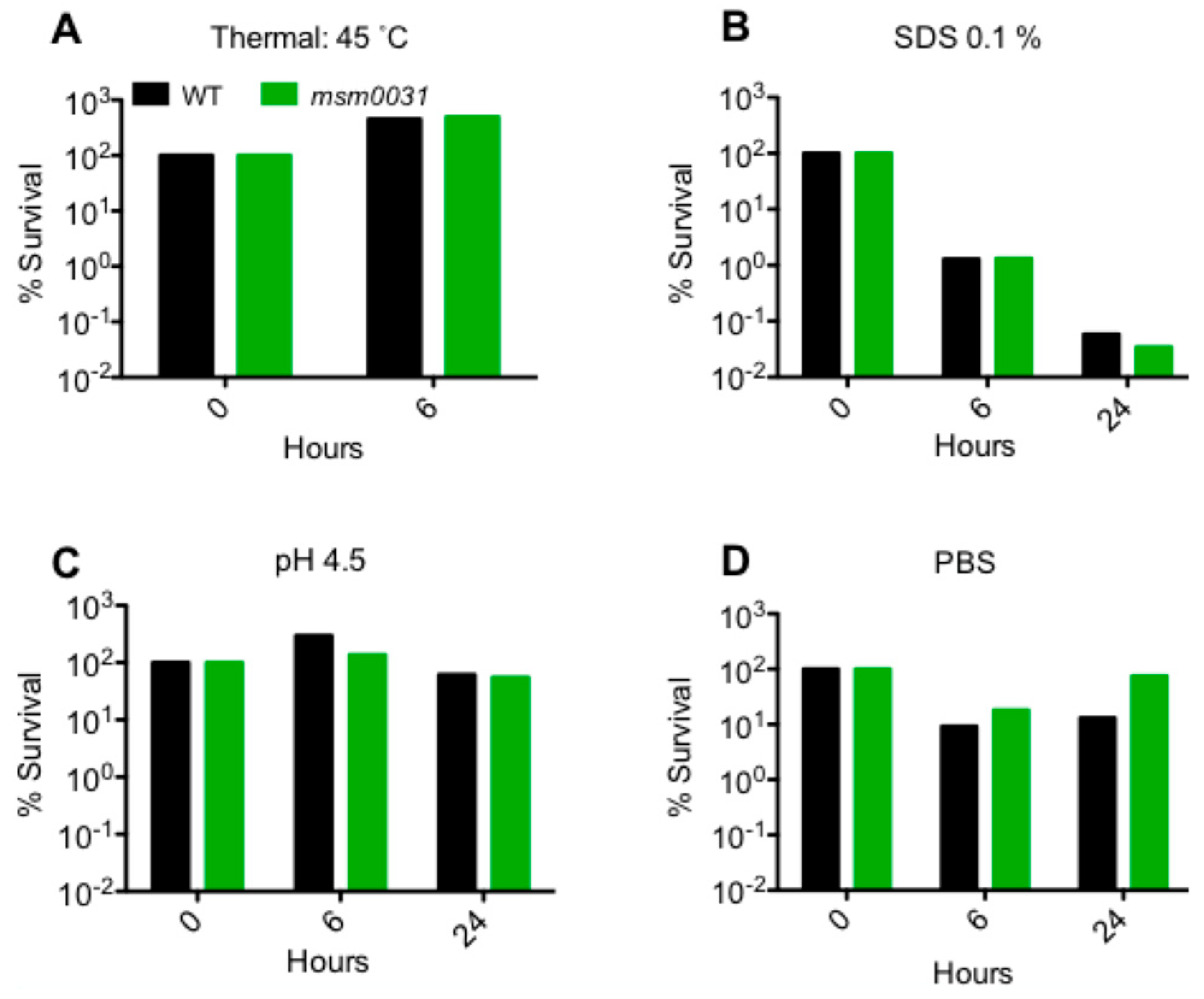
3.3. Stress Responses
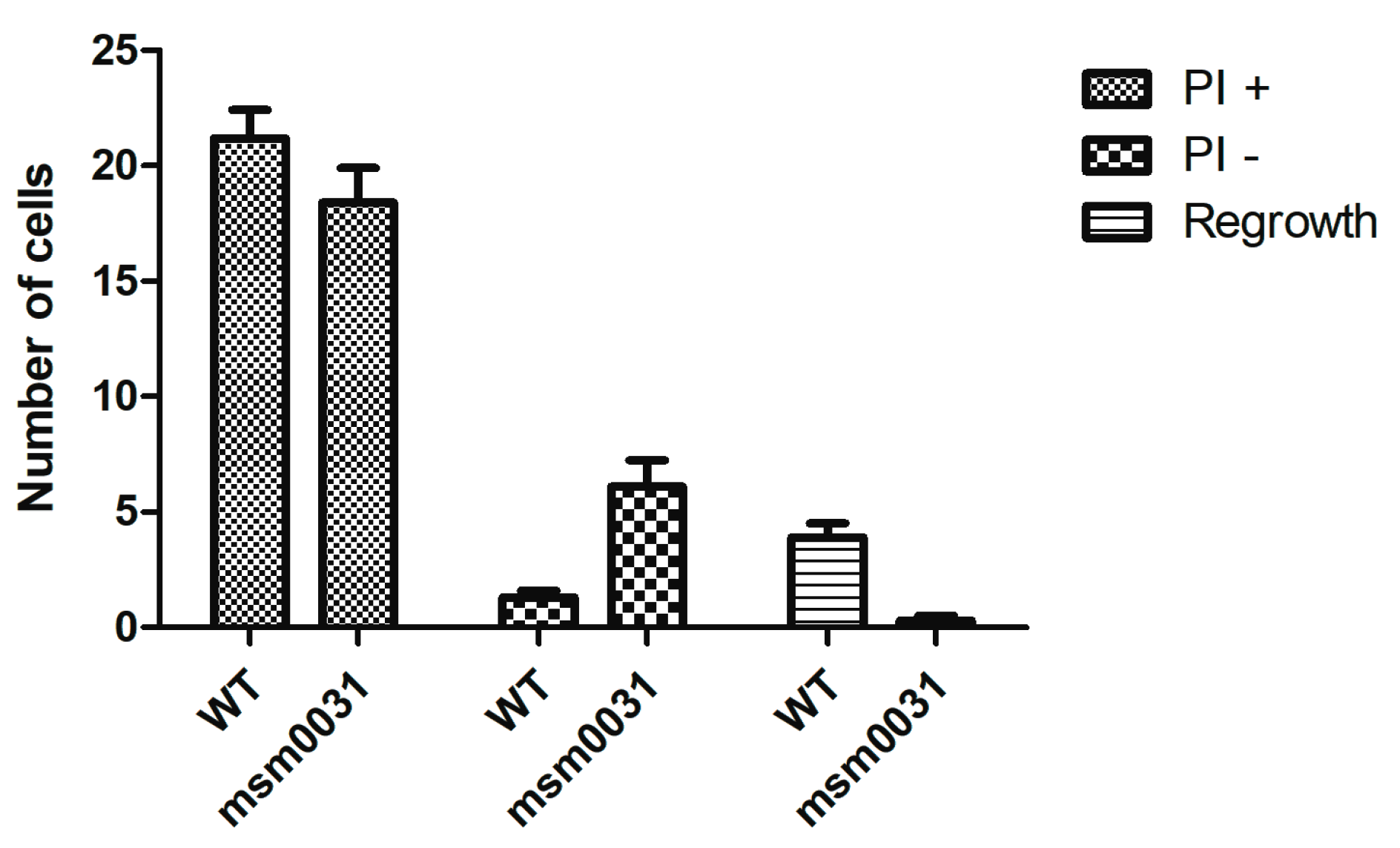
3.4. On-Chip Behavior of the msm0031 Transposon Mutant and Wild-Type M. smegmatis Cells
4. Discussion
Supplementary Materials
Funding
Acknowledgments
Conflicts of Interest
References
- Schneider, E.K.; Reyes-Ortega, F.; Velkov, T.; Li, J. Antibiotic-non-antibiotic combinations for combating extremely drug-resistant Gram-negative ‘superbugs’. Essays Biochem. 2017, 61, 115–125. [Google Scholar] [CrossRef] [PubMed]
- WHO. WHO GAP AMR Newsletter; The World Health Organization (WHO): Geneve, Switzerland, 2017; Volume 27. [Google Scholar]
- Maltezu, H.C.; Theodoriduo, M.; Daikos, G.L. Antimicrobial resistance and the current refugee crisis. J. Glob. Antimicrob. Resist. 2017, 10, 75–79. [Google Scholar] [CrossRef] [PubMed]
- Dhar, N.; McKinney, J.D. Microbial phenotypic heterogeneity and antibiotic tolerance. Curr. Opin. Microbiol. 2007, 10, 30–38. [Google Scholar] [CrossRef] [PubMed]
- Weibel, D.B.; DiLuzio, W.L.; Whitesides, G.M. Microfabrication meets microbiology. Nat. Rev. Microbiol. 2007, 5, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Rusconi, R.; Garren, M.; Stocker, R. Microfluidics expanding the frontiers of microbial ecology. Annu. Rev. Biophys. 2014, 43, 65–91. [Google Scholar] [CrossRef] [PubMed]
- Yawata, Y.; Nguyen, J.; Stocker, R.; Rusconi, R. Microfluidic Studies of Biofilm Formation in Dynamic Environments. J. Bacteriol. 2016, 198, 2589–2595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roggo, C.; van der Meer, J.R. Miniaturized and integrated whole cell living bacterial sensors in field applicable autonomous devices. Curr. Opin. Biotechnol. 2017, 45, 24–33. [Google Scholar] [CrossRef] [PubMed]
- Brehm-Stecher, B.F.; Johnson, E.A. Single-cell microbiology: Tools, technologies, and applications. Microbiol. Mol. Biol. Rev. 2004, 268, 538–599. [Google Scholar] [CrossRef] [PubMed]
- Wakamato, Y.; Dhar, N.; Chait, R.; Schnieder, K.; Signorino-Gelo, F.; Liebler, S.; McKinney, J.D. Dynamic persistence of antibiotic-stressed mycobacteria. Science 2013, 339, 91–95. [Google Scholar] [CrossRef] [PubMed]
- Stewart, E.J. Growing Unculturable Bacteria. J. Bacteriol. 2012, 194, 4151–4160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gu, H.; Hou, S.; Yongyat, C.; De Tore, S.; Ren, D. Patterned Biofilm Formation Reveals a Mechanism for Structural Heterogeneity in Bacterial Biofilms. Langmuir 2013, 29, 11145–11153. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.P.; Be’er, A.; Florin, E.-L.; Swinney, H.L. Collective motion and density fluctuations in bacterial colonies. Proc. Natl. Acad. Sci. USA 2010, 107, 13626–13630. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esteban, J.; García-Coca, M. Mycobacterium Biofilms. Front. Microbiol. 2017, 8, 2651. [Google Scholar] [CrossRef] [PubMed]
- Totani, T.; Nishiuchi, Y.; Tateishi, Y.; Yoshida, Y.; Kitanaka, H.; Niki, M.; Kaneko, Y.; Matsumoto, S. Effects of nutritional and ambient oxygen condition on biofilm formation in Mycobacterium avium subsp. hominissuis via altered glycolipid expression. Sci. Rep. 2017, 7, 41775. [Google Scholar] [CrossRef] [PubMed]
- Lebeaux, D.; Chauhan, A.; Rendueles, O.; Beloin, C. From in vitro to in vivo models of bacterial biofilm-related infections. Pathogens 2013, 2, 288–356. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Lambert, G.; Liao, D.; Kim, H.; Robin, K.; Tung, C.-K.; Pourmand, N.; Austin, R.H. Acceleration of Emergence of Bacterial Antibiotic Resistance in Connected Microenvironments. Science 2011, 333, 1764–1767. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Bos, J.; Tarnopolskiy, G.; Sturm, J.G.; Kim, H.; Pourmand, N.; Austin, R.H. You cannot tell a book by looking at the cover: Cryptic complexity in bacterial evolution. Biomicrofluidics 2014, 9, 052004. [Google Scholar] [CrossRef] [PubMed]
- Lambert, G.; Vyawahare, S.; Austin, R.H. Bacteria and game theory: The rise and fall of cooperation in spatially heterogeneous environments. Interface Focus 2014, 4, 20140029. [Google Scholar] [CrossRef] [PubMed]
- Mathis, R.; Ackermann, M. Response of single bacterial cells to stress gives rise to complex history dependence at the population level. Proc. Natl. Acad. Sci. USA 2016, 113, 4224–4229. [Google Scholar] [CrossRef] [PubMed]
- Mathis, R.; Ackermann, M. Asymmetric cellular memory in bacteria exposed to antibiotics. BMC Evolut. Biol. 2017, 17, 73. [Google Scholar] [CrossRef] [PubMed]
- Schreiber, F.; Littmann, S.; Lavik, G.; Escrig, S.; Meiborn, A.; Kuypers, M.M.; Ackermann, M. Phenotypic heterogeneity driven by nutrient limitation promotes growth in fluctuating environments. Nat. Microbiol. 2016, 1, 16055. [Google Scholar] [CrossRef] [PubMed]
- Hall, A.R.; Angst, D.C.; Schiessl, K.T.; Ackermann, M. Costs of antibiotic resistance—Separating trait effects and selective effects. Evol. Appl. 2015, 8, 261–272. [Google Scholar] [CrossRef] [PubMed]
- Benoît, Z.; Chami, M.; Houssin, C.; Dubochet, J.; Griffiths, G.; Daffé, M. Direct visualization of the outer membrane of mycobacteria and corynebacteria in their native state. J. Bacteriol. 2008, 90, 5672–5680. [Google Scholar] [CrossRef]
- Hoffmann, C.; Leis, A.; Niederweis, M.; Plitzko, J.M.; Engelhardt, H. Disclosure of the mycobacterial outer membrane: Cryo-electron tomography and vitreous sections reveal the lipid bilayer structure. Proc. Natl. Acad. Sci. USA 2008, 105, 3963–3967. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reyrat, J.M.; Kahn, D. Mycobacterium smegmatis: An absurd model for tuberculosis? Trends Microbiol. 2001, 9, 472–474. [Google Scholar] [CrossRef]
- Tyai, J.S.; Sharma, D. Mycobacterium smegmatis and tuberculosis. Tends Microbiol. 2002, 10, 68–69. [Google Scholar] [CrossRef]
- Haenni, M.; Moreillon, P. Fitness Cost and Impaired Survival in Penicillin-Resistant Streptococcus gordonii Isolates Selected in the Laboratory. Antimicrob. Agents Chemother. 2008, 52, 337–339. [Google Scholar] [CrossRef] [PubMed]
- Elitas, M. Isoniazid Killing of Mycobacterium smegmatis NADH Pyrophosphatase Mutant at Single-Cell Level using Microfluidics and Time-Lapse Microscopy. Sci. Rep. 2017, 7, 10770. [Google Scholar] [CrossRef] [PubMed]
- Patru, M.M.; Pavelka, M.S., Jr. A Role for the class a penicillin-binding protein PonA2 in the survival of Mycobacterium smegmatis under conditions of nonreplication. J. Bacteriol. 2010, 192, 3043–3054. [Google Scholar] [CrossRef] [PubMed]
- Bansal, A.; Kar, D.; Murugan, R.A.; Mallick, S.; Dutta, M.; Pandey, S.D.; Chowdhury, C.; Gosh, A. A putative low-molecular mass (LMM) penicillin-binding protein (PBP) of Mycobacterium smegmatis exhibits prominent physiological characters of DD-Carboxypeptidase and beta-lactamase. Microbiology 2015, 161, 1081–1091. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Enany, S.; Yoshida, Y.; Tateishi, Y.; Ozeki, Y.; Nishiyama, A.; Savitskaya, A.; Yamaguchi, T.; Ohara, Y.; Yamamoto, T.; Ato, M.; et al. Mycobacterial DNA-binding protein 1 is critical for long term survival of Mycobacterium smegmatis and simultaneously coordinates cellular functions. Sci. Rep. 2017, 7, 6810. [Google Scholar] [CrossRef] [PubMed]
- Kieser, K.J.; Baranowski, C.; Chao, M.C.; Long, J.E.; Sassetti, C.M.; Waldor, M.K.; Sacchettini, J.C.; Ioerger, T.R.; Rubin, E.J. Peptidoglycan synthesis in Mycobacterium tuberculosis is organized into networks with varying drug susceptibility. Proc. Natl. Acad. Sci. USA 2015, 112, 13087–13092. [Google Scholar] [CrossRef] [PubMed]
- Flores, A.R.; Parson, L.M.; Pavelka, M.S., Jr. Characterization of novel Mycobacterium tuberculosis and Mycobacterium smegmatis mutants hypersusceptible to beta-lactam antibiotics. J. Bacteriol. 2005, 187, 1892–1900. [Google Scholar] [CrossRef] [PubMed]
- Wendel, S.O.; Perera, A.S.; Pfromm, P.H.; Czermak, P.; Bossmann, S.H. Adaptation of Mycobacterium smegmatis to an industrial scale medium and isolation of the Mycobaterial PorinMspA. Open Microbiol. J. 2013, 7, 92–98. [Google Scholar] [CrossRef] [PubMed]
- Belisle, J.T.; Sonnenberg, M.G. Isolation of genomic DNA from mycobacteria. Methods Mol. Biol. 1998, 101, 31–44. [Google Scholar] [CrossRef] [PubMed]
- Gebhard, S.; Humpel, A.; McLellan, A.D.; Cook, G.M. The alternative sigma factor SigF of Mycobacterium smegmatis is required for survival of heat shock, acidic pH and oxidative stress. Microbiology 2008, 154, 2786–2795. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marrakchi, H.; Lanéelle, G.; Quémard, A. InhA, a target of the antituberculous drug isoniazid, is involved in a mycobacterial fatty acid elongation system, FAS-II. Microbiology 2000, 146, 289–296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Agrawal, S.; Panchagnula, R. In vitro analysis of rifampicin and its effect on quality control tests of rifampicin containing dosage forms. Pharmazie 2004, 59, 775–781. [Google Scholar] [PubMed]
- Singh, S.; Mariappan, T.T.; Sharda, N.; Singh, B. Degradation of rifampicin, isoniazid and pyrazinamide from prepared mixtures and marketed single and combinational products under acid conditions. Pharm. Pharmacol. Commun. 2000, 6, 491–494. [Google Scholar] [CrossRef]
- Quan, S.; Venter, H.; Dabbs, E.R. Ribosylative inactivation of rifampin by Mycobacterium Smegmatis is a principal contributor to its low susceptibility to this antibiotic. Antimicrob. Agents Chemother. 1997, 41, 2456–2460. [Google Scholar] [CrossRef] [PubMed]


| Antibiotics | FSR ± SE WT | FSR ± SE msm0031 |
|---|---|---|
| INH (50 µg/mL) | 1 ± 0.160 | 0.024 ± 0.040 *** |
| EMB (5 µg/mL) | 1 ± 0.630 | 0.028 ± 0.050 ** |
| RIF (200 µg/mL) | 1 ± 0.001 | 1.670 ± 0.003 |
| ETH (200 µg/mL) | 1 ± 3.000 | 0.040 ± 3.700 * |
© 2018 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elitas, M. On-Chip Isoniazid Exposure of Mycobacterium smegmatis Penicillin-Binding Protein (PBP) Mutant Using Time-Lapse Fluorescent Microscopy. Micromachines 2018, 9, 561. https://doi.org/10.3390/mi9110561
Elitas M. On-Chip Isoniazid Exposure of Mycobacterium smegmatis Penicillin-Binding Protein (PBP) Mutant Using Time-Lapse Fluorescent Microscopy. Micromachines. 2018; 9(11):561. https://doi.org/10.3390/mi9110561
Chicago/Turabian StyleElitas, Meltem. 2018. "On-Chip Isoniazid Exposure of Mycobacterium smegmatis Penicillin-Binding Protein (PBP) Mutant Using Time-Lapse Fluorescent Microscopy" Micromachines 9, no. 11: 561. https://doi.org/10.3390/mi9110561