Update on Immunohistochemistry for the Diagnosis of Lung Cancer
Abstract
:1. Introduction
2. Adenocarcinoma vs. Squamous Cell Carcinoma
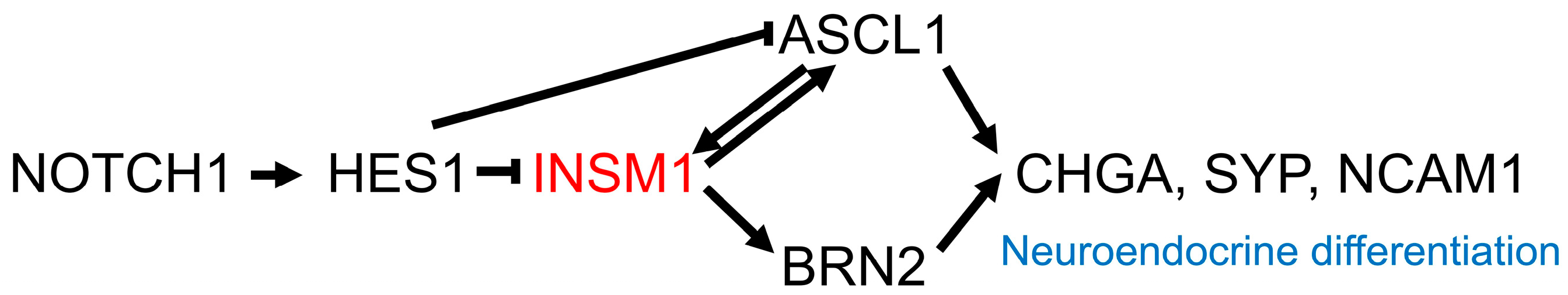
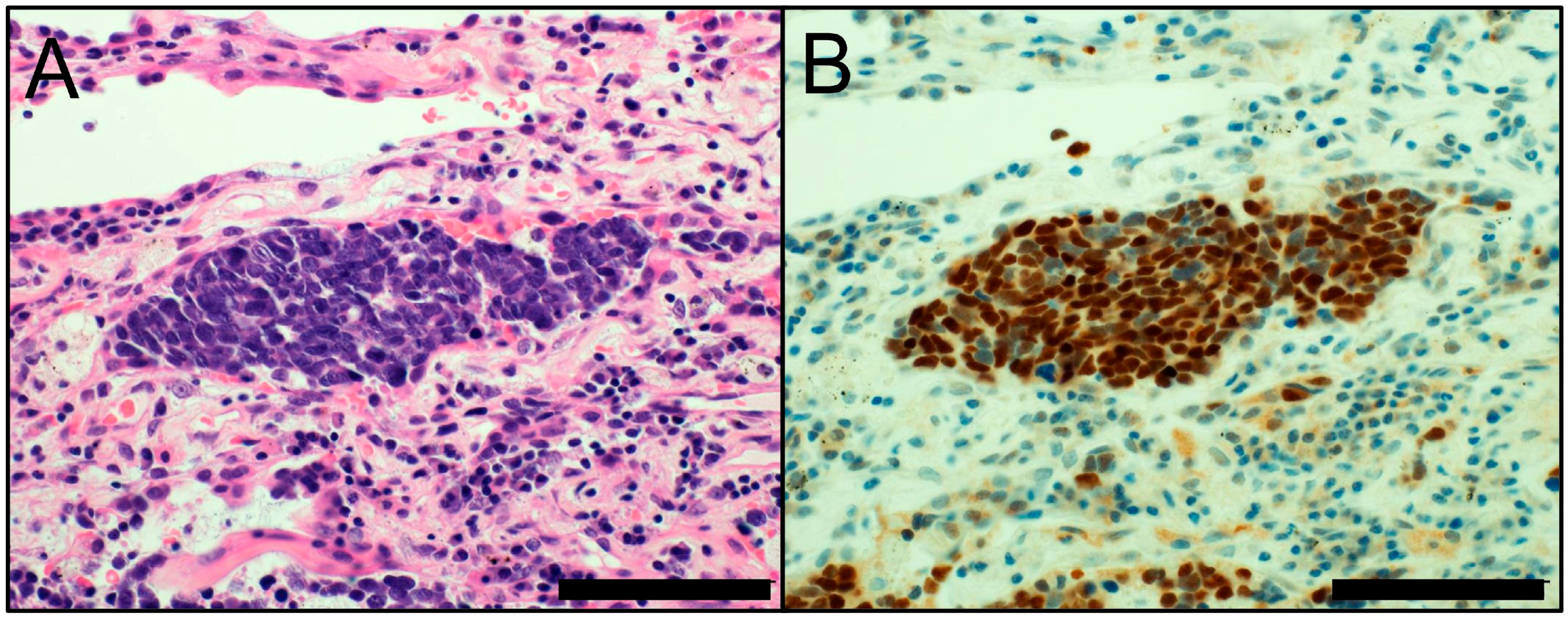
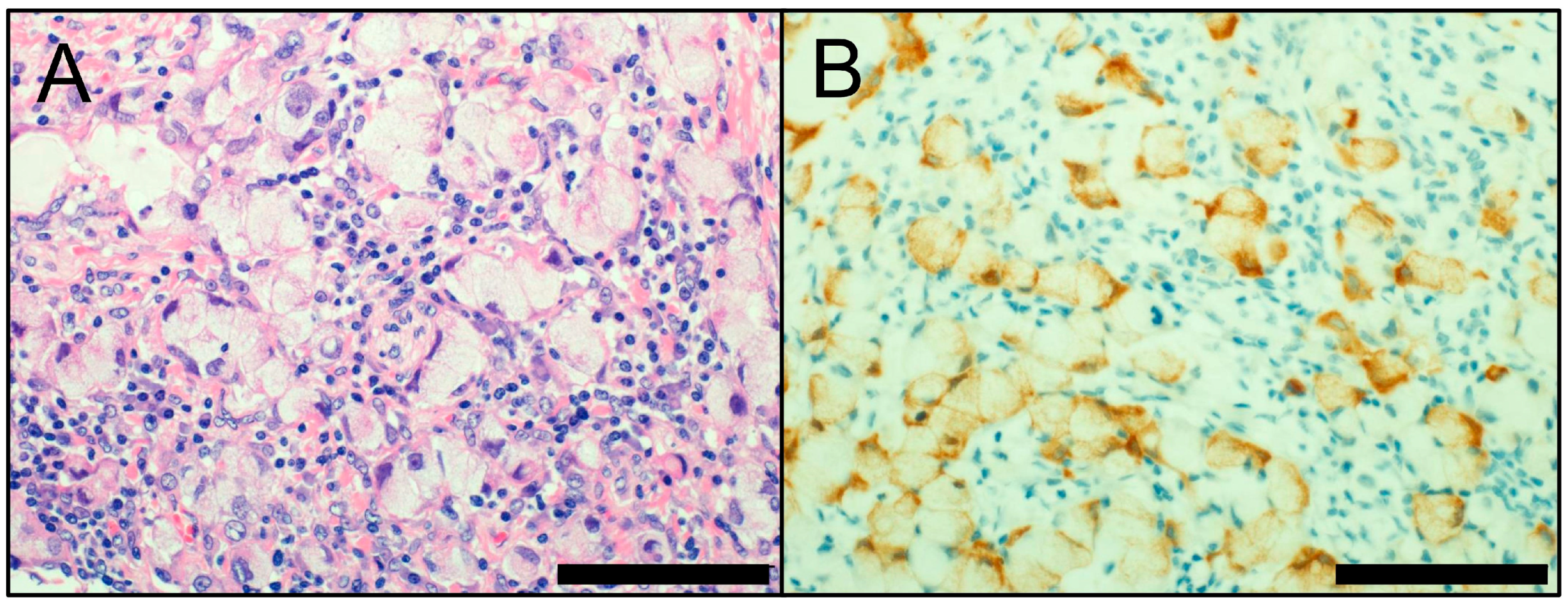
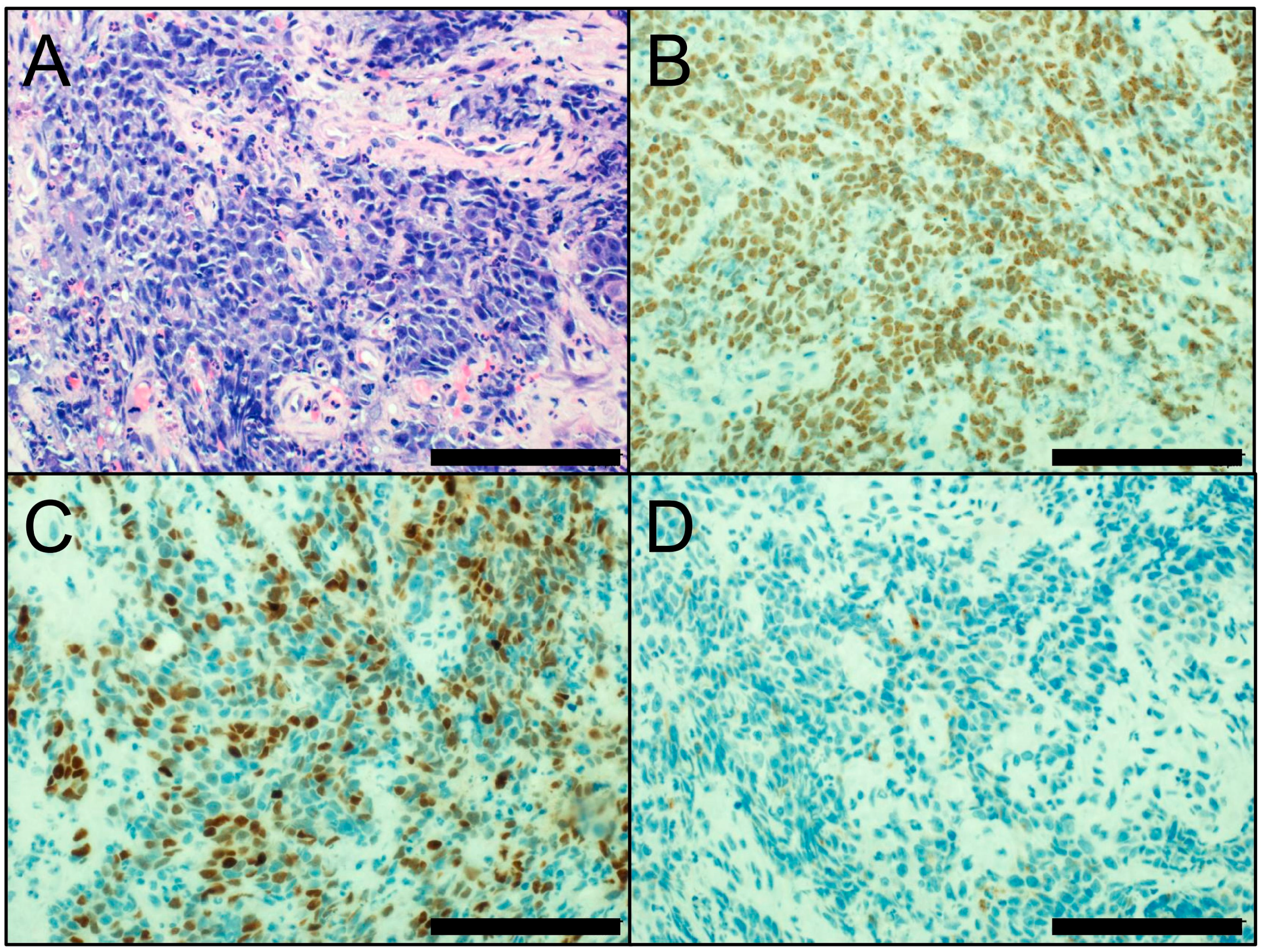
3. Neuroendocrine Markers
4. ALK, ROS1, and EGFR
5. PD-L1 (CD274)
6. Lung Carcinoma vs. Malignant Mesothelioma
7. NUT Carcinoma
8. Conclusions and Future Directions
Acknowledgments
Conflicts of Interest
Abbreviations
| FDA | Food and Drug Administration |
| FISH | fluorescence in situ hybridization |
| HE | hematoxylin-eosin |
| HGNET | high-grade neuroendocrine tumor |
| IASLC | International Association for the Study of Lung Cancer |
| INSM1 | insulinoma-associated protein 1 |
| LCNEC | large cell neuroendocrine carcinoma |
| MM | malignant mesothelioma |
| NSCLC | non-small cell lung carcinoma |
| SCLC | small cell lung carcinoma |
| SqCC | squamous cell carcinoma |
| TKI | tyrosine kinase inhibitor |
| WHO | World Health Organization |
References
- Cagle, P.T.; Allen, T.C.; Bernicker, E.H.; Ge, Y.; Haque, A.; Barrios, R. Impact of recent developments in lung cancer on the practice of pathology. Arch. Pathol. Lab. Med. 2016, 140, 322–325. [Google Scholar] [CrossRef] [PubMed]
- Thunnissen, E.; Allen, T.C.; Adam, J.; Aisner, D.L.; Beasley, M.B.; Borczuk, A.C.; Cagle, P.T.; Capelozzi, V.L.; Cooper, W.; Hariri, L.P.; et al. Immunohistochemistry of pulmonary biomarkers: A perspective from members of the pulmonary pathology society. Arch. Pathol. Lab. Med. 2018, 142. [Google Scholar] [CrossRef] [PubMed]
- Mino-Kenudson, M. Immunohistochemistry for predictive biomarkers in non-small cell lung cancer. Transl. Lung Cancer Res. 2017, 6, 570–587. [Google Scholar] [CrossRef] [PubMed]
- Woo, J.S.; Reddy, O.L.; Koo, M.; Xiong, Y.; Li, F.; Xu, H. Application of Immunohistochemistry in the Diagnosis of Pulmonary and Pleural Neoplasms. Arch. Pathol. Lab. Med. 2017, 141, 1195–1213. [Google Scholar] [CrossRef] [PubMed]
- Rossi, G.; Ragazzi, M.; Tamagnini, I.; Mengoli, M.C.; Vincenzi, G.; Barbieri, F.; Piccioli, S.; Bisagni, A.; Vavala, T.; Righi, L.; et al. Does immunohistochemistry represent a robust alternative technique in determining drugable predictive gene alterations in non-small cell lung cancer? Curr. Drug Targets 2017, 18, 13–26. [Google Scholar] [CrossRef] [PubMed]
- Travis, W.D.; Brambilla, E.; Burke, A.P.; Marx, A.; Nicholson, A.G. WHO Classification of Tumours of the Lung, Pleura, Thymus and Heart, 4th ed.; International Agency for Research on Cancer (IARC): Lyon, France, 2015. [Google Scholar]
- The Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature 2014, 511, 543–550. [Google Scholar]
- The Cancer Genome Atlas Research Network. Comprehensive genomic characterization of squamous cell lung cancers. Nature 2012, 489, 519–525. [Google Scholar]
- George, J.; Lim, J.S.; Jang, S.J.; Cun, Y.; Ozretic, L.; Kong, G.; Leenders, F.; Lu, X.; Fernandez-Cuesta, L.; Bosco, G.; et al. Comprehensive genomic profiles of small cell lung cancer. Nature 2015, 524, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Peifer, M.; Fernandez-Cuesta, L.; Sos, M.L.; George, J.; Seidel, D.; Kasper, L.H.; Plenker, D.; Leenders, F.; Sun, R.; Zander, T.; et al. Integrative genome analyses identify key somatic driver mutations of small-cell lung cancer. Nat. Genet. 2012, 44, 1104–1110. [Google Scholar] [CrossRef] [PubMed]
- Rudin, C.M.; Durinck, S.; Stawiski, E.W.; Poirier, J.T.; Modrusan, Z.; Shames, D.S.; Bergbower, E.A.; Guan, Y.; Shin, J.; Guillory, J.; et al. Comprehensive genomic analysis identifies SOX2 as a frequently amplified gene in small-cell lung cancer. Nat. Genet. 2012, 44, 1111–1116. [Google Scholar] [CrossRef] [PubMed]
- Rizvi, N.A.; Hellmann, M.D.; Snyder, A.; Kvistborg, P.; Makarov, V.; Havel, J.J.; Lee, W.; Yuan, J.; Wong, P.; Ho, T.S.; et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 2015, 348, 124–128. [Google Scholar] [CrossRef] [PubMed]
- Hwang, D.H.; Sholl, L.M.; Rojas-Rudilla, V.; Hall, D.L.; Shivdasani, P.; Garcia, E.P.; MacConaill, L.E.; Vivero, M.; Hornick, J.L.; Kuo, F.C.; et al. KRAS and NKX2-1 mutations in invasive mucinous adenocarcinoma of the lung. J. Thorac. Oncol. 2016, 11, 496–503. [Google Scholar] [CrossRef] [PubMed]
- Polley, E.; Kunkel, M.; Evans, D.; Silvers, T.; Delosh, R.; Laudeman, J.; Ogle, C.; Reinhart, R.; Selby, M.; Connelly, J.; et al. Small Cell Lung Cancer Screen of Oncology Drugs, Investigational Agents, and Gene and microRNA Expression. J. Natl. Cancer Inst. 2016, 108. [Google Scholar] [CrossRef] [PubMed]
- Thunnissen, E.; van der Oord, K.; den Bakker, M. Prognostic and predictive biomarkers in lung cancer. A review. Virchows Arch. 2014, 464, 347–358. [Google Scholar] [CrossRef] [PubMed]
- Sabir, S.R.; Yeoh, S.; Jackson, G.; Bayliss, R. EML4-ALK Variants: Biological and Molecular Properties, and the Implications for Patients. Cancers 2017, 9, 118. [Google Scholar] [CrossRef] [PubMed]
- Inamura, K. Diagnostic and Therapeutic Potential of MicroRNAs in Lung Cancer. Cancers 2017, 9, 49. [Google Scholar] [CrossRef] [PubMed]
- Travis, W.D.; Brambilla, E.; Noguchi, M.; Nicholson, A.G.; Geisinger, K.R.; Yatabe, Y.; Beer, D.G.; Powell, C.A.; Riely, G.J.; Van Schil, P.E.; et al. International association for the study of lung cancer/american thoracic society/european respiratory society international multidisciplinary classification of lung adenocarcinoma. J. Thorac. Oncol. 2011, 6, 244–285. [Google Scholar] [CrossRef] [PubMed]
- Inamura, K. Lung Cancer: Understanding Its Molecular Pathology and the 2015 WHO Classification. Front. Oncol. 2017, 7, 193. [Google Scholar] [CrossRef] [PubMed]
- Rekhtman, N.; Ang, D.C.; Sima, C.S.; Travis, W.D.; Moreira, A.L. Immunohistochemical algorithm for differentiation of lung adenocarcinoma and squamous cell carcinoma based on large series of whole-tissue sections with validation in small specimens. Mod. Pathol. 2011, 24, 1348–1359. [Google Scholar] [CrossRef] [PubMed]
- Kadota, K.; Nitadori, J.; Rekhtman, N.; Jones, D.R.; Adusumilli, P.S.; Travis, W.D. Reevaluation and reclassification of resected lung carcinomas originally diagnosed as squamous cell carcinoma using immunohistochemical analysis. Am. J. Surg. Pathol. 2015, 39, 1170–1180. [Google Scholar] [CrossRef] [PubMed]
- Micke, P.; Mattsson, J.S.; Djureinovic, D.; Nodin, B.; Jirstrom, K.; Tran, L.; Jonsson, P.; Planck, M.; Botling, J.; Brunnstrom, H. The Impact of the Fourth Edition of the WHO Classification of Lung Tumours on Histological Classification of Resected Pulmonary NSCCs. J. Thorac. Oncol. 2016, 11, 862–872. [Google Scholar] [CrossRef] [PubMed]
- Matoso, A.; Singh, K.; Jacob, R.; Greaves, W.O.; Tavares, R.; Noble, L.; Resnick, M.B.; Delellis, R.A.; Wang, L.J. Comparison of thyroid transcription factor-1 expression by 2 monoclonal antibodies in pulmonary and nonpulmonary primary tumors. Appl. Immunohistochem. Mol. Morphol. 2010, 18, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Comperat, E.; Zhang, F.; Perrotin, C.; Molina, T.; Magdeleinat, P.; Marmey, B.; Regnard, J.F.; Audouin, J.; Camilleri-Broet, S. Variable sensitivity and specificity of TTF-1 antibodies in lung metastatic adenocarcinoma of colorectal origin. Mod. Pathol. 2005, 18, 1371–1376. [Google Scholar] [CrossRef] [PubMed]
- Nonaka, D. A study of DeltaNp63 expression in lung non-small cell carcinomas. Am. J. Surg. Pathol. 2012, 36, 895–899. [Google Scholar] [CrossRef] [PubMed]
- Bishop, J.A.; Teruya-Feldstein, J.; Westra, W.H.; Pelosi, G.; Travis, W.D.; Rekhtman, N. p40 (DeltaNp63) is superior to p63 for the diagnosis of pulmonary squamous cell carcinoma. Mod. Pathol. 2012, 25, 405–415. [Google Scholar] [CrossRef] [PubMed]
- Fujino, K.; Motooka, Y.; Hassan, W.A.; Ali Abdalla, M.O.; Sato, Y.; Kudoh, S.; Hasegawa, K.; Niimori-Kita, K.; Kobayashi, H.; Kubota, I.; et al. Insulinoma-Associated Protein 1 Is a Crucial Regulator of Neuroendocrine Differentiation in Lung Cancer. Am. J. Pathol. 2015, 185, 3164–3177. [Google Scholar] [CrossRef] [PubMed]
- Jia, S.; Wildner, H.; Birchmeier, C. Insm1 controls the differentiation of pulmonary neuroendocrine cells by repressing Hes1. Dev. Biol. 2015, 408, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Goto, Y.; De Silva, M.G.; Toscani, A.; Prabhakar, B.S.; Notkins, A.L.; Lan, M.S. A novel human insulinoma-associated cDNA, IA-1, encodes a protein with “zinc-finger” DNA-binding motifs. J. Biol. Chem. 1992, 267, 15252–15257. [Google Scholar] [PubMed]
- Rooper, L.M.; Sharma, R.; Li, Q.K.; Illei, P.B.; Westra, W.H. INSM1 Demonstrates Superior Performance to the Individual and Combined Use of Synaptophysin, Chromogranin and CD56 for Diagnosing Neuroendocrine Tumors of the Thoracic Cavity. Am. J. Surg. Pathol. 2017, 41, 1561–1569. [Google Scholar] [CrossRef] [PubMed]
- Shaw, A.T.; Hsu, P.P.; Awad, M.M.; Engelman, J.A. Tyrosine kinase gene rearrangements in epithelial malignancies. Nat. Rev. Cancer 2013, 13, 772–787. [Google Scholar] [CrossRef] [PubMed]
- Soda, M.; Choi, Y.L.; Enomoto, M.; Takada, S.; Yamashita, Y.; Ishikawa, S.; Fujiwara, S.; Watanabe, H.; Kurashina, K.; Hatanaka, H.; et al. Identification of the transforming EML4-ALK fusion gene in non-small-cell lung cancer. Nature 2007, 448, 561–566. [Google Scholar] [CrossRef] [PubMed]
- Inamura, K.; Takeuchi, K.; Togashi, Y.; Nomura, K.; Ninomiya, H.; Okui, M.; Satoh, Y.; Okumura, S.; Nakagawa, K.; Soda, M.; et al. EML4-ALK fusion is linked to histological characteristics in a subset of lung cancers. J. Thorac. Oncol. 2008, 3, 13–17. [Google Scholar] [CrossRef] [PubMed]
- Shaw, A.T.; Yeap, B.Y.; Mino-Kenudson, M.; Digumarthy, S.R.; Costa, D.B.; Heist, R.S.; Solomon, B.; Stubbs, H.; Admane, S.; McDermott, U.; et al. Clinical features and outcome of patients with non-small-cell lung cancer who harbor EML4-ALK. J. Clin. Oncol. 2009, 27, 4247–4253. [Google Scholar] [CrossRef] [PubMed]
- Inamura, K.; Takeuchi, K.; Togashi, Y.; Hatano, S.; Ninomiya, H.; Motoi, N.; Mun, M.Y.; Sakao, Y.; Okumura, S.; Nakagawa, K.; et al. EML4-ALK lung cancers are characterized by rare other mutations, a TTF-1 cell lineage, an acinar histology, and young onset. Mod. Pathol. 2009, 22, 508–515. [Google Scholar] [CrossRef] [PubMed]
- Kwak, E.L.; Bang, Y.J.; Camidge, D.R.; Shaw, A.T.; Solomon, B.; Maki, R.G.; Ou, S.H.; Dezube, B.J.; Janne, P.A.; Costa, D.B.; et al. Anaplastic lymphoma kinase inhibition in non-small-cell lung cancer. N. Engl. J. Med. 2010, 363, 1693–1703. [Google Scholar] [CrossRef] [PubMed]
- Shaw, A.T.; Kim, D.W.; Mehra, R.; Tan, D.S.; Felip, E.; Chow, L.Q.; Camidge, D.R.; Vansteenkiste, J.; Sharma, S.; De Pas, T.; et al. Ceritinib in ALK-rearranged non-small-cell lung cancer. N. Engl. J. Med. 2014, 370, 1189–1197. [Google Scholar] [CrossRef] [PubMed]
- Solomon, B.J.; Mok, T.; Kim, D.W.; Wu, Y.L.; Nakagawa, K.; Mekhail, T.; Felip, E.; Cappuzzo, F.; Paolini, J.; Usari, T.; et al. First-line crizotinib versus chemotherapy in ALK-positive lung cancer. N. Engl. J. Med. 2014, 371, 2167–2177. [Google Scholar] [CrossRef] [PubMed]
- Hofman, P. ALK in Non-Small Cell Lung Cancer (NSCLC) Pathobiology, Epidemiology, Detection from Tumor Tissue and Algorithm Diagnosis in a Daily Practice. Cancers 2017, 9, 107. [Google Scholar] [CrossRef] [PubMed]
- Conklin, C.M.; Craddock, K.J.; Have, C.; Laskin, J.; Couture, C.; Ionescu, D.N. Immunohistochemistry is a reliable screening tool for identification of ALK rearrangement in non-small-cell lung carcinoma and is antibody dependent. J. Thorac. Oncol. 2013, 8, 45–51. [Google Scholar] [CrossRef] [PubMed]
- Van der Wekken, A.J.; Pelgrim, R.; ’t Hart, N.; Werner, N.; Mastik, M.F.; Hendriks, L.; van der Heijden, E.; Looijen-Salamon, M.; de Langen, A.J.; Staal-van den Brekel, J.; et al. Dichotomous ALK-IHC Is a Better Predictor for ALK Inhibition Outcome than Traditional ALK-FISH in Advanced Non-Small Cell Lung Cancer. Clin. Cancer Res. 2017, 23, 4251–4258. [Google Scholar] [CrossRef] [PubMed]
- Thorne-Nuzzo, T.; Williams, C.; Catallini, A.; Clements, J.; Singh, S.; Amberson, J.; Dickinson, K.; Gatalica, Z.; Ho, S.N.; Loftin, I.; et al. A sensitive ALK immunohistochemistry companion diagnostic test identifies patients eligible for treatment with Crizotinib. J. Thorac. Oncol. 2017, 12, 804–813. [Google Scholar] [CrossRef] [PubMed]
- Marchetti, A.; Di Lorito, A.; Pace, M.V.; Iezzi, M.; Felicioni, L.; D’Antuono, T.; Filice, G.; Guetti, L.; Mucilli, F.; Buttitta, F. ALK Protein Analysis by IHC Staining after Recent Regulatory Changes: A Comparison of Two Widely Used Approaches, Revision of the Literature, and a New Testing Algorithm. J. Thorac. Oncol. 2016, 11, 487–495. [Google Scholar] [CrossRef] [PubMed]
- Wynes, M.W.; Sholl, L.M.; Dietel, M.; Schuuring, E.; Tsao, M.S.; Yatabe, Y.; Tubbs, R.R.; Hirsch, F.R. An international interpretation study using the ALK IHC antibody D5F3 and a sensitive detection kit demonstrates high concordance between ALK IHC and ALK FISH and between evaluators. J. Thorac. Oncol. 2014, 9, 631–638. [Google Scholar] [CrossRef] [PubMed]
- Bergethon, K.; Shaw, A.T.; Ou, S.H.; Katayama, R.; Lovly, C.M.; McDonald, N.T.; Massion, P.P.; Siwak-Tapp, C.; Gonzalez, A.; Fang, R.; et al. ROS1 rearrangements define a unique molecular class of lung cancers. J. Clin. Oncol. 2012, 30, 863–870. [Google Scholar] [CrossRef] [PubMed]
- Davies, K.D.; Le, A.T.; Theodoro, M.F.; Skokan, M.C.; Aisner, D.L.; Berge, E.M.; Terracciano, L.M.; Cappuzzo, F.; Incarbone, M.; Roncalli, M.; et al. Identifying and targeting ROS1 gene fusions in non-small cell lung cancer. Clin. Cancer Res. 2012, 18, 4570–4579. [Google Scholar] [CrossRef] [PubMed]
- Shaw, A.T.; Ou, S.H.; Bang, Y.J.; Camidge, D.R.; Solomon, B.J.; Salgia, R.; Riely, G.J.; Varella-Garcia, M.; Shapiro, G.I.; Costa, D.B.; et al. Crizotinib in ROS1-rearranged non-small-cell lung cancer. N. Engl. J. Med. 2014, 371, 1963–1971. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, A.; Tsuta, K.; Wakai, S.; Arai, Y.; Asamura, H.; Shibata, T.; Furuta, K.; Kohno, T.; Kushima, R. Immunohistochemical detection of ROS1 is useful for identifying ROS1 rearrangements in lung cancers. Mod. Pathol. 2014, 27, 711–720. [Google Scholar] [CrossRef] [PubMed]
- Lim, S.M.; Kim, H.R.; Lee, J.S.; Lee, K.H.; Lee, Y.G.; Min, Y.J.; Cho, E.K.; Lee, S.S.; Kim, B.S.; Choi, M.Y.; et al. Open-label, multicenter, phase II study of ceritinib in patients with non-small-cell lung cancer harboring ROS1 rearrangement. J. Clin. Oncol. 2017, 35, 2613–2618. [Google Scholar] [CrossRef] [PubMed]
- Boyle, T.A.; Masago, K.; Ellison, K.E.; Yatabe, Y.; Hirsch, F.R. ROS1 immunohistochemistry among major genotypes of non-small-cell lung cancer. Clin. Lung Cancer 2015, 16, 106–111. [Google Scholar] [CrossRef] [PubMed]
- Mescam-Mancini, L.; Lantuejoul, S.; Moro-Sibilot, D.; Rouquette, I.; Souquet, P.J.; Audigier-Valette, C.; Sabourin, J.C.; Decroisette, C.; Sakhri, L.; Brambilla, E.; et al. On the relevance of a testing algorithm for the detection of ROS1-rearranged lung adenocarcinomas. Lung Cancer 2014, 83, 168–173. [Google Scholar] [CrossRef] [PubMed]
- Bubendorf, L.; Buttner, R.; Al-Dayel, F.; Dietel, M.; Elmberger, G.; Kerr, K.; Lopez-Rios, F.; Marchetti, A.; Oz, B.; Pauwels, P.; et al. Testing for ROS1 in non-small cell lung cancer: A review with recommendations. Virchows Arch. 2016, 469, 489–503. [Google Scholar] [CrossRef] [PubMed]
- Sholl, L.M.; Sun, H.; Butaney, M.; Zhang, C.; Lee, C.; Janne, P.A.; Rodig, S.J. ROS1 immunohistochemistry for detection of ROS1-rearranged lung adenocarcinomas. Am. J. Surg. Pathol. 2013, 37, 1441–1449. [Google Scholar] [CrossRef] [PubMed]
- Cha, Y.J.; Lee, J.S.; Kim, H.R.; Lim, S.M.; Cho, B.C.; Lee, C.Y.; Shim, H.S. Screening of ROS1 rearrangements in lung adenocarcinoma by immunohistochemistry and comparison with ALK rearrangements. PLoS ONE 2014, 9, e103333. [Google Scholar] [CrossRef] [PubMed]
- Shigematsu, H.; Lin, L.; Takahashi, T.; Nomura, M.; Suzuki, M.; Wistuba, I.I.; Fong, K.M.; Lee, H.; Toyooka, S.; Shimizu, N.; et al. Clinical and biological features associated with epidermal growth factor receptor gene mutations in lung cancers. J. Natl. Cancer Inst. 2005, 97, 339–346. [Google Scholar] [CrossRef] [PubMed]
- Inamura, K.; Ninomiya, H.; Ishikawa, Y.; Matsubara, O. Is the epidermal growth factor receptor status in lung cancers reflected in clinicopathologic features? Arch. Pathol. Lab. Med. 2010, 134, 66–72. [Google Scholar] [PubMed]
- Leighl, N.B.; Rekhtman, N.; Biermann, W.A.; Huang, J.; Mino-Kenudson, M.; Ramalingam, S.S.; West, H.; Whitlock, S.; Somerfield, M.R. Molecular testing for selection of patients with lung cancer for epidermal growth factor receptor and anaplastic lymphoma kinase tyrosine kinase inhibitors: American Society of Clinical Oncology endorsement of the College of American Pathologists/International Association for the study of lung cancer/association for molecular pathology guideline. J. Clin. Oncol. 2014, 32, 3673–3679. [Google Scholar] [PubMed]
- Lindeman, N.I.; Cagle, P.T.; Beasley, M.B.; Chitale, D.A.; Dacic, S.; Giaccone, G.; Jenkins, R.B.; Kwiatkowski, D.J.; Saldivar, J.S.; Squire, J.; et al. Molecular testing guideline for selection of lung cancer patients for EGFR and ALK tyrosine kinase inhibitors: Guideline from the College of American Pathologists, International Association for the Study of Lung Cancer, and Association for Molecular Pathology. J. Thorac. Oncol. 2013, 8, 823–859. [Google Scholar] [PubMed]
- Yu, J.; Kane, S.; Wu, J.; Benedettini, E.; Li, D.; Reeves, C.; Innocenti, G.; Wetzel, R.; Crosby, K.; Becker, A.; et al. Mutation-specific antibodies for the detection of EGFR mutations in non-small-cell lung cancer. Clin. Cancer Res. 2009, 15, 3023–3028. [Google Scholar] [CrossRef] [PubMed]
- Tumeh, P.C.; Harview, C.L.; Yearley, J.H.; Shintaku, I.P.; Taylor, E.J.; Robert, L.; Chmielowski, B.; Spasic, M.; Henry, G.; Ciobanu, V.; et al. PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 2014, 515, 568–571. [Google Scholar] [CrossRef] [PubMed]
- Simon, S.; Labarriere, N. PD-1 expression on tumor-specific T cells: Friend or foe for immunotherapy? Oncoimmunology 2017, 7, e1364828. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Boyle, T.A.; Zhou, C.; Rimm, D.L.; Hirsch, F.R. PD-L1 Expression in Lung Cancer. J. Thorac. Oncol. 2016, 11, 964–975. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, L.H.; Kummel, A.; Gorlich, D.; Mohr, M.; Brockling, S.; Mikesch, J.H.; Grunewald, I.; Marra, A.; Schultheis, A.M.; Wardelmann, E.; et al. PD-1 and PD-L1 Expression in NSCLC Indicate a Favorable Prognosis in Defined Subgroups. PLoS ONE 2015, 10, e0136023. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, C.Y.; Lin, M.W.; Chang, Y.L.; Wu, C.T.; Yang, P.C. Programmed cell death-ligand 1 expression in surgically resected stage I pulmonary adenocarcinoma and its correlation with driver mutations and clinical outcomes. Eur. J. Cancer 2014, 50, 1361–1369. [Google Scholar] [CrossRef] [PubMed]
- Cooper, W.A.; Tran, T.; Vilain, R.E.; Madore, J.; Selinger, C.I.; Kohonen-Corish, M.; Yip, P.; Yu, B.; O’Toole, S.A.; McCaughan, B.C.; et al. PD-L1 expression is a favorable prognostic factor in early stage non-small cell carcinoma. Lung Cancer 2015, 89, 181–188. [Google Scholar] [CrossRef] [PubMed]
- Tang, Y.; Fang, W.; Zhang, Y.; Hong, S.; Kang, S.; Yan, Y.; Chen, N.; Zhan, J.; He, X.; Qin, T.; et al. The association between PD-L1 and EGFR status and the prognostic value of PD-L1 in advanced non-small cell lung cancer patients treated with EGFR-TKIs. Oncotarget 2015, 6, 14209–14219. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.Y.; Lin, M.W.; Chang, Y.L.; Wu, C.T.; Yang, P.C. Programmed cell death-ligand 1 expression is associated with a favourable immune microenvironment and better overall survival in stage I pulmonary squamous cell carcinoma. Eur. J. Cancer 2016, 57, 91–103. [Google Scholar] [CrossRef] [PubMed]
- Ishii, H.; Azuma, K.; Kawahara, A.; Yamada, K.; Imamura, Y.; Tokito, T.; Kinoshita, T.; Kage, M.; Hoshino, T. Significance of programmed cell death-ligand 1 expression and its association with survival in patients with small cell lung cancer. J. Thorac. Oncol. 2015, 10, 426–430. [Google Scholar] [CrossRef] [PubMed]
- Inamura, K.; Yokouchi, Y.; Kobayashi, M.; Ninomiya, H.; Sakakibara, R.; Nishio, M.; Okumura, S.; Ishikawa, Y. Relationship of tumor PD-L1 (CD274) expression with lower mortality in lung high-grade neuroendocrine tumor. Cancer Med. 2017, 6, 2347–2356. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.L.; Yang, C.Y.; Huang, Y.L.; Wu, C.T.; Yang, P.C. High PD-L1 expression is associated with stage IV disease and poorer overall survival in 186 cases of small cell lung cancers. Oncotarget 2017, 8, 18021–18030. [Google Scholar] [CrossRef] [PubMed]
- Borghaei, H.; Paz-Ares, L.; Horn, L.; Spigel, D.R.; Steins, M.; Ready, N.E.; Chow, L.Q.; Vokes, E.E.; Felip, E.; Holgado, E.; et al. Nivolumab versus Docetaxel in Advanced Nonsquamous Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2015, 373, 1627–1639. [Google Scholar] [CrossRef] [PubMed]
- Brahmer, J.; Reckamp, K.L.; Baas, P.; Crino, L.; Eberhardt, W.E.; Poddubskaya, E.; Antonia, S.; Pluzanski, A.; Vokes, E.E.; Holgado, E.; et al. Nivolumab versus Docetaxel in Advanced Squamous-Cell Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2015, 373, 123–135. [Google Scholar] [CrossRef] [PubMed]
- Horn, L.; Spigel, D.R.; Vokes, E.E.; Holgado, E.; Ready, N.; Steins, M.; Poddubskaya, E.; Borghaei, H.; Felip, E.; Paz-Ares, L.; et al. Nivolumab Versus Docetaxel in Previously Treated Patients With Advanced Non-Small-Cell Lung Cancer: Two-Year Outcomes From Two Randomized, Open-Label, Phase III Trials (CheckMate 017 and CheckMate 057). J. Clin. Oncol. 2017, 35, 3924–3933. [Google Scholar] [CrossRef] [PubMed]
- Garon, E.B.; Rizvi, N.A.; Hui, R.; Leighl, N.; Balmanoukian, A.S.; Eder, J.P.; Patnaik, A.; Aggarwal, C.; Gubens, M.; Horn, L.; et al. Pembrolizumab for the treatment of non-small-cell lung cancer. N. Engl. J. Med. 2015, 372, 2018–2028. [Google Scholar] [CrossRef] [PubMed]
- Hofman, P. PD-L1 immunohistochemistry for non-small cell lung carcinoma: Which strategy should be adopted? Expert Rev. Mol. Diagn. 2017, 17, 1097–1108. [Google Scholar] [CrossRef] [PubMed]
- Weber, J.S.; Kudchadkar, R.R.; Yu, B.; Gallenstein, D.; Horak, C.E.; Inzunza, H.D.; Zhao, X.; Martinez, A.J.; Wang, W.; Gibney, G.; et al. Safety, efficacy, and biomarkers of nivolumab with vaccine in ipilimumab-refractory or -naive melanoma. J. Clin. Oncol. 2013, 31, 4311–4318. [Google Scholar] [CrossRef] [PubMed]
- Planchard, D.; Yokoi, T.; McCleod, M.J.; Fischer, J.R.; Kim, Y.C.; Ballas, M.; Shi, K.; Soria, J.C. A Phase III Study of Durvalumab (MEDI4736) With or Without Tremelimumab for Previously Treated Patients with Advanced NSCLC: Rationale and Protocol Design of the ARCTIC Study. Clin. Lung Cancer 2016, 17, 232–236. [Google Scholar] [CrossRef] [PubMed]
- Kaufman, H.L.; Russell, J.; Hamid, O.; Bhatia, S.; Terheyden, P.; D’Angelo, S.P.; Shih, K.C.; Lebbe, C.; Linette, G.P.; Milella, M.; et al. Avelumab in patients with chemotherapy-refractory metastatic Merkel cell carcinoma: A multicentre, single-group, open-label, phase 2 trial. Lancet Oncol. 2016, 17, 1374–1385. [Google Scholar] [CrossRef]
- Antonia, S.J.; Villegas, A.; Daniel, D.; Vicente, D.; Murakami, S.; Hui, R.; Yokoi, T.; Chiappori, A.; Lee, K.H.; de Wit, M.; et al. Durvalumab after Chemoradiotherapy in Stage III Non-Small-Cell Lung Cancer. N. Engl. J. Med. 2017, 377, 1919–1929. [Google Scholar] [CrossRef] [PubMed]
- Gulley, J.L.; Rajan, A.; Spigel, D.R.; Iannotti, N.; Chandler, J.; Wong, D.J.L.; Leach, J.; Edenfield, W.J.; Wang, D.; Grote, H.J.; et al. Avelumab for patients with previously treated metastatic or recurrent non-small-cell lung cancer (JAVELIN Solid Tumor): Dose-expansion cohort of a multicentre, open-label, phase 1b trial. Lancet Oncol. 2017, 18, 599–610. [Google Scholar] [CrossRef]
- Ilie, M.; Khambata-Ford, S.; Copie-Bergman, C.; Huang, L.; Juco, J.; Hofman, V.; Hofman, P. Use of the 22C3 anti-PD-L1 antibody to determine PD-L1 expression in multiple automated immunohistochemistry platforms. PLoS ONE 2017, 12, e0183023. [Google Scholar]
- Hirsch, F.R.; McElhinny, A.; Stanforth, D.; Ranger-Moore, J.; Jansson, M.; Kulangara, K.; Richardson, W.; Towne, P.; Hanks, D.; Vennapusa, B.; et al. PD-L1 Immunohistochemistry Assays for Lung Cancer: Results from Phase 1 of the Blueprint PD-L1 IHC Assay Comparison Project. J. Thorac. Oncol. 2017, 12, 208–222. [Google Scholar] [CrossRef] [PubMed]
- Rimm, D.L.; Han, G.; Taube, J.M.; Yi, E.S.; Bridge, J.A.; Flieder, D.B.; Homer, R.; West, W.W.; Wu, H.; Roden, A.C.; et al. A Prospective, Multi-institutional, Pathologist-Based Assessment of 4 Immunohistochemistry Assays for PD-L1 Expression in Non-Small Cell Lung Cancer. JAMA Oncol. 2017, 3, 1051–1058. [Google Scholar] [CrossRef] [PubMed]
- Brunnstrom, H.; Johansson, A.; Westbom-Fremer, S.; Backman, M.; Djureinovic, D.; Patthey, A.; Isaksson-Mettavainio, M.; Gulyas, M.; Micke, P. PD-L1 immunohistochemistry in clinical diagnostics of lung cancer: Inter-pathologist variability is higher than assay variability. Mod. Pathol. 2017, 30, 1411–1421. [Google Scholar] [CrossRef] [PubMed]
- Marchetti, A.; Barberis, M.; Franco, R.; De Luca, G.; Pace, M.V.; Staibano, S.; Volante, M.; Buttitta, F.; Guerini-Rocco, E.; Righi, L.; et al. Multicenter Comparison of 22C3 PharmDx (Agilent) and SP263 (Ventana) Assays to Test PD-L1 Expression for NSCLC Patients to Be Treated with Immune Checkpoint Inhibitors. J. Thorac. Oncol. 2017, 12, 1654–1663. [Google Scholar] [CrossRef] [PubMed]
- Ratcliffe, M.J.; Sharpe, A.; Midha, A.; Barker, C.; Scott, M.; Scorer, P.; Al-Masri, H.; Rebelatto, M.C.; Walker, J. Agreement between Programmed Cell Death Ligand-1 Diagnostic Assays across Multiple Protein Expression Cutoffs in Non-Small Cell Lung Cancer. Clin. Cancer Res. 2017, 23, 3585–3591. [Google Scholar] [CrossRef] [PubMed]
- Cigognetti, M.; Lonardi, S.; Fisogni, S.; Balzarini, P.; Pellegrini, V.; Tironi, A.; Bercich, L.; Bugatti, M.; Rossi, G.; Murer, B.; et al. BAP1 (BRCA1-associated protein 1) is a highly specific marker for differentiating mesothelioma from reactive mesothelial proliferations. Mod. Pathol. 2015, 28, 1043–1057. [Google Scholar] [CrossRef] [PubMed]
- Hwang, H.C.; Pyott, S.; Rodriguez, S.; Cindric, A.; Carr, A.; Michelsen, C.; Thompson, K.; Tse, C.H.; Gown, A.M.; Churg, A. BAP1 Immunohistochemistry and p16 FISH in the Diagnosis of Sarcomatous and Desmoplastic Mesotheliomas. Am. J. Surg. Pathol. 2016, 40, 714–718. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Hiroshima, K.; Yusa, T.; Ozaki, D.; Koh, E.; Sekine, Y.; Matsumoto, S.; Nabeshima, K.; Sato, A.; Tsujimura, T.; et al. Usefulness of p16/CDKN2A fluorescence in situ hybridization and BAP1 immunohistochemistry for the diagnosis of biphasic mesothelioma. Ann. Diagn. Pathol. 2017, 26, 31–37. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Hiroshima, K.; Matsumoto, S.; Nabeshima, K.; Yusa, T.; Ozaki, D.; Fujino, M.; Yamakawa, H.; Nakatani, Y.; Tada, Y.; et al. Diagnostic usefulness of p16/CDKN2A FISH in distinguishing between sarcomatoid mesothelioma and fibrous pleuritis. Am. J. Clin. Pathol. 2013, 139, 39–46. [Google Scholar] [CrossRef] [PubMed]
- Husain, A.N.; Colby, T.V.; Ordonez, N.G.; Allen, T.C.; Attanoos, R.L.; Beasley, M.B.; Butnor, K.J.; Chirieac, L.R.; Churg, A.M.; Dacic, S.; et al. Guidelines for Pathologic Diagnosis of Malignant Mesothelioma 2017 Update of the Consensus Statement From the International Mesothelioma Interest Group. Arch. Pathol. Lab. Med. 2018, 142, 89–108. [Google Scholar] [CrossRef] [PubMed]
- Kuraoka, M.; Amatya, V.J.; Kushitani, K.; Mawas, A.S.; Miyata, Y.; Okada, M.; Kishimoto, T.; Inai, K.; Nishisaka, T.; Sueda, T.; et al. Identification of DAB2 and Intelectin-1 as Novel Positive Immunohistochemical Markers of Epithelioid Mesothelioma by Transcriptome Microarray Analysis for Its Differentiation From Pulmonary Adenocarcinoma. Am. J. Surg. Pathol. 2017, 41, 1045–1052. [Google Scholar] [CrossRef] [PubMed]
- Amatya, V.J.; Kushitani, K.; Mawas, A.S.; Miyata, Y.; Okada, M.; Kishimoto, T.; Inai, K.; Takeshima, Y. MUC4, a novel immunohistochemical marker identified by gene expression profiling, differentiates pleural sarcomatoid mesothelioma from lung sarcomatoid carcinoma. Mod. Pathol. 2017, 30, 672–681. [Google Scholar] [CrossRef] [PubMed]
- Sholl, L.M.; Nishino, M.; Pokharel, S.; Mino-Kenudson, M.; French, C.A.; Janne, P.A.; Lathan, C. Primary Pulmonary NUT Midline Carcinoma: Clinical, Radiographic, and Pathologic Characterizations. J. Thorac. Oncol. 2015, 10, 951–959. [Google Scholar] [CrossRef] [PubMed]
- Bauer, D.E.; Mitchell, C.M.; Strait, K.M.; Lathan, C.S.; Stelow, E.B.; Luer, S.C.; Muhammed, S.; Evans, A.G.; Sholl, L.M.; Rosai, J.; et al. Clinicopathologic features and long-term outcomes of NUT midline carcinoma. Clin. Cancer Res. 2012, 18, 5773–5779. [Google Scholar] [CrossRef] [PubMed]
- French, C.A.; Kutok, J.L.; Faquin, W.C.; Toretsky, J.A.; Antonescu, C.R.; Griffin, C.A.; Nose, V.; Vargas, S.O.; Moschovi, M.; Tzortzatou-Stathopoulou, F.; et al. Midline carcinoma of children and young adults with NUT rearrangement. J. Clin. Oncol. 2004, 22, 4135–4139. [Google Scholar] [CrossRef] [PubMed]
- French, C.A.; Miyoshi, I.; Kubonishi, I.; Grier, H.E.; Perez-Atayde, A.R.; Fletcher, J.A. BRD4-NUT fusion oncogene: A novel mechanism in aggressive carcinoma. Cancer Res. 2003, 63, 304–307. [Google Scholar] [PubMed]
- French, C.A.; Ramirez, C.L.; Kolmakova, J.; Hickman, T.T.; Cameron, M.J.; Thyne, M.E.; Kutok, J.L.; Toretsky, J.A.; Tadavarthy, A.K.; Kees, U.R.; et al. BRD-NUT oncoproteins: A family of closely related nuclear proteins that block epithelial differentiation and maintain the growth of carcinoma cells. Oncogene 2008, 27, 2237–2242. [Google Scholar] [CrossRef] [PubMed]
- French, C.A.; Rahman, S.; Walsh, E.M.; Kuhnle, S.; Grayson, A.R.; Lemieux, M.E.; Grunfeld, N.; Rubin, B.P.; Antonescu, C.R.; Zhang, S.; et al. NSD3-NUT fusion oncoprotein in NUT midline carcinoma: Implications for a novel oncogenic mechanism. Cancer Discov. 2014, 4, 928–941. [Google Scholar] [CrossRef] [PubMed]
- French, C.A. Pathogenesis of NUT midline carcinoma. Annu. Rev. Pathol. 2012, 7, 247–265. [Google Scholar] [CrossRef] [PubMed]
- French, C.A. The importance of diagnosing NUT midline carcinoma. Head Neck Pathol. 2013, 7, 11–16. [Google Scholar] [CrossRef] [PubMed]
- Stathis, A.; Zucca, E.; Bekradda, M.; Gomez-Roca, C.; Delord, J.P.; de La Motte Rouge, T.; Uro-Coste, E.; de Braud, F.; Pelosi, G.; French, C.A. Clinical Response of Carcinomas Harboring the BRD4-NUT Oncoprotein to the Targeted Bromodomain Inhibitor OTX015/MK-8628. Cancer Discov. 2016, 6, 492–500. [Google Scholar] [CrossRef] [PubMed]
- Stathis, A.; Bertoni, F. BET Proteins as Targets for Anticancer Treatment. Cancer Discov. 2018, 8, 24–36. [Google Scholar] [CrossRef] [PubMed]
- Haack, H.; Johnson, L.A.; Fry, C.J.; Crosby, K.; Polakiewicz, R.D.; Stelow, E.B.; Hong, S.M.; Schwartz, B.E.; Cameron, M.J.; Rubin, M.A.; et al. Diagnosis of NUT midline carcinoma using a NUT-specific monoclonal antibody. Am. J. Surg. Pathol. 2009, 33, 984–991. [Google Scholar] [CrossRef] [PubMed]
- Zhu, B.; Laskin, W.; Chen, Y.; French, C.A.; Cameron, M.J.; Nayar, R.; Lin, X. NUT midline carcinoma: A neoplasm with diagnostic challenges in cytology. Cytopathology 2011, 22, 414–417. [Google Scholar] [CrossRef] [PubMed]
- Evans, A.G.; French, C.A.; Cameron, M.J.; Fletcher, C.D.; Jackman, D.M.; Lathan, C.S.; Sholl, L.M. Pathologic characteristics of NUT midline carcinoma arising in the mediastinum. Am. J. Surg. Pathol. 2012, 36, 1222–1227. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.W.; Roh, J.; Park, C.S. Immunohistochemistry for Pathologists: Protocols, Pitfalls, and Tips. J. Pathol. Transl. Med. 2016, 50, 411–418. [Google Scholar] [CrossRef] [PubMed]
- Bass, B.P.; Engel, K.B.; Greytak, S.R.; Moore, H.M. A review of preanalytical factors affecting molecular, protein, and morphological analysis of formalin-fixed, paraffin-embedded (FFPE) tissue: How well do you know your FFPE specimen? Arch. Pathol. Lab. Med. 2014, 138, 1520–1530. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Inamura, K. Update on Immunohistochemistry for the Diagnosis of Lung Cancer. Cancers 2018, 10, 72. https://doi.org/10.3390/cancers10030072
Inamura K. Update on Immunohistochemistry for the Diagnosis of Lung Cancer. Cancers. 2018; 10(3):72. https://doi.org/10.3390/cancers10030072
Chicago/Turabian StyleInamura, Kentaro. 2018. "Update on Immunohistochemistry for the Diagnosis of Lung Cancer" Cancers 10, no. 3: 72. https://doi.org/10.3390/cancers10030072




