A Fluidic Device for Immunomagnetic Separation of Foodborne Bacteria Using Self-Assembled Magnetic Nanoparticle Chains
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Design and Fabrication of the Fluidic Chip
2.3. Modification of the Immune Magnetic Nanoparticles
2.4. Immunomagnetic Separation of the Target Bacteria in the Fluidic Chip
2.5. Preparation and Enumeration of the Target Bacteria
2.6. Calculation of Separation Efficiency
3. Results and Discussions
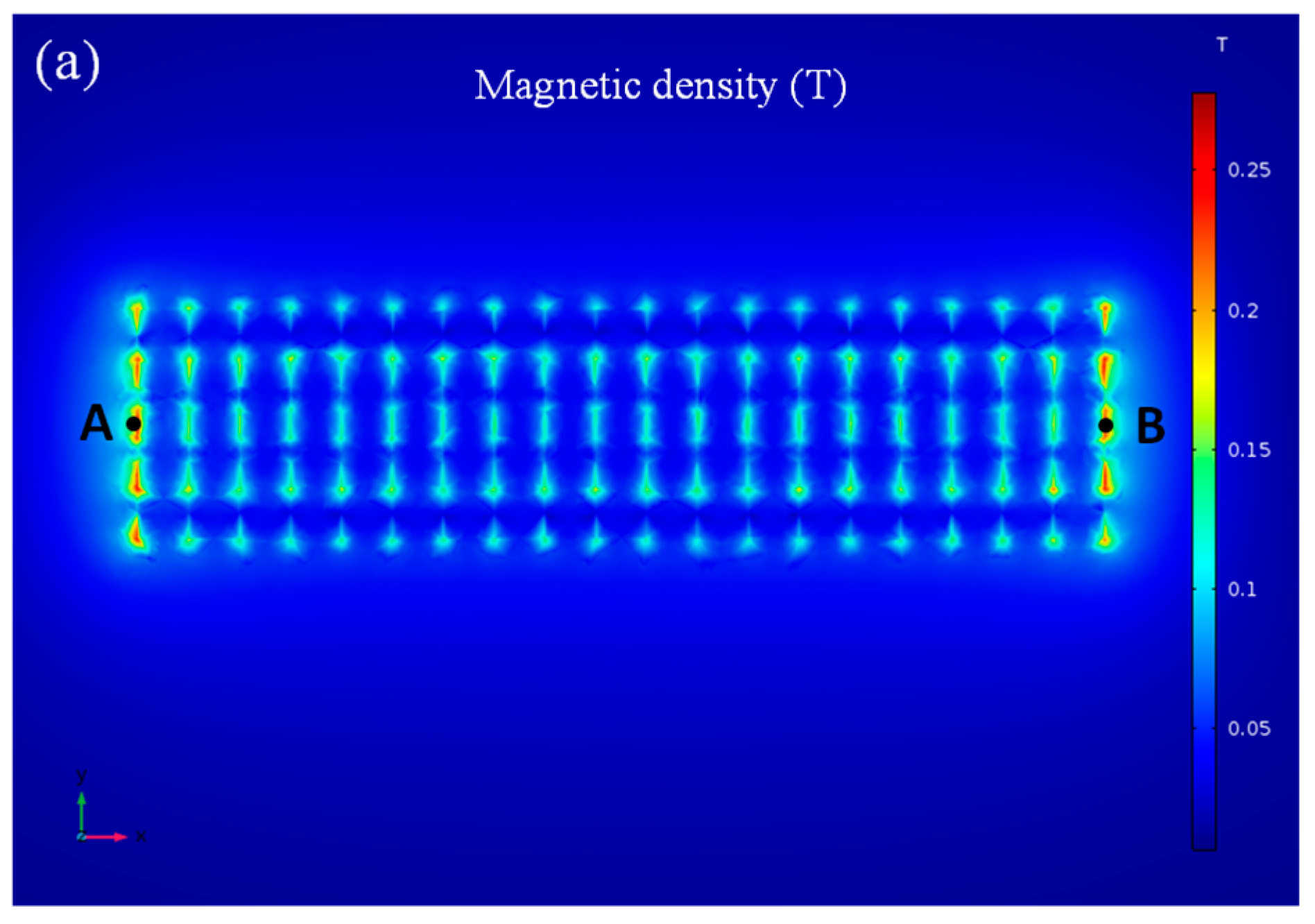
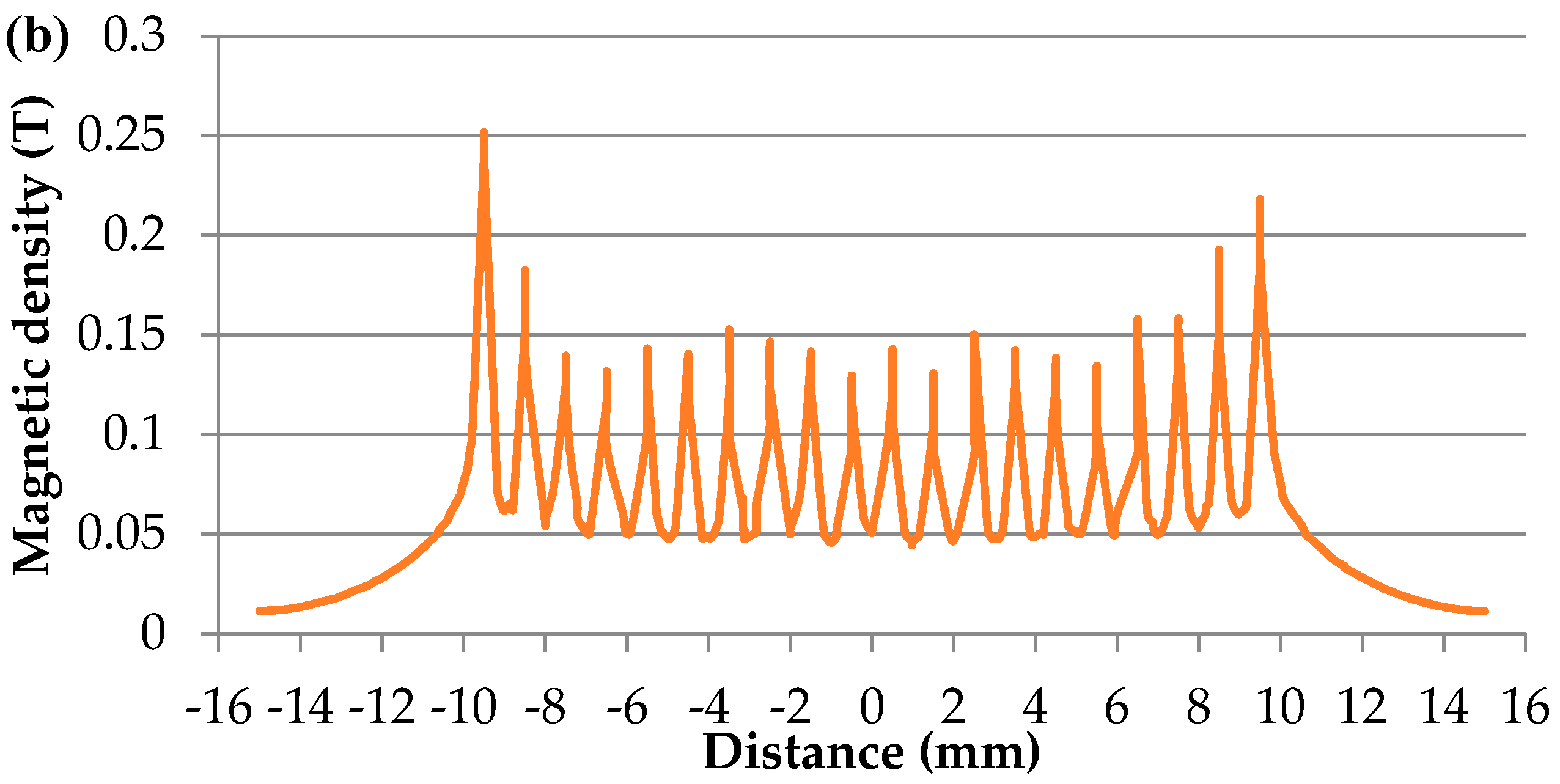
3.1. Simulation of the Dot-Array Magnetic Field
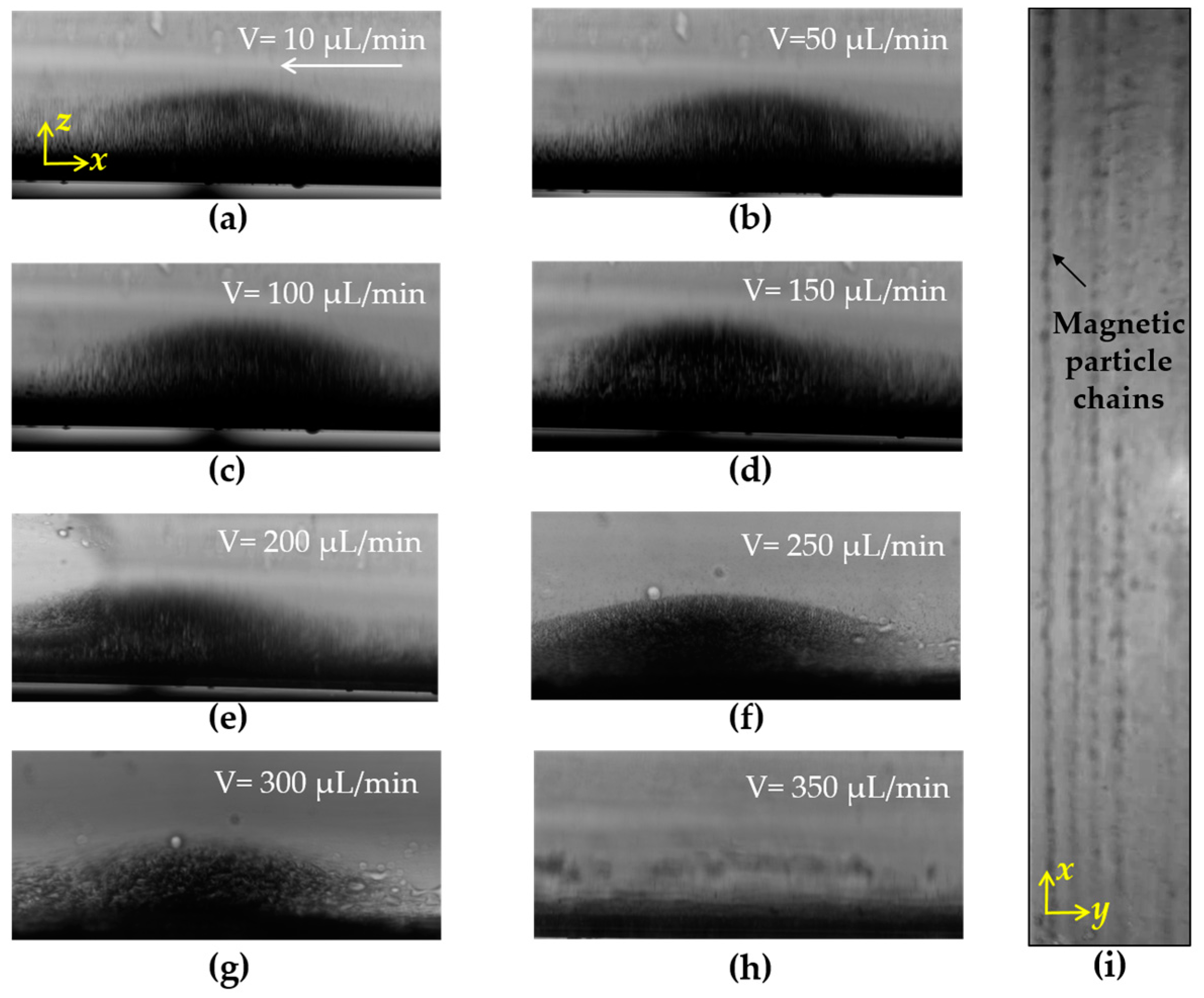
3.2. Forming of the Magnetic Nanoparticle Chains
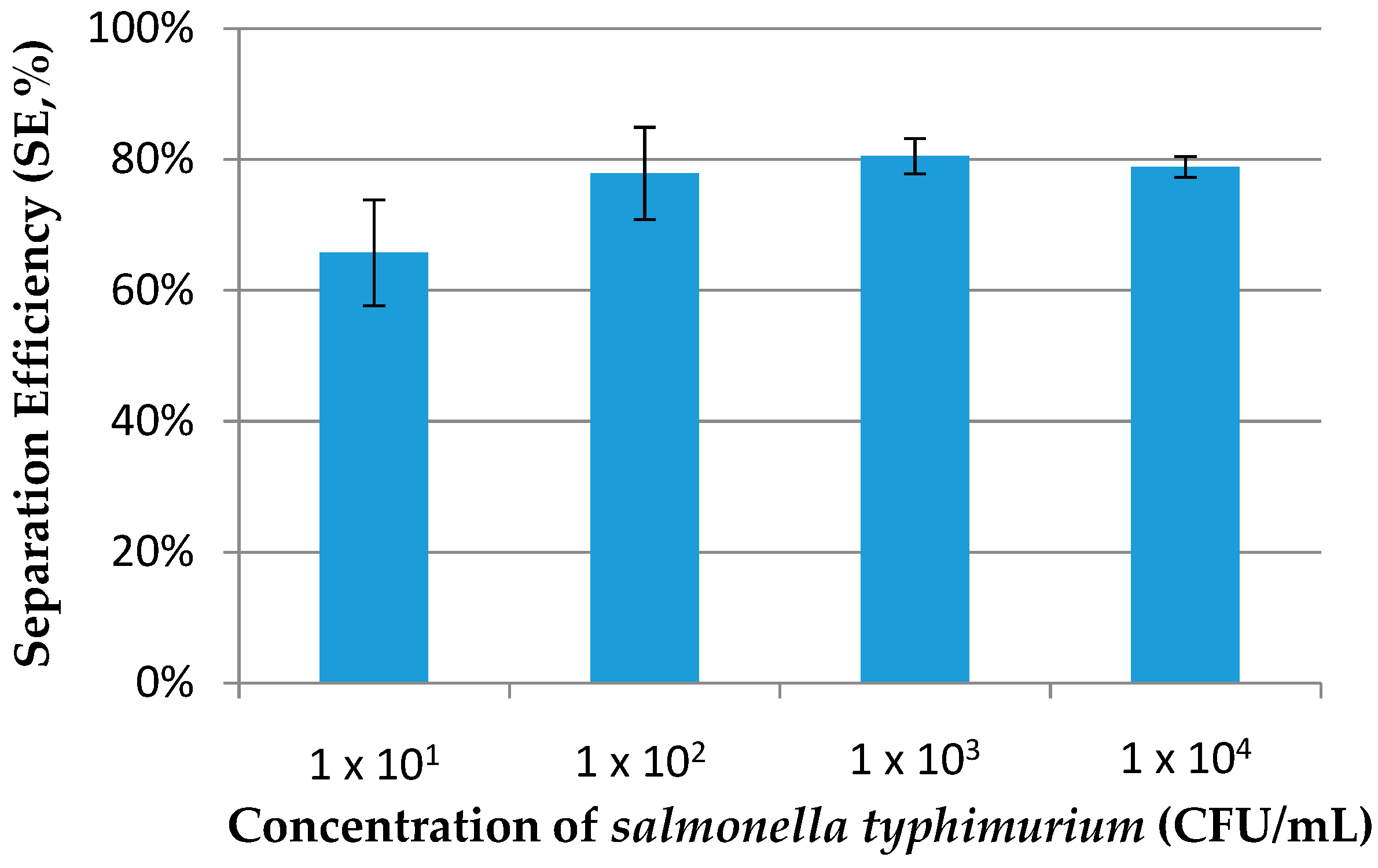
3.3. Immunomagnetic Separation of Salmonella Typhimurium in the Fluidic Chip
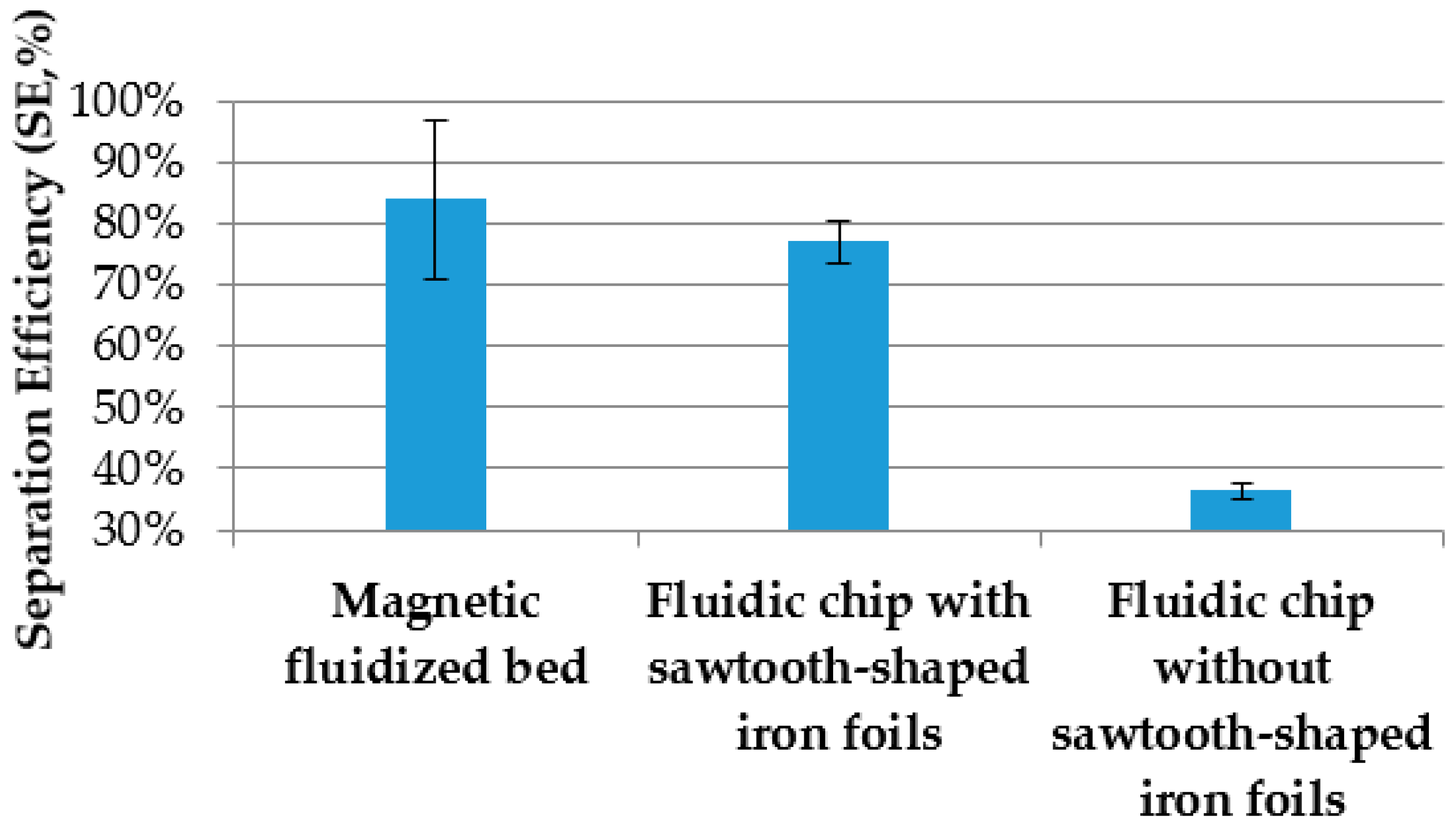
3.4. Comparsion of the Separation with and without Sawtooth-Shaped Iron Foils
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chockalingam, A.M.; Babu, H.K.R.R.; Chittor, R.; Tiwari, J.P. Gum arabic modified Fe3O4 nanoparticles cross linked with collagen for isolation of bacteria. J. Nanobiotechnol. 2010, 8, 30. [Google Scholar] [CrossRef] [PubMed]
- Luo, K.; Jeong, K.-B.; You, S.-M.; Lee, D.-H.; Jung, J.-Y.; Kim, Y.-R. Surface-engineered starch magnetic microparticles for highly effective separation of a broad range of bacteria. ACS Sustain. Chem. Eng. 2018, 6, 13524–13531. [Google Scholar] [CrossRef]
- Liu, J.; Shang, H.; Wang, X.; Liu, C. A portable membrane-based analysis system for rapid and sensitive detection of bacteria. In Proceedings of the 2010 8th World Congress on Intelligent Control and Automation, Jinan, China, 7–9 July 2010; pp. 2951–2954. [Google Scholar]
- Liu, B.C.; Chen, T.I.; Liu, C.H. A cell separation chip using micro-structures filter and multi-frequencies dielectrophoresis. In Proceedings of the 13th International Conference on Solid-State Sensors, Actuators and Microsystems, 2005. Digest of Technical Papers. TRANSDUCERS '05, Seoul, Korea, 5–9 June 2005. [Google Scholar]
- Ji, H.S.; Park, B.H.; Oh, S.J.; Choi, G.; Kim, D.H.; Lee, E.Y.; Seo, T.S. Development of a high-throughput centrifugal loop-mediated isothermal amplification microdevice for multiplex foodborne pathogenic bacteria detection. Sens. Actuators B Chem. 2017, 246, 146–153. [Google Scholar]
- Czilwik, G.; Messinger, T.; Strohmeier, O.; Wadle, S.; Von, S.F.; Paust, N.; Roth, G.; Zengerle, R.; Saarinen, P.; Niittymãki, J. Rapid and fully automated bacterial pathogen detection on a centrifugal-microfluidic labdisk using highly sensitive nested PCR with integrated sample preparation. Lab Chip 2015, 15, 3749–3759. [Google Scholar] [CrossRef] [PubMed]
- Litvinov, J.; Moen, S.T.; Koh, C.Y.; Singh, A.K. Centrifugal sedimentation immunoassays for multiplexed detection of enteric bacteria in ground water. Biomicrofluidics 2016, 10, 014103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mao, Y.; Huang, X.; Xiong, S.; Xu, H.; Aguilar, Z.P.; Xiong, Y. Large-volume immunomagnetic separation combined with multiplex PCR assay for simultaneous detection of listeria monocytogenes and listeria ivanovii in lettuce. Food Control. 2016, 59, 601–608. [Google Scholar] [CrossRef]
- Lin, J.; Li, M.; Li, Y.; Chen, Q. A high gradient and strength bioseparator with nano-sized immunomagnetic particles for specific separation and efficient concentration of E. coli O157:H7. J. Magn. Magn. Mater. 2015, 378, 206–213. [Google Scholar] [CrossRef]
- Zheng, Q.; Mikå-Krajnik, M.; Yang, Y.; Xu, W.; Yuk, H.G. Real-time PCR method combined with immunomagnetic separation for detecting healthy and heat-injured salmonella typhimurium on raw duck wings. Int. J. Food Microbiol. 2014, 186, 6–13. [Google Scholar] [CrossRef] [PubMed]
- Guyer, C.; Tao, S.; Alex, M. Magnetic micro-particle immune separation combined with enzyme linked immunosorbent assay for quick detection of salmonella typhimurium in raw eggs. Nano Res. Appl. 2016, 2, 1. [Google Scholar]
- Najafi, R.; Mukherjee, S.; Jr, H.J.; Sharma, A.; Banerjee, P. Development of a rapid capture-cum-detection method for Escherichia coli O157 from apple juice comprising nano-immunomagnetic separation in tandem with surface enhanced Raman scattering. Int. J. Food Microbiol. 2014, 189, 89–97. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Cai, R.; Yuan, Y.; Niu, C.; Hu, Z.; Yue, T. An immunomagnetic separation-real-time PCR system for the detection of alicyclobacillus acidoterrestris in fruit products. Int. J. Food Microbiol. 2014, 175, 30–35. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.; Xu, F.; Xu, H.; Aguilar, Z.P.; Niu, R.; Yuan, Y.; Sun, J.; You, X.; Lai, W.; Xiong, Y. Magnetic nano-beads based separation combined with propidium monoazide treatment and multiplex PCR assay for simultaneous detection of viable salmonella typhimurium, Escherichia coli O157:H7 and listeria monocytogenes in food products. Food Microbiol. 2013, 34, 418–424. [Google Scholar] [CrossRef] [PubMed]
- Hyeon, J.Y.; Deng, X. Rapid detection of salmonella in raw chicken breast using real-time PCR combined with immunomagnetic separation and whole genome amplification. Food Microbiol. 2017, 63, 111–116. [Google Scholar] [CrossRef] [PubMed]
- Jeníková, G.; Pazlarová, J.; Demnerová, K. Detection of salmonella in food samples by the combination of immunomagnetic separation and PCR assay. Int. Microbiol. Off. J. Span. Soc. Microbiol. 2000, 3, 225–229. [Google Scholar]
- Wang, Z.; Yue, T.; Yuan, Y.; Cai, R.; Niu, C.; Guo, C. Development and evaluation of an immunomagnetic separation—ELISA for the detection of Alicyclobacillus spp. in apple juice. Int. J. Food Microbiol. 2013, 166, 28–33. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Hou, N.; Jin, M.; Qiu, Z.; Wang, J.; Zhang, B.; Wang, X.; Wang, J.; Zhou, D.; Li, J. A novel enzyme-linked immunosorbent assay for detection of Escherichia coli O157:H7 using immunomagnetic and beacon gold nanoparticles. Gut Pathog. 2014, 6, 14. [Google Scholar] [CrossRef] [PubMed]
- Huang, Z.; Xia, S.Q.; Liu, D.F.; Liu, C.W.; Lai, W.H. Fluorescent microspheres lateral flow assay based on immunomagnetic separation for detection of S. Choleraesuis. Chin. J. Anal. Chem. 2017, 45, 217–223. [Google Scholar] [CrossRef]
- Cui, X.; Xiong, Q.-R.; Xiong, Y.-H.; Shan, S.; Lai, W.-H. Establishing of a method combined immunomagnetic separation with colloidal gold lateral flow assay and its application in rapid detection of Escherichia coli O157:H7. Chin. J. Anal. Chem. 2013, 41, 1812–1816. [Google Scholar] [CrossRef]
- Lee, J.J.; Jeong, K.J.; Hashimoto, M.; Kwon, A.H.; Rwei, A.; Shankarappa, S.A.; Tsui, J.H.; Kohane, D.S. Synthetic ligand-coated magnetic nanoparticles for microfluidic bacterial separation from blood. Nano Lett. 2014, 14, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Karle, M.; Miwa, J.; Czilwik, G.; Auwärter, V.; Roth, G.; Zengerle, R.; Von, S.F. Continuous microfluidic DNA extraction using phase-transfer magnetophoresis. Lab Chip 2010, 10, 3284–3290. [Google Scholar] [CrossRef] [PubMed]
- Lee, W.; Kwon, D.; Chung, B.; Jung, G.Y.; Au, A.; Folch, A.; Jeon, S. Ultrarapid detection of pathogenic bacteria using a 3D immunomagnetic flow assay. Anal. Chem. 2014, 86, 6683–6688. [Google Scholar] [CrossRef] [PubMed]
- Nagaoka, Y.; Morimoto, H.; Maekawa, T. Dynamics of disklike clusters formed in a magnetorheological fluid under a rotational magnetic field. Phys. Rev. E Stat. Nonlinear Soft Matter Phys. 2005, 71, 032502. [Google Scholar] [CrossRef] [PubMed]
- Ukai, T.; Maekawa, T. Patterns formed by paramagnetic particles in a horizontal layer of a magnetorheological fluid subjected to a dc magnetic field. Phys. Rev. E Stat. Nonlinear Soft Matter Phys. 2004, 69, 032501. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Lawrence, E.M.; Wu, A.; Ivey, M.L.; Flores, G.A.; Javier, K.; Bibette, J.; Richard, J. Field-induced structures in ferrofluid emulsions. Phys. Rev. Lett. 1995, 74, 2828–2831. [Google Scholar] [CrossRef] [PubMed]
- Le Nel, A.; Minc, N.; Smadja, C.; Slovakova, M.; Bilkova, Z.; Peyrin, J.M.; Viovy, J.L.; Taverna, M. Controlled proteolysis of normal and pathological prion protein in a microfluidic chip. Lab Chip 2008, 8, 294–301. [Google Scholar] [CrossRef] [PubMed]
- Mohamadi, R.M.; Svobodova, Z.; Bilkova, Z.; Otto, M.; Taverna, M.; Descroix, S.; Viovy, J.L. An integrated microfluidic chip for immunocapture, preconcentration and separation of beta-amyloid peptides. Biomicrofluidics 2015, 9, 054117. [Google Scholar] [CrossRef] [PubMed]
- Saliba, A.E.; Saias, L.; Psychari, E.; Minc, N.; Simon, D.; Bidard, F.C.; Mathiot, C.; Pierga, J.Y.; Fraisier, V.; Salamero, J.; et al. Microfluidic sorting and multimodal typing of cancer cells in self-assembled magnetic arrays. Proc. Natl. Acad. Sci. USA 2010, 107, 14524–14529. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Armbrecht, L.; Dincer, C.; Kling, A.; Horak, J.; Kieninger, J.; Urban, G. Self-assembled magnetic bead chains for sensitivity enhancement of microfluidic electrochemical biosensor platforms. Lab Chip 2015, 15, 4314–4321. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alexandre, L.; Pereiro, I.; Bendali, A.; Tabnaoui, S.; Srbova, J.; Bilkova, Z.; Deegan, S.; Joshi, L.; Viovy, J.L.; Malaquin, L.; et al. A microfluidic fluidized bed to capture, amplify and detect bacteria from raw samples. Methods Cell Biol. 2018, 147, 59–75. [Google Scholar] [PubMed]
- Bouguelia, S.; Roupioz, Y.; Slimani, S.; Mondani, L.; Casabona, M.G.; Durmort, C.; Vernet, T.; Calemczuk, R.; Livache, T. On-chip microbial culture for the specific detection of very low levels of bacteria. Lab Chip 2013, 13, 4024–4032. [Google Scholar] [CrossRef] [PubMed]
- Pereiro, I.; Bendali, A.; Tabnaoui, S.; Alexandre, L.; Srbova, J.; Bilkova, Z.; Deegan, S.; Joshi, L.; Viovy, J.L.; Malaquin, L.; et al. A new microfluidic approach for the one-step capture, amplification and label-free quantification of bacteria from raw samples. Chem. Sci. 2017, 8, 1329–1336. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Srbova, J.; Krulisova, P.; Holubova, L.; Pereiro, I.; Bendali, A.; Hamiot, A.; Podzemna, V.; Macak, J.; Dupuy, B.; Descroix, S. Advanced immunocapture of milk-borne salmonella by microfluidic magnetically stabilized fluidized bed. Electrophoresis 2017, 39, 526–533. [Google Scholar] [CrossRef] [PubMed]
- Pereiro, I.; Tabnaoui, S.; Fermigier, M.; Du, R.O.; Descroix, S.; Viovy, J.L.; Malaquin, L. Magnetic fluidized bed for solid phase extraction in microfluidic systems. Lab Chip 2017, 17, 1603–1615. [Google Scholar] [CrossRef] [PubMed] [Green Version]



© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cai, G.; Wang, S.; Zheng, L.; Lin, J. A Fluidic Device for Immunomagnetic Separation of Foodborne Bacteria Using Self-Assembled Magnetic Nanoparticle Chains. Micromachines 2018, 9, 624. https://doi.org/10.3390/mi9120624
Cai G, Wang S, Zheng L, Lin J. A Fluidic Device for Immunomagnetic Separation of Foodborne Bacteria Using Self-Assembled Magnetic Nanoparticle Chains. Micromachines. 2018; 9(12):624. https://doi.org/10.3390/mi9120624
Chicago/Turabian StyleCai, Gaozhe, Siyuan Wang, Lingyan Zheng, and Jianhan Lin. 2018. "A Fluidic Device for Immunomagnetic Separation of Foodborne Bacteria Using Self-Assembled Magnetic Nanoparticle Chains" Micromachines 9, no. 12: 624. https://doi.org/10.3390/mi9120624




