Predictive and Prognostic Relevance of Tumor-Infiltrating Immune Cells: Tailoring Personalized Treatments against Different Cancer Types
Abstract
:Simple Summary
Abstract
1. Introduction
2. Technological and Computational Interventions for Assessment and Quantification of TIICs
2.1. Conventional Approaches and Their Limitations
2.2. Machine Learning-Based Computational Approaches for the Establishment of ICI Scores
2.3. Spatially Variant Immune Infiltration Scoring using SpatialVizScore
2.4. Nonnegative Tensor Factorization using ProTICS
3. Heterogeneity in the Predictive and Prognostic Relevance of TIICs in Different Cancer Types
3.1. Predictive and Prognostic Powers of Tumor Infiltrating Lymphocytes: T-Regs, CD4+ T-Helper, CD8+ Cytotoxic T-Cells, and B-Cells
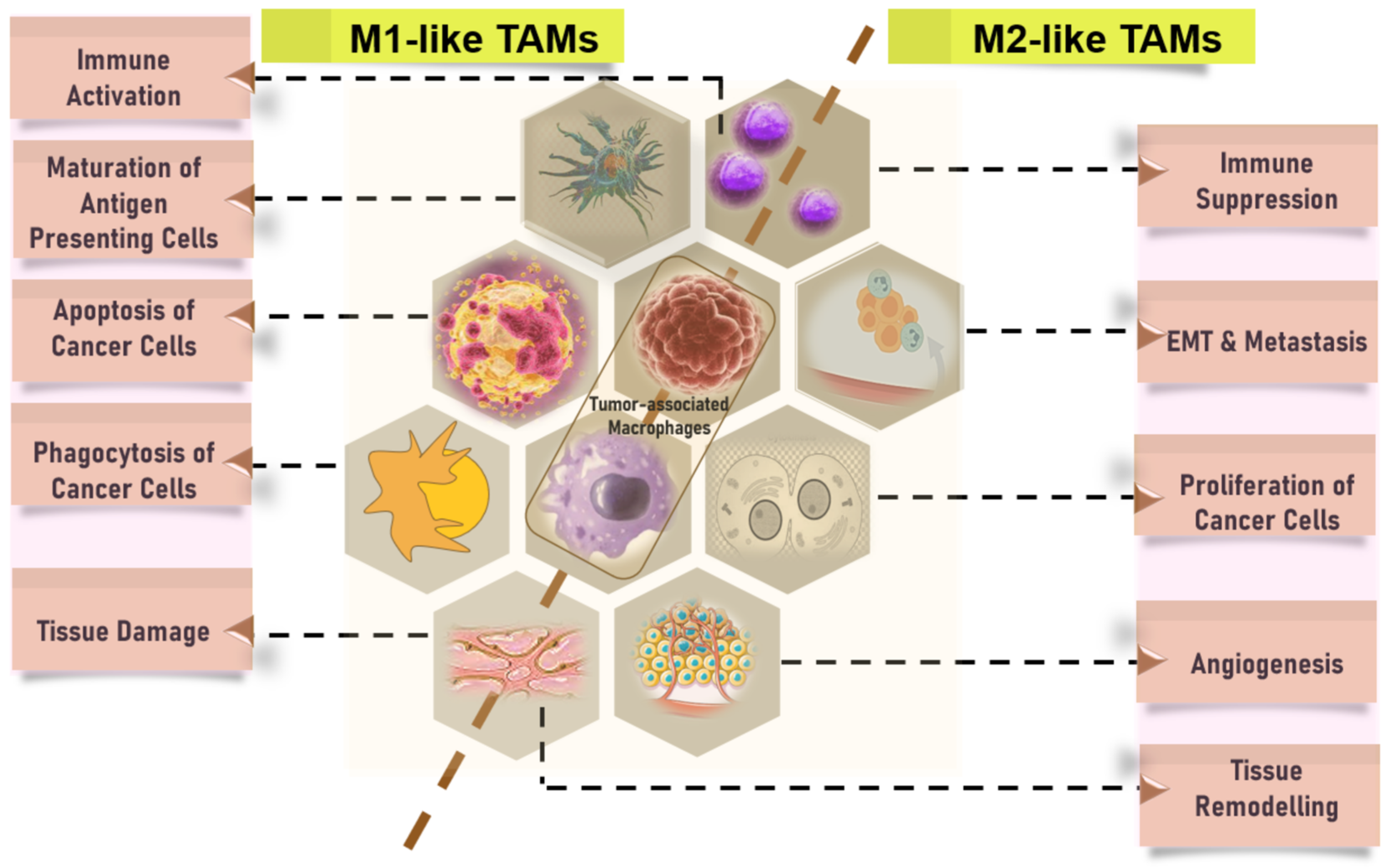
3.1.1. CD68 is a Pan-Macrophage Marker Expressed in M1 and M2 Macrophages, while CD163 is M2-Specific
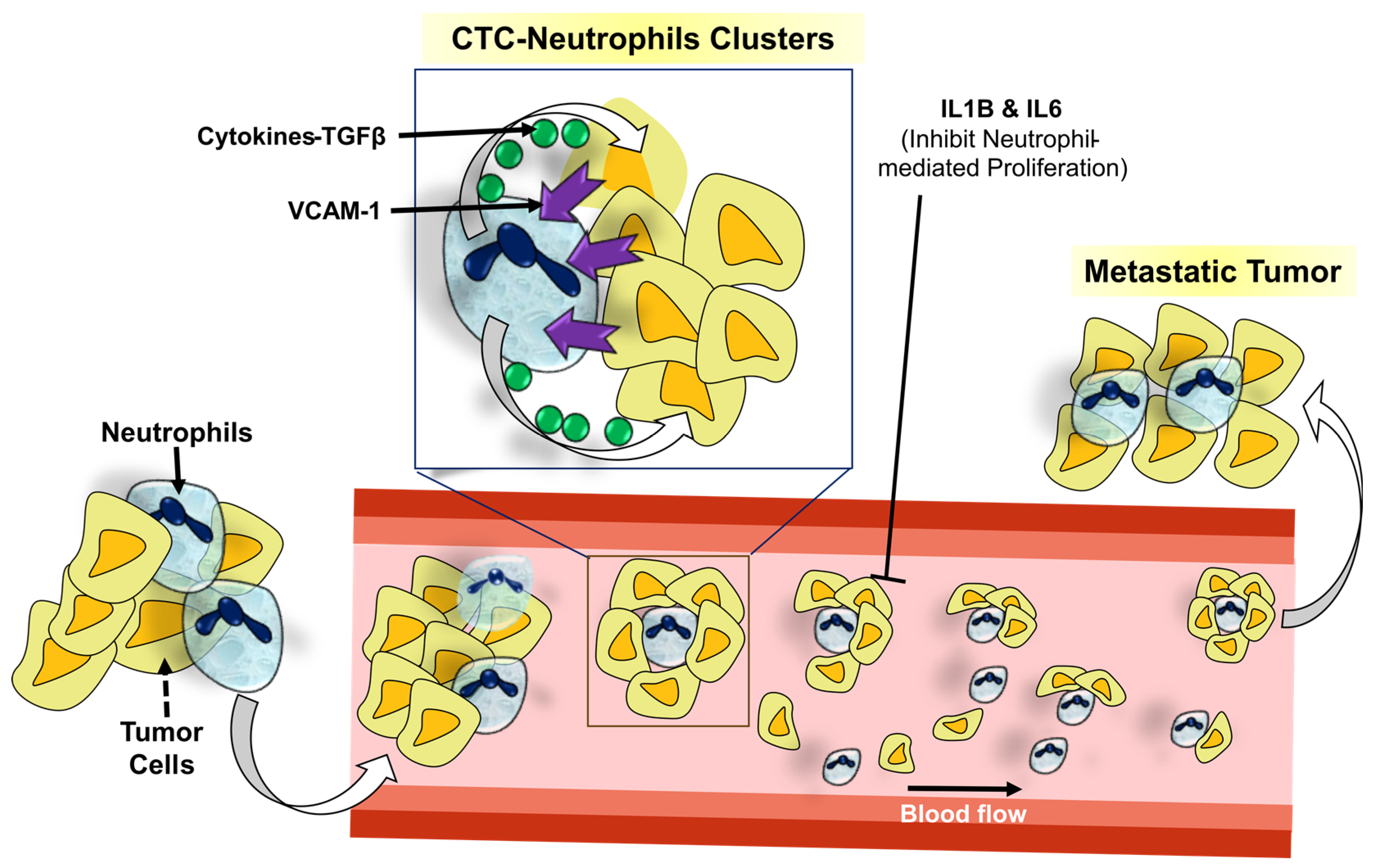
3.1.2. Predictive and Prognostic Relevance of Other Immune Cells: Neutrophils, Eosinophils, Mast Cells, and CAFs
3.1.3. The Prognostic Value of TMB and the Relationship between TMB and Tumor Immune Infiltration
4. Common Prognostic Biomarkers for the Presence of Immune Cell Infiltration across Different Cancers
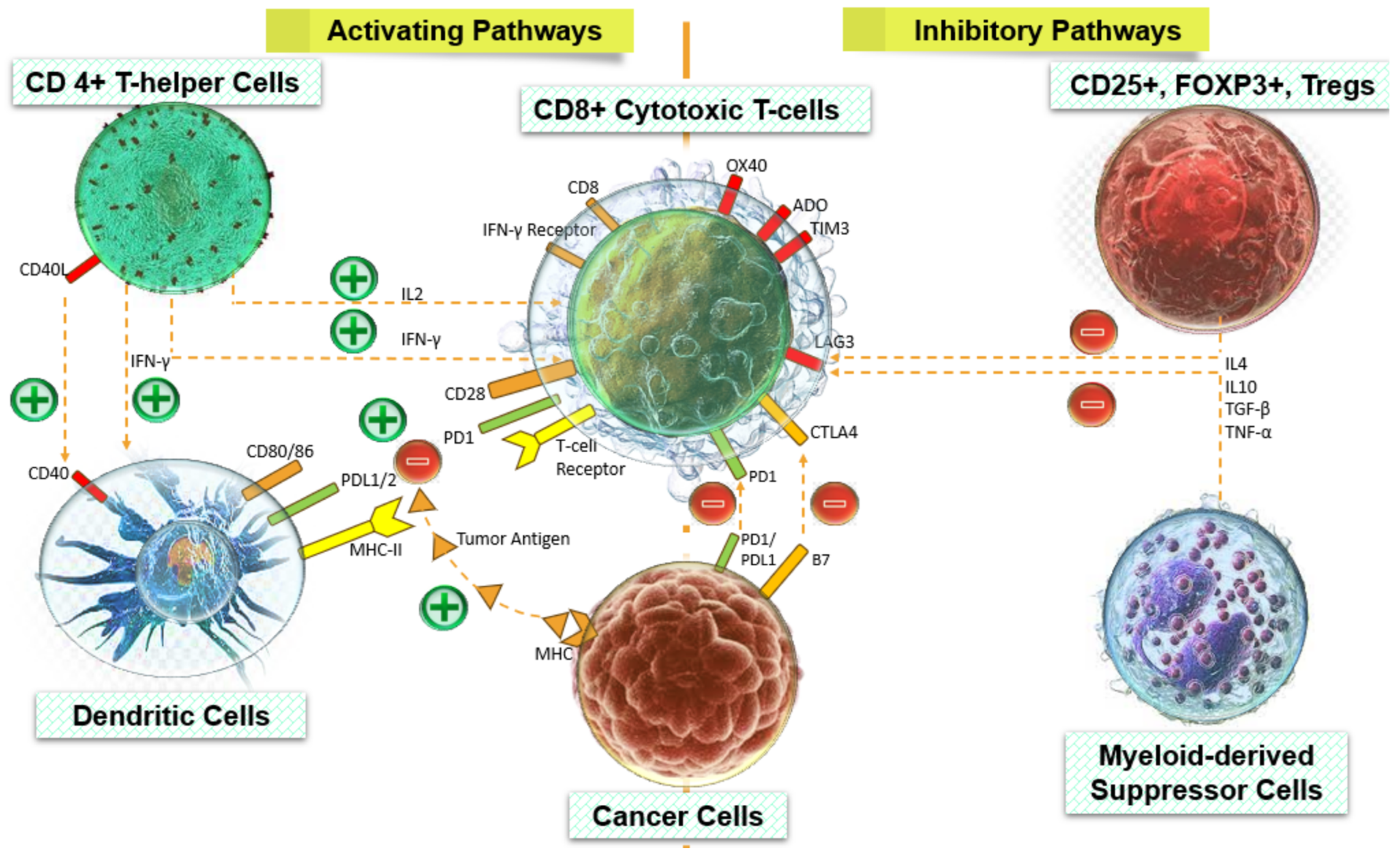
5. Predictive and Prognostic Relevance Correlate to Activated Signaling Circuits that Rely on Infiltrating Immune Cells
6. Relationship between Immune Cells and Cancer Treatment Modalities
7. Discussion
8. Future Perspectives: Implications in Therapeutics and Unanswered Questions
9. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
References
- Chand, D.T.; Dhabhai, B.; Agarwal, D.; Gupta, R.; Nagda, G.; Meena, A.R.; Dhakar, R.; Menon, A.; Mathur, R.; Mona; et al. Mechanistic basis of co-stimulatory CD40-CD40L ligation mediated regulation of immune responses in cancer and autoimmune disorders. Immunobiology 2020, 225, 151899. [Google Scholar]
- Thorsson, V.; Gibbs, D.L.; Brown, S.D.; Wolf, D.; Bortone, D.S.; Ou Yang, T.H.; Porta-Pardo, E.; Gao, G.F.; Plaisier, C.L.; Eddy, J.A.; et al. The Immune Landscape of Cancer. Immunity 2018, 48, 812–830.e14. [Google Scholar] [CrossRef]
- Singh, M.; Thakur, M.; Mishra, M.; Yadav, M.; Vibhuti, R.; Menon, A.M.; Nagda, G.; Dwivedi, V.P.; Dakal, T.C.; Yadav, V. Gene regulation of intracellular adhesion molecule-1 (ICAM-1): A molecule with multiple functions. Immunol. Lett. 2021, 240, 123–136. [Google Scholar] [CrossRef] [PubMed]
- Grivennikov, S.I.; Greten, F.R.; Karin, M. Immunity, Inflammation, and Cancer. Cell 2010, 140, 883–899. [Google Scholar] [CrossRef] [PubMed]
- Turley, S.J.; Cremasco, V.; Astarita, J.L. Immunological hallmarks of stromal cells in the tumour microenvironment. Nat. Rev. Immunol. 2015, 15, 669–682. [Google Scholar] [CrossRef]
- Mao, X.; Xu, J.; Wang, W.; Liang, C.; Hua, J.; Liu, J.; Zhang, B.; Meng, Q.; Yu, X.; Shi, S. Crosstalk between cancer-associated fibroblasts and immune cells in the tumor microenvironment: New findings and future perspectives. Mol. Cancer 2021, 20, 131. [Google Scholar] [CrossRef]
- Hudson, W.H.; Wieland, A. Technology meets TILs: Deciphering T cell function in the -omics era. Cancer Cell 2023, 41, 41–57. [Google Scholar] [CrossRef]
- Lin, B.; Du, L.; Li, H.; Zhu, X.; Cui, L.; Li, X. Tumor-infiltrating lymphocytes: Warriors fight against tumors powerfully. Biomed. Pharmacother. 2020, 132, 110873. [Google Scholar] [CrossRef]
- Dieci, M.V.; Miglietta, F.; Guarneri, V. Immune Infiltrates in Breast Cancer: Recent Updates and Clinical Implications. Cells. 2021, 10, 223. [Google Scholar] [CrossRef]
- Tran Janco, J.M.; Lamichhane, P.; Karyampudi, L.; Knutson, K.L. Tumor-infiltrating dendritic cells in cancer pathogenesis. J. Immunol. 2015, 194, 2985–2991. [Google Scholar] [CrossRef]
- Gajewski, T.F.; Schreiber, H.; Fu, Y.X. Innate and adaptive immune cells in the tumor microenvironment. Nat. Immunol. 2013, 14, 1014–1022. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.S.; Liu, W.; Ly, D.; Xu, H.; Qu, L.; Zhang, L. Tumor-infiltrating B cells: Their role and application in anti-tumor immunity in lung cancer. Cell. Mol. Immunol. 2019, 16, 6–18. [Google Scholar] [CrossRef] [PubMed]
- Stanton, S.E.; Disis, M.L. Clinical significance of tumor-infiltrating lymphocytes in breast cancer. J. Immunother. Cancer 2016, 4, 59. [Google Scholar] [CrossRef]
- Attrill, G.H.; Ferguson, P.M.; Palendira, U.; Long, G.V.; Wilmott, J.S.; Scolyer, R.A. The tumour immune landscape and its implications in cutaneous melanoma. Pigment Cell Melanoma Res. 2021, 34, 529–549. [Google Scholar] [CrossRef] [PubMed]
- McRitchie, B.R.; Akkaya, B. Exhaust the exhausters: Targeting regulatory T cells in the tumor microenvironment. Front. Immunol. 2022, 13, 940052. [Google Scholar] [CrossRef] [PubMed]
- Agahozo, M.C.; Hammerl, D.; Debets, R.; Kok, M.; van Deurzen, C.H.M. Tumor-infiltrating lymphocytes and ductal carcinoma in situ of the breast: Friends or foes? Mod. Pathol. 2018, 31, 1012–1025. [Google Scholar] [CrossRef] [PubMed]
- Denkert, C.; von Minckwitz, G.; Darb-Esfahani, S.; Lederer, B.; Heppner, B.I.; Weber, K.E.; Budczies, J.; Huober, J.; Klauschen, F.; Furlanetto, J.; et al. Tumour-infiltrating lymphocytes and prognosis in different subtypes of breast cancer: A pooled analysis of 3771 patients treated with neoadjuvant therapy. Lancet Oncol. 2018, 19, 40–50. [Google Scholar] [CrossRef]
- Denkert, C.; von Minckwitz, G.; Darb-Esfahani, S.; Lederer, B.; Heppner, B.I.; Weber, K.E.; Budczies, J.; Huober, J.; Klauschen, F.; Furlanetto, J.; et al. Biological processes associated with breast cancer clinical outcome depend on the molecular subtypes. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2008, 14, 5158–5165. [Google Scholar]
- Ligeiro, D.; Rao, M.; Maia, A.; Castillo, M.; Beltran, A.; Maeurer, M. B Cells in the Gastrointestinal Tumor Microenvironment with a Focus on Pancreatic Cancer: Opportunities for Precision Medicine? Adv. Exp. Med. Biol. 2020, 1273, 175–195. [Google Scholar] [PubMed]
- Miyake, M.; Hori, S.; Owari, T.; Oda, Y.; Tatsumi, Y.; Nakai, Y.; Fujii, T.; Fujimoto, K. Clinical Impact of Tumor-Infiltrating Lymphocytes and PD-L1-Positive Cells as Prognostic and Predictive Biomarkers in Urological Malignancies and Retroperitoneal Sarcoma. Cancers 2020, 12, 3153. [Google Scholar] [CrossRef]
- Santoiemma, P.P.; Powell, D.J., Jr. Tumor infiltrating lymphocytes in ovarian cancer. Cancer Biol. Ther. 2015, 16, 807–820. [Google Scholar] [CrossRef] [PubMed]
- Liu, D.; Heij, L.R.; Czigany, Z.; Dahl, E.; Lang, S.A.; Ulmer, T.F.; Luedde, T.; Neumann, U.P.; Bednarsch, J. The role of tumor-infiltrating lymphocytes in cholangiocarcinoma. J. Exp. Clin. Cancer Res. CR 2022, 41, 127. [Google Scholar] [CrossRef] [PubMed]
- De Meulenaere, A.; Vermassen, T.; Aspeslagh, S.; Vandecasteele, K.; Rottey, S.; Ferdinande, L. TILs in Head and Neck Cancer: Ready for Clinical Implementation and Why (Not)? Head Neck Pathol. 2017, 11, 354–363. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Shin, D.H.; Lee, Y.J.; Lee, S.J.; Hwang, C.S.; Kim, A.; Park, W.Y.; Lee, J.H.; Choi, K.U.; Kim, J.Y.; et al. PD-L1 expression and infiltration by CD4+ and FoxP3+ T cells are increased in Xp11 translocation renal cell carcinoma and indicate poor prognosis. Histopathology 2020, 76, 714–721. [Google Scholar] [CrossRef] [PubMed]
- Petitprez, F.; Fossati, N.; Vano, Y.; Freschi, M.; Becht, E.; Lucianò, R.; Calderaro, J.; Guédet, T.; Lacroix, L.; Rancoita, P.M.V.; et al. PD-L1 Expression and CD8+ T-cell Infiltrate are Associated with Clinical Progression in Patients with Node-positive Prostate Cancer. Eur. Urol. Focus 2019, 5, 192–196. [Google Scholar] [CrossRef] [PubMed]
- Giraldo, N.A.; Becht, E.; Vano, Y.; Petitprez, F.; Lacroix, L.; Validire, P.; Sanchez-Salas, R.; Ingels, A.; Oudard, S.; Moatti, A.; et al. Tumor-Infiltrating and Peripheral Blood T-cell Immunophenotypes Predict Early Relapse in Localized Clear Cell Renal Cell Carcinoma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2017, 23, 4416–4428. [Google Scholar] [CrossRef] [PubMed]
- Becht, E.; Giraldo, N.A.; Beuselinck, B.; Job, S.; Marisa, L.; Vano, Y.; Oudard, S.; Zucman-Rossi, J.; Laurent-Puig, P.; Sautès-Fridman, C.; et al. Prognostic and theranostic impact of molecular subtypes and immune classifications in renal cell cancer (RCC) and colorectal cancer (CRC). Oncoimmunology 2015, 4, e1049804. [Google Scholar] [CrossRef] [PubMed]
- Scott, D.W.; Chan, F.C.; Hong, F.; Rogic, S.; Tan, K.L.; Meissner, B.; Ben-Neriah, S.; Boyle, M.; Kridel, R.; Telenius, A.; et al. Gene expression-based model using formalin-fixed paraffin-embedded biopsies predicts overall survival in advanced-stage classical Hodgkin lymphoma. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2013, 31, 692–700. [Google Scholar] [CrossRef] [PubMed]
- Muris, J.J.; Meijer, C.J.; Cillessen, S.A.; Vos, W.; Kummer, J.A.; Bladergroen, B.A.; Bogman, M.J.; MacKenzie, M.A.; Jiwa, N.M.; Siegenbeek van Heukelom, L.H.; et al. Prognostic significance of activated cytotoxic T-lymphocytes in primary nodal diffuse large B-cell lymphomas. Leukemia 2004, 18, 589–596. [Google Scholar] [CrossRef] [PubMed]
- Fortis, S.P.; Sofopoulos, M.; Sotiriadou, N.N.; Haritos, C.; Vaxevanis, C.K.; Anastasopoulou, E.A.; Janssen, N.; Arnogiannaki, N.; Ardavanis, A.; Pawelec, G.; et al. Differential intratumoral distributions of CD8 and CD163 immune cells as prognostic biomarkers in breast cancer. J. Immunother. Cancer 2017, 5, 39. [Google Scholar] [CrossRef] [PubMed]
- Ancuta, E.; Ancuţa, C.; Zugun-Eloae, F.; Iordache, C.; Chirieac, R.; Carasevici, E. Predictive value of cellular immune response in cervical cancer. Rom. J. Morphol. Embryol. 2009, 50, 651–655. [Google Scholar] [PubMed]
- Pan, Y.; Yu, Y.; Wang, X.; Zhang, T. Tumor-Associated Macrophages in Tumor Immunity. Front. Immunol. 2020, 11, 583084. [Google Scholar] [CrossRef] [PubMed]
- Murray, P.J.; Allen, J.E.; Biswas, S.K.; Fisher, E.A.; Gilroy, D.W.; Goerdt, S.; Gordon, S.; Hamilton, J.A.; Ivashkiv, L.B.; Lawrence, T.; et al. Macrophage activation and polarization: Nomenclature and experimental guidelines. Immunity 2014, 41, 14–20. [Google Scholar] [CrossRef] [PubMed]
- Galdiero, M.R.; Garlanda, C.; Jaillon, S.; Marone, G.; Mantovani, A. Tumor associated macrophages and neutrophils in tumor progression. J. Cell. Physiol. 2013, 228, 1404–1412. [Google Scholar] [CrossRef] [PubMed]
- Ruffell, B.; Affara, N.I.; Coussens, L.M. Differential macrophage programming in the tumor microenvironment. Trends Immunol. 2012, 33, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Mei, J.; Xiao, Z.; Guo, C.; Pu, Q.; Ma, L.; Liu, C.; Lin, F.; Liao, H.; You, Z.; Liu, L. Prognostic impact of tumor-associated macrophage infiltration in non-small cell lung cancer: A systemic review and meta-analysis. Oncotarget 2016, 7, 34217–34228. [Google Scholar] [CrossRef] [PubMed]
- Fujimura, T.; Aiba, S. Significance of Immunosuppressive Cells as a Target for Immunotherapies in Melanoma and Non-Melanoma Skin Cancers. Biomolecules 2020, 10, 1087. [Google Scholar] [CrossRef] [PubMed]
- Tiainen, S.; Masarwah, A.; Oikari, S.; Rilla, K.; Hämäläinen, K.; Sudah, M.; Sutela, A.; Vanninen, R.; Ikonen, J.; Tammi, R.; et al. Microenvironment and breast cancer survival: Combined effects of breast fat, M2 macrophages and hyaluronan create a dismal prognosis. Breast Cancer Res. Treat. 2020, 179, 565–575. [Google Scholar] [CrossRef] [PubMed]
- Garvin, S.; Oda, H.; Arnesson, L.G.; Lindström, A.; Shabo, I. Tumor cell expression of CD163 is associated to postoperative radiotherapy and poor prognosis in patients with breast cancer treated with breast-conserving surgery. J. Cancer Res. Clin. Oncol. 2018, 144, 1253–1263. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.J.; Han, L.F.; Wu, X.G.; Wei, W.F.; Wu, L.F.; Yi, H.Y.; Yan, R.M.; Bai, X.Y.; Zhong, M.; Yu, Y.H.; et al. Clinical Significance of CD163+ and CD68+ Tumor-associated Macrophages in High-risk HPV-related Cervical Cancer. J. Cancer 2017, 8, 3868–3875. [Google Scholar] [CrossRef] [PubMed]
- Reinartz, S.; Schumann, T.; Finkernagel, F.; Wortmann, A.; Jansen, J.M.; Meissner, W.; Krause, M.; Schwörer, A.M.; Wagner, U.; Müller-Brüsselbach, S.; et al. Mixed-polarization phenotype of ascites-associated macrophages in human ovarian carcinoma: Correlation of CD163 expression, cytokine levels and early relapse. Int. J. Cancer 2014, 134, 32–42. [Google Scholar] [CrossRef] [PubMed]
- Lan, C.; Huang, X.; Lin, S.; Huang, H.; Cai, Q.; Wan, T.; Lu, J.; Liu, J. Expression of M2-polarized macrophages is associated with poor prognosis for advanced epithelial ovarian cancer. Technol. Cancer Res. Treat. 2013, 12, 259–267. [Google Scholar] [CrossRef] [PubMed]
- Denkert, C.; von Minckwitz, G.; Brase, J.C.; Sinn, B.V.; Gade, S.; Kronenwett, R.; Pfitzner, B.M.; Salat, C.; Loi, S.; Schmitt, W.D.; et al. Tumor-infiltrating lymphocytes and response to neoadjuvant chemotherapy with or without carboplatin in human epidermal growth factor receptor 2-positive and triple-negative primary breast cancers. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2015, 33, 983–991. [Google Scholar] [CrossRef] [PubMed]
- Loi, S.; Michiels, S.; Salgado, R.; Sirtaine, N.; Jose, V.; Fumagalli, D.; Kellokumpu-Lehtinen, P.L.; Bono, P.; Kataja, V.; Desmedt, C.; et al. Tumor infiltrating lymphocytes are prognostic in triple negative breast cancer and predictive for trastuzumab benefit in early breast cancer: Results from the FinHER trial. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2014, 25, 1544–1550. [Google Scholar] [CrossRef] [PubMed]
- Loi, S.; Sirtaine, N.; Piette, F.; Salgado, R.; Viale, G.; Van Eenoo, F.; Rouas, G.; Francis, P.; Crown, J.P.; Hitre, E.; et al. Prognostic and predictive value of tumor-infiltrating lymphocytes in a phase III randomized adjuvant breast cancer trial in node-positive breast cancer comparing the addition of docetaxel to doxorubicin with doxorubicin-based chemotherapy: BIG 02-98. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2013, 31, 860–867. [Google Scholar] [CrossRef] [PubMed]
- Denkert, C.; Loibl, S.; Noske, A.; Roller, M.; Müller, B.M.; Komor, M.; Budczies, J.; Darb-Esfahani, S.; Kronenwett, R.; Hanusch, C.; et al. Tumor-associated lymphocytes as an independent predictor of response to neoadjuvant chemotherapy in breast cancer. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2010, 28, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Zhou, R.; Zhang, J.; Zeng, D.; Sun, H.; Rong, X.; Shi, M.; Bin, J.; Liao, Y.; Liao, W. Immune cell infiltration as a biomarker for the diagnosis and prognosis of stage I-III colon cancer. Cancer Immunol. Immunother. 2019, 68, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Lan, S.; Zheng, Q. Prognostic significance of infiltrating immune cell subtypes in invasive ductal carcinoma of the breast. Tumori 2018, 104, 196–201. [Google Scholar] [CrossRef] [PubMed]
- Finotello, F.; Trajanoski, Z. Quantifying tumor-infiltrating immune cells from transcriptomics data. Cancer Immunol. Immunother. 2018, 67, 1031–1040. [Google Scholar] [CrossRef] [PubMed]
- Balermpas, P.; Michel, Y.; Wagenblast, J.; Seitz, O.; Weiss, C.; Rödel, F.; Rödel, C.; Fokas, E. Tumour-infiltrating lymphocytes predict response to definitive chemoradiotherapy in head and neck cancer. Br. J. Cancer 2014, 110, 501–509. [Google Scholar] [CrossRef]
- Balermpas, P.; Rödel, F.; Rödel, C.; Krause, M.; Linge, A.; Lohaus, F.; Baumann, M.; Tinhofer, I.; Budach, V.; Gkika, E.; et al. CD8+ tumour-infiltrating lymphocytes in relation to HPV status and clinical outcome in patients with head and neck cancer after postoperative chemoradiotherapy: A multicentre study of the German cancer consortium radiation oncology group (DKTK-ROG). Int. J. Cancer 2016, 138, 171–181. [Google Scholar] [CrossRef] [PubMed]
- Pagès, F.; Kirilovsky, A.; Mlecnik, B.; Asslaber, M.; Tosolini, M.; Bindea, G.; Lagorce, C.; Wind, P.; Marliot, F.; Bruneval, P.; et al. In situ cytotoxic and memory T cells predict outcome in patients with early-stage colorectal cancer. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2009, 27, 5944–5951. [Google Scholar] [CrossRef] [PubMed]
- Lei, Y.; Xie, Y.; Tan, Y.S.; Prince, M.E.; Moyer, J.S.; Nör, J.; Wolf, G.T. Telltale tumor infiltrating lymphocytes (TIL) in oral, head & neck cancer. Oral Oncol. 2016, 61, 159–165. [Google Scholar] [PubMed]
- Salama, P.; Phillips, M.; Grieu, F.; Morris, M.; Zeps, N.; Joseph, D.; Platell, C.; Iacopetta, B. Tumor-Infiltrating FOXP3+ T Regulatory Cells Show Strong Prognostic Significance in Colorectal Cancer. J. Clin. Oncol. 2009, 27, 186–192. [Google Scholar] [CrossRef] [PubMed]
- Noy, R.; Pollard, J.W. Tumor-associated macrophages: From mechanisms to therapy. Immunity 2014, 41, 49–61. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Xu, F.; Zhang, J.; Wang, L.; Zheng, Y.; Wu, X.; Wang, J.; Huang, Q.; Lai, M. Tumor-associated macrophages remodeling EMT and predicting survival in colorectal carcinoma. Oncoimmunology 2018, 7, e1380765. [Google Scholar] [CrossRef]
- Sinicrope, F.A.; Rego, R.L.; Ansell, S.M.; Knutson, K.L.; Foster, N.R.; Sargent, D.J. Intraepithelial Effector (CD3+)/Regulatory (FoxP3+) T-Cell Ratio Predicts a Clinical Outcome of Human Colon Carcinoma. Gastroenterology 2009, 137, 1270–1279. [Google Scholar] [CrossRef] [PubMed]
- Charoentong, P.; Finotello, F.; Angelova, M.; Mayer, C.; Efremova, M.; Rieder, D.; Hackl, H.; Trajanoski, Z. Pan-cancer Immunogenomic Analyses Reveal Genotype-Immunophenotype Relationships and Predictors of Response to Checkpoint Blockade. Cell Rep. 2017, 18, 248–262. [Google Scholar] [CrossRef] [PubMed]
- Karpinski, P.; Rossowska, J.; Malgorzata Sasiadek, M. Immunological landscape of consensus clusters in colorectal cancer. Oncotarget 2017, 8, 105299–105311. [Google Scholar] [CrossRef] [PubMed]
- Aran, D.; Hu, Z.; Butte, A.J. xCell: Digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017, 18, 220. [Google Scholar] [CrossRef]
- Clough, E.; Barrett, T. The Gene Expression Omnibus Database. Methods Mol. Biol. 2016, 1418, 93–110. [Google Scholar] [PubMed]
- Conroy, J.M.; Pabla, S.; Glenn, S.T.; Burgher, B.; Nesline, M.; Papanicolau-Sengos, A.; Andreas, J.; Giamo, V.; Lenzo, F.L.; Hyland, F.C.L.; et al. Analytical Validation of a Next-Generation Sequencing Assay to Monitor Immune Responses in Solid Tumors. J. Mol. Diagn. 2018, 20, 95–109. [Google Scholar] [CrossRef] [PubMed]
- Weinstein, J.N.; Collisson, E.A.; Mills, G.B.; Shaw, K.R.; Ozenberger, B.A.; Ellrott, K.; Shmulevich, I.; Sander, C.; Stuart, J.M. The Cancer Genome Atlas Pan-Cancer analysis project. Nat. Genet. 2013, 45, 1113–1120. [Google Scholar] [CrossRef] [PubMed]
- Hedberg, M.L.; Goh, G.; Chiosea, S.I.; Bauman, J.E.; Freilino, M.L.; Zeng, Y.; Wang, L.; Diergaarde, B.B.; Gooding, W.E.; Lui, V.W.; et al. Genetic landscape of metastatic and recurrent head and neck squamous cell carcinoma. J. Clin. Investig. 2016, 126, 169–180. [Google Scholar] [CrossRef] [PubMed]
- Subramanian, A.; Tamayo, P.; Mootha, V.K.; Mukherjee, S.; Ebert, B.L.; Gillette, M.A.; Paulovich, A.; Pomeroy, S.L.; Golub, T.R.; Lander, E.S.; et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 2005, 102, 15545–15550. [Google Scholar] [CrossRef] [PubMed]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef] [PubMed]
- Yoshihara, K.; Shahmoradgoli, M.; Martínez, E.; Vegesna, R.; Kim, H.; Torres-Garcia, W.; Treviño, V.; Shen, H.; Laird, P.W.; Levine, D.A.; et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat. Commun. 2013, 4, 2612. [Google Scholar] [CrossRef] [PubMed]
- Barbie, D.A.; Tamayo, P.; Boehm, J.S.; Kim, S.Y.; Moody, S.E.; Dunn, I.F.; Schinzel, A.C.; Sandy, P.; Meylan, E.; Scholl, C.; et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 2009, 462, 108–112. [Google Scholar] [CrossRef] [PubMed]
- Ye, L.; Zhang, T.; Kang, Z.; Guo, G.; Sun, Y.; Lin, K.; Huang, Q.; Shi, X.; Ni, Z.; Ding, N.; et al. Tumor-Infiltrating Immune Cells Act as a Marker for Prognosis in Colorectal Cancer. Front. Immunol. 2019, 10, 2368. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, M.; Kuwata, T.; Setsuda, A.; Tokunaga, M.; Kaito, A.; Sugita, S.; Tonouchi, A.; Kinoshita, T.; Nagino, M. Molecular and pathological analyses of gastric stump cancer by next-generation sequencing and immunohistochemistry. Sci. Rep. 2021, 11, 4165. [Google Scholar] [CrossRef] [PubMed]
- Hao, Y.; Yan, M.; Heath, B.R.; Lei, Y.L.; Xie, Y. Fast and robust deconvolution of tumor infiltrating lymphocyte from expression profiles using least trimmed squares. PLoS Comput. Biol. 2019, 15, e1006976. [Google Scholar] [CrossRef] [PubMed]
- Vallania, F.; Tam, A.; Lofgren, S.; Schaffert, S.; Azad, T.D.; Bongen, E.; Haynes, W.; Alsup, M.; Alonso, M.; Davis, M.; et al. Leveraging heterogeneity across multiple datasets increases cell-mixture deconvolution accuracy and reduces biological and technical biases. Nat. Commun. 2018, 9, 4735. [Google Scholar] [CrossRef] [PubMed]
- Bruni, D.; Angell, H.K.; Galon, J. The immune contexture and Immunoscore in cancer prognosis and therapeutic efficacy. Nat. Rev. Cancer 2020, 20, 662–680. [Google Scholar] [CrossRef] [PubMed]
- Allam, M.; Hu, T.; Lee, J.; Aldrich, J.; Badve, S.S.; Gökmen-Polar, Y.; Bhave, M.; Ramalingam, S.S.; Schneider, F.; Coskun, A.F. Spatially variant immune infiltration scoring in human cancer tissues. NPJ Precis. Oncol. 2022, 6, 60. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Zhang, Y.; Shang, X.; Zhang, Z. ProTICS reveals prognostic impact of tumor infiltrating immune cells in different molecular subtypes. Brief. Bioinform. 2021, 22, bbab164. [Google Scholar] [CrossRef] [PubMed]
- Gregory, A.D.; Houghton, A.M. Tumor-associated neutrophils: New targets for cancer therapy. Cancer Res. 2011, 71, 2411–2416. [Google Scholar] [CrossRef] [PubMed]
- Di Giacomo, A.M.; Calabrò, L.; Danielli, R.; Fonsatti, E.; Bertocci, E.; Pesce, I.; Fazio, C.; Cutaia, O.; Giannarelli, D.; Miracco, C.; et al. Long-term survival and immunological parameters in metastatic melanoma patients who responded to ipilimumab 10 mg/kg within an expanded access programme. Cancer Immunol. Immunother. 2013, 62, 1021–1028. [Google Scholar] [CrossRef] [PubMed]
- Wei, B.; Kong, W.; Mou, X.; Wang, S. Comprehensive analysis of tumor immune infiltration associated with endogenous competitive RNA networks in lung adenocarcinoma. Pathol. Res. Pract. 2019, 215, 159–170. [Google Scholar] [CrossRef] [PubMed]
- Zuo, S.; Wei, M.; Wang, S.; Dong, J.; Wei, J. Pan-Cancer Analysis of Immune Cell Infiltration Identifies a Prognostic Immune-Cell Characteristic Score (ICCS) in Lung Adenocarcinoma. Front. Immunol. 2020, 11, 1218. [Google Scholar] [CrossRef] [PubMed]
- Pimenoff, V.N.; Tous, S.; Benavente, Y.; Alemany, L.; Quint, W.; Bosch, F.X.; Bravo, I.G.; de Sanjosé, S. Distinct geographic clustering of oncogenic human papillomaviruses multiple infections in cervical cancers: Results from a worldwide cross-sectional study. Int. J. Cancer 2019, 144, 2478–2488. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Jiang, G.; Gonzalez-Rivas, D.; Zhang, P. An individualized immune prognostic signature in lung adenocarcinoma. Cancer Cell Int. 2020, 20, 156. [Google Scholar] [CrossRef] [PubMed]
- Edin, S.; Kaprio, T.; Hagström, J.; Larsson, P.; Mustonen, H.; Böckelman, C.; Strigård, K.; Gunnarsson, U.; Haglund, C.; Palmqvist, R. The Prognostic Importance of CD20+ B lymphocytes in Colorectal Cancer and the Relation to Other Immune Cell subsets. Sci. Rep. 2019, 9, 19997. [Google Scholar] [CrossRef] [PubMed]
- Berntsson, J.; Nodin, B.; Eberhard, J.; Micke, P.; Jirström, K. Prognostic impact of tumour-infiltrating B cells and plasma cells in colorectal cancer. Int. J. Cancer 2016, 139, 1129–1139. [Google Scholar] [CrossRef]
- Xia, J.; Xie, Z.; Niu, G.; Lu, Z.; Wang, Z.; Xing, Y.; Ren, J.; Hu, Z.; Hong, R.; Cao, Z.; et al. Single-cell landscape and clinical outcomes of infiltrating B cells in colorectal cancer. Immunology 2023, 168, 135–151. [Google Scholar] [CrossRef] [PubMed]
- Ma, J.; Zheng, B.; Goswami, S.; Meng, L.; Zhang, D.; Cao, C.; Li, T.; Zhu, F.; Ma, L.; Zhang, Z.; et al. PD1Hi CD8+ T cells correlate with exhausted signature and poor clinical outcome in hepatocellular carcinoma. J. Immunother. Cancer 2019, 7, 331. [Google Scholar] [CrossRef] [PubMed]
- Granier, C.; Dariane, C.; Combe, P.; Verkarre, V.; Urien, S.; Badoual, C.; Roussel, H.; Mandavit, M.; Ravel, P.; Sibony, M.; et al. Tim-3 Expression on Tumor-Infiltrating PD-1+CD8+ T Cells Correlates with Poor Clinical Outcome in Renal Cell Carcinoma. Cancer Res. 2017, 77, 1075–1082. [Google Scholar] [CrossRef] [PubMed]
- Kansy, B.A.; Concha-Benavente, F.; Srivastava, R.M.; Jie, H.B.; Shayan, G.; Lei, Y.; Moskovitz, J.; Moy, J.; Li, J.; Brandau, S.; et al. PD-1 Status in CD8+ T Cells Associates with Survival and Anti-PD-1 Therapeutic Outcomes in Head and Neck Cancer. Cancer Res. 2017, 77, 6353–6364. [Google Scholar] [CrossRef] [PubMed]
- Muenst, S.; Soysal, S.D.; Gao, F.; Obermann, E.C.; Oertli, D.; Gillanders, W.E. The presence of programmed death 1 (PD-1)-positive tumor-infiltrating lymphocytes is associated with poor prognosis in human breast cancer. Breast Cancer Res. Treat. 2013, 139, 667–676. [Google Scholar] [CrossRef] [PubMed]
- van den Broek, M.E.; Kägi, D.; Ossendorp, F.; Toes, R.; Vamvakas, S.; Lutz, W.K.; Melief, C.J.; Zinkernagel, R.M.; Hengartner, H. Decreased tumor surveillance in perforin-deficient mice. J. Exp. Med. 1996, 184, 1781–1790. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Jiang, P.; Wei, S.; Xu, X.; Wang, J. Regulatory T cells in tumor microenvironment: New mechanisms, potential therapeutic strategies and future prospects. Mol. Cancer 2020, 19, 116. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Toker, A.; Liu, Z.Q.; Ohashi, P.S. Turning the Tide Against Regulatory T Cells. Front. Oncol. 2019, 9, 279. [Google Scholar] [CrossRef] [PubMed]
- Cao, X.; Cai, S.F.; Fehniger, T.A.; Song, J.; Collins, L.I.; Piwnica-Worms, D.R.; Ley, T.J. Granzyme B and perforin are important for regulatory T cell-mediated suppression of tumor clearance. Immunity 2007, 27, 635–646. [Google Scholar] [CrossRef] [PubMed]
- Deaglio, S.; Dwyer, K.M.; Gao, W.; Friedman, D.; Usheva, A.; Erat, A.; Chen, J.F.; Enjyoji, K.; Linden, J.; Oukka, M.; et al. Adenosine generation catalyzed by CD39 and CD73 expressed on regulatory T cells mediates immune suppression. J. Exp. Med. 2007, 204, 1257–1265. [Google Scholar] [CrossRef]
- Garnelo, M.; Tan, A.; Her, Z.; Yeong, J.; Lim, C.J.; Chen, J.; Lim, K.H.; Weber, A.; Chow, P.; Chung, A.; et al. Interaction between tumour-infiltrating B cells and T cells controls the progression of hepatocellular carcinoma. Gut 2017, 66, 342–351. [Google Scholar] [CrossRef] [PubMed]
- Forssell, J.; Öberg Ak Henriksson, M.L.; Stenling, R.; Jung, A.; Palmqvist, R. High Macrophage Infiltration along the Tumor Front Correlates with Improved Survival in Colon Cancer. Clin. Cancer Res. 2007, 13, 1472–1479. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.-w.; Liu, L.; Gong, C.-y.; Shi, H.-s.; Zeng, Y.-h.; Wang, X.-z.; Zhao, Y.-w.; Wei, Y.-q. Prognostic Significance of Tumor-Associated Macrophages in Solid Tumor: A Meta-Analysis of the Literature. PLoS ONE 2012, 7, e50946. [Google Scholar] [CrossRef] [PubMed]
- Shabo, I.; Stål, O.; Olsson, H.; Doré, S.; Svanvik, J. Breast cancer expression of CD163, a macrophage scavenger receptor, is related to early distant recurrence and reduced patient survival. Int. J. Cancer 2008, 123, 780–786. [Google Scholar] [CrossRef] [PubMed]
- Shabo, I.; Olsson, H.; Sun, X.F.; Svanvik, J. Expression of the macrophage antigen CD163 in rectal cancer cells is associated with early local recurrence and reduced survival time. Int. J. Cancer 2009, 125, 1826–1831. [Google Scholar] [CrossRef] [PubMed]
- Biswas, S.K.; Mantovani, A. Macrophage plasticity and interaction with lymphocyte subsets: Cancer as a paradigm. Nat. Immunol. 2010, 11, 889–896. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Sozzani, S.; Locati, M.; Allavena, P.; Sica, A. Macrophage polarization: Tumor-associated macrophages as a paradigm for polarized M2 mononuclear phagocytes. Trends Immunol. 2002, 23, 549–555. [Google Scholar] [CrossRef] [PubMed]
- Mosser, D.M.; Edwards, J.P. Exploring the full spectrum of macrophage activation. Nat. Rev. Immunol. 2008, 8, 958–969. [Google Scholar] [CrossRef] [PubMed]
- Sica, A.; Mantovani, A. Macrophage plasticity and polarization: In vivo veritas. J. Clin. Investig. 2012, 122, 787–795. [Google Scholar] [CrossRef] [PubMed]
- Wang, B.; Liu, H.; Dong, X.; Wu, S.; Zeng, H.; Liu, Z.; Wan, D.; Dong, W.; He, W.; Chen, X.; et al. High CD204+ tumor-infiltrating macrophage density predicts a poor prognosis in patients with urothelial cell carcinoma of the bladder. Oncotarget 2015, 6, 20204–20214. [Google Scholar] [CrossRef]
- Shigeoka, M.; Urakawa, N.; Nakamura, T.; Nishio, M.; Watajima, T.; Kuroda, D.; Komori, T.; Kakeji, Y.; Semba, S.; Yokozaki, H. Tumor associated macrophage expressing CD204 is associated with tumor aggressiveness of esophageal squamous cell carcinoma. Cancer Sci. 2013, 104, 1112–1119. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, M.; Mitsunaga, S.; Yoshikawa, K.; Kato, Y.; Gotohda, N.; Takahashi, S.; Konishi, M.; Ikeda, M.; Kojima, M.; Ochiai, A.; et al. Prognostic impact of M2 macrophages at neural invasion in patients with invasive ductal carcinoma of the pancreas. Eur. J. Cancer 2014, 50, 1900–1908. [Google Scholar] [CrossRef] [PubMed]
- Ohtaki, Y.; Ishii, G.; Nagai, K.; Ashimine, S.; Kuwata, T.; Hishida, T.; Nishimura, M.; Yoshida, J.; Takeyoshi, I.; Ochiai, A. Stromal macrophage expressing CD204 is associated with tumor aggressiveness in lung adenocarcinoma. J. Thorac. Oncol. Off. Publ. Int. Assoc. Study Lung Cancer 2010, 5, 1507–1515. [Google Scholar] [CrossRef] [PubMed]
- Kubota, K.; Moriyama, M.; Furukawa, S.; Rafiul, H.; Maruse, Y.; Jinno, T.; Tanaka, A.; Ohta, M.; Ishiguro, N.; Yamauchi, M.; et al. CD163+CD204+ tumor-associated macrophages contribute to T cell regulation via interleukin-10 and PD-L1 production in oral squamous cell carcinoma. Sci. Rep. 2017, 7, 1755. [Google Scholar] [CrossRef] [PubMed]
- Sato, J.; Kitano, S.; Motoi, N.; Ino, Y.; Yamamoto, N.; Watanabe, S.; Ohe, Y.; Hiraoka, N. CD20+ tumor-infiltrating immune cells and CD204+ M2 macrophages are associated with prognosis in thymic carcinoma. Cancer Sci. 2020, 111, 1921–1932. [Google Scholar] [CrossRef] [PubMed]
- Skytthe, M.K.; Graversen, J.H.; Moestrup, S.K. Targeting of CD163+ Macrophages in Inflammatory and Malignant Diseases. Int. J. Mol. Sci. 2020, 21, 5497. [Google Scholar] [CrossRef] [PubMed]
- Dunn, G.P.; Old, L.J.; Schreiber, R.D. The Three Es of Cancer Immunoediting. Annu. Rev. Immunol. 2004, 22, 329–360. [Google Scholar] [CrossRef]
- Hiraoka, N.; Onozato, K.; Kosuge, T.; Hirohashi, S. Prevalence of FOXP3+ Regulatory T Cells Increases During the Progression of Pancreatic Ductal Adenocarcinoma and Its Premalignant Lesions. Clin. Cancer Res. 2006, 12, 5423–5434. [Google Scholar] [CrossRef] [PubMed]
- Sica, A.; Bronte, V. Altered macrophage differentiation and immune dysfunction in tumor development. J. Clin. Investig. 2007, 117, 1155–1166. [Google Scholar] [CrossRef] [PubMed]
- Prizment, A.E.; Vierkant, R.A.; Smyrk, T.C.; Tillmans, L.S.; Lee, J.J.; Sriramarao, P.; Nelson, H.H.; Lynch, C.F.; Thibodeau, S.N.; Church, T.R.; et al. Tumor eosinophil infiltration and improved survival of colorectal cancer patients: Iowa Women’s Health Study. Mod. Pathol. 2016, 29, 516–527. [Google Scholar] [CrossRef] [PubMed]
- Somasundaram, R.; Connelly, T.; Choi, R.; Choi, H.; Samarkina, A.; Li, L.; Gregorio, E.; Chen, Y.; Thakur, R.; Abdel-Mohsen, M.; et al. Tumor-infiltrating mast cells are associated with resistance to anti-PD-1 therapy. Nat. Commun. 2021, 12, 346. [Google Scholar] [CrossRef] [PubMed]
- Geng, S.-K.; Fu, S.-M.; Ma, S.-H.; Fu, Y.-P.; Zhang, H.-W. Tumor infiltrating neutrophil might play a major role in predicting the clinical outcome of breast cancer patients treated with neoadjuvant chemotherapy. BMC Cancer 2021, 21, 68. [Google Scholar] [CrossRef] [PubMed]
- Yan, M.; Zheng, M.; Niu, R.; Yang, X.; Tian, S.; Fan, L.; Li, Y.; Zhang, S. Roles of tumor-associated neutrophils in tumor metastasis and its clinical applications. Front. Cell Dev. Biol. 2022, 10, 938289. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Zhao, Y.; Wang, X.; Chen, N.; Mao, F.; Teng, Y.; Wang, T.; Peng, L.; Zhang, J.; Cheng, P.; et al. Increased intratumoral mast cells foster immune suppression and gastric cancer progression through TNF-α-PD-L1 pathway. J. Immunother. Cancer 2019, 7, 54. [Google Scholar] [CrossRef] [PubMed]
- Demkow, U. Neutrophil Extracellular Traps (NETs) in Cancer Invasion, Evasion and Metastasis. Cancers 2021, 13, 4495. [Google Scholar] [CrossRef] [PubMed]
- Chalmers, Z.R.; Connelly, C.F.; Fabrizio, D.; Gay, L.; Ali, S.M.; Ennis, R.; Schrock, A.; Campbell, B.; Shlien, A.; Chmielecki, J.; et al. Analysis of 100,000 human cancer genomes reveals the landscape of tumor mutational burden. Genome Med. 2017, 9, 34. [Google Scholar] [CrossRef] [PubMed]
- Luchini, C.; Bibeau, F.; Ligtenberg, M.J.L.; Singh, N.; Nottegar, A.; Bosse, T.; Miller, R.; Riaz, N.; Douillard, J.Y.; Andre, F.; et al. ESMO recommendations on microsatellite instability testing for immunotherapy in cancer, and its relationship with PD-1/PD-L1 expression and tumour mutational burden: A systematic review-based approach. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2019, 30, 1232–1243. [Google Scholar] [CrossRef] [PubMed]
- Marabelle, A.; Fakih, M.; Lopez, J.; Shah, M.; Shapira-Frommer, R.; Nakagawa, K.; Chung, H.C.; Kindler, H.L.; Lopez-Martin, J.A.; Miller, W.H., Jr.; et al. Association of tumour mutational burden with outcomes in patients with advanced solid tumours treated with pembrolizumab: Prospective biomarker analysis of the multicohort, open-label, phase 2 KEYNOTE-158 study. Lancet Oncol. 2020, 21, 1353–1365. [Google Scholar] [CrossRef] [PubMed]
- Schrock, A.B.; Ouyang, C.; Sandhu, J.; Sokol, E.; Jin, D.; Ross, J.S.; Miller, V.A.; Lim, D.; Amanam, I.; Chao, J.; et al. Tumor mutational burden is predictive of response to immune checkpoint inhibitors in MSI-high metastatic colorectal cancer. Ann. Oncol. Off. J. Eur. Soc. Med. Oncol. 2019, 30, 1096–1103. [Google Scholar] [CrossRef] [PubMed]
- Samstein, R.M.; Lee, C.H.; Shoushtari, A.N.; Hellmann, M.D.; Shen, R.; Janjigian, Y.Y.; Barron, D.A.; Zehir, A.; Jordan, E.J.; Omuro, A.; et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat. Genet. 2019, 51, 202–206. [Google Scholar] [CrossRef] [PubMed]
- Strickler, J.H.; Hanks, B.A.; Khasraw, M. Tumor Mutational Burden as a Predictor of Immunotherapy Response: Is More Always Better? Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2021, 27, 1236–1241. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Chen, S.; Hu, Y.; Huang, W. Landscape of Immune Microenvironment Under Immune Cell Infiltration Pattern in Breast Cancer. Front. Immunol. 2021, 12, 711433. [Google Scholar] [CrossRef] [PubMed]
- Wu, W.; Wang, X.; Le, W.; Lu, C.; Li, H.; Zhu, Y.; Chen, X.; An, W.; Xu, C.; Wu, Q.; et al. Immune microenvironment infiltration landscape and immune-related subtypes in prostate cancer. Front. Immunol. 2022, 13, 1001297. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Tang, D.; Gu, C.; Wang, B.; Yao, Y.; Wang, R.; Zhang, H.; Gao, W. Characterization of the Immune Microenvironmental Landscape of Lung Squamous Cell Carcinoma with Immune Cell Infiltration. Dis. Markers 2022, 2022, 2361507. [Google Scholar] [CrossRef]
- Yarchoan, M.; Hopkins, A.; Jaffee, E.M. Tumor Mutational Burden and Response Rate to PD-1 Inhibition. N. Engl. J. Med. 2017, 377, 2500–2501. [Google Scholar] [CrossRef]
- Gubin, M.M.; Artyomov, M.N.; Mardis, E.R.; Schreiber, R.D. Tumor neoantigens: Building a framework for personalized cancer immunotherapy. J. Clin. Investig. 2015, 125, 3413–3421. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Xu, J.; Du, C.; Wu, Y.; Xia, D.; Lv, W.; Hu, J. The Predictive Value of Tumor Mutation Burden on Efficacy of Immune Checkpoint Inhibitors in Cancers: A Systematic Review and Meta-Analysis. Front. Oncol. 2019, 9, 1161. [Google Scholar] [CrossRef] [PubMed]
- Sholl, L.M.; Hirsch, F.R.; Hwang, D.; Botling, J.; Lopez-Rios, F.; Bubendorf, L.; Mino-Kenudson, M.; Roden, A.C.; Beasley, M.B.; Borczuk, A.; et al. The Promises and Challenges of Tumor Mutation Burden as an Immunotherapy Biomarker: A Perspective from the International Association for the Study of Lung Cancer Pathology Committee. J. Thorac. Oncol. Off. Publ. Int. Assoc. Study Lung Cancer 2020, 15, 1409–1424. [Google Scholar] [CrossRef] [PubMed]
- Büttner, R.; Gosney, J.R.; Skov, B.G.; Adam, J.; Motoi, N.; Bloom, K.J.; Dietel, M.; Longshore, J.W.; López-Ríos, F.; Penault-Llorca, F.; et al. Programmed Death-Ligand 1 Immunohistochemistry Testing: A Review of Analytical Assays and Clinical Implementation in Non-Small-Cell Lung Cancer. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2017, 35, 3867–3876. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Jin, C.; Dong, X.; Wen, J.; Xia, E.; Wang, Q.; Wang, O. Pan-cancer analysis of the prognostic and immunological role of PSMB8. Sci. Rep. 2021, 11, 20492. [Google Scholar] [CrossRef] [PubMed]
- Xu, W.X.; Zhang, J.; Hua, Y.T.; Yang, S.J.; Wang, D.D.; Tang, J.H. An Integrative Pan-Cancer Analysis Revealing LCN2 as an Oncogenic Immune Protein in Tumor Microenvironment. Front. Oncol. 2020, 10, 605097. [Google Scholar] [CrossRef] [PubMed]
- Wen, J.; Mao, X.; Cheng, Q.; Liu, Z.; Liu, F. A pan-cancer analysis revealing the role of TIGIT in tumor microenvironment. Sci. Rep. 2021, 11, 22502. [Google Scholar] [CrossRef] [PubMed]
- Lin, T.; Zhang, Y.; Lin, Z.; Peng, L. Roles of HMGBs in Prognosis and Immunotherapy: A Pan-Cancer Analysis. Front. Genet. 2021, 12, 764245. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Gui, Y.; Yuan, T.; Liao, G.; Bian, C.; Jiang, Q.; Huang, S.; Liu, B.; Wu, D. Overexpression of high mobility group box 1 with poor prognosis in patients after radical prostatectomy. BJU Int. 2012, 110, E1125–E1130. [Google Scholar] [CrossRef]
- Liu, H.; Yang, M.; Dong, Z. HSPB11 is a Prognostic Biomarker Associated with Immune Infiltrates in Hepatocellular Carcinoma. Int. J. Gen. Med. 2022, 15, 4017–4027. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Li, L.; Wu, X.; Yu, J.; Ma, H.; Zhang, R.; Li, Y.; Wang, W. Loss of SDC1 Expression Is Associated with Poor Prognosis of Colorectal Cancer Patients in Northern China. Dis. Markers 2019, 2019, 3768708. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.J.; Li, J.P.; Zeng, S.H.; Han, M.; Liu, S.L.; Zou, X. DZIP1 Expression as a Prognostic Marker in Gastric Cancer: A Bioinformatics-Based Analysis. Pharmacogenomics Pers. Med. 2021, 14, 1151–1168. [Google Scholar] [CrossRef]
- Peng, Y.; Li, H.; Fu, Y.; Guo, S.; Qu, C.; Zhang, Y.; Zong, B.; Liu, S. JAM2 predicts a good prognosis and inhibits invasion and migration by suppressing EMT pathway in breast cancer. Int. Immunopharmacol. 2022, 103, 108430. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Fang, L.; Zhang, Z.P.; Yan, Z.L. TPM1 is a Novel Predictive Biomarker for Gastric Cancer Diagnosis and Prognosis. Clin. Lab. 2020, 66, 489–494. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.; Huang, Z.; Zhao, X.; Wang, Y.; Zhao, H.; Zhong, Z.; Wang, L. Overexpression of high mobility group box 1 contributes to progressive clinicopathological features and poor prognosis of human bladder urothelial carcinoma. OncoTargets Ther. 2018, 11, 2111–2120. [Google Scholar] [CrossRef] [PubMed]
- Bao, G.; Qiao, Q.; Zhao, H.; He, X. Prognostic value of HMGB1 overexpression in resectable gastric adenocarcinomas. World J. Surg. Oncol. 2010, 8, 52. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.H.; Shen, X.; Wei, Y.; Chen, Y.; Chai, H.; Xia, L.; Leng, W. A Pan-Cancer Analysis Reveals the Prognostic and Immunotherapeutic Value of Stanniocalcin-2 (STC2). Front. Genet. 2022, 13, 927046. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.X.; Luo, J.H.; Wu, L.Q. FN1 overexpression is correlated with unfavorable prognosis and immune infiltrates in breast cancer. Front. Genet. 2022, 13, 913659. [Google Scholar] [CrossRef]
- de la Cruz-Merino, L.; Palazón-Carrión, N.; Henao-Carrasco, F.; Nogales-Fernández, E.; Álamo-de la Gala, M.; Vallejo-Benítez, A.; Chiesa, M.; Sánchez-Margalet, V.; GEICAM; GÉTICA. New horizons in breast cancer: The promise of immunotherapy. Clin. Transl. Oncol. 2019, 21, 117–125. [Google Scholar] [CrossRef] [PubMed]
- Lecerf, C.; Kamal, M.; Vacher, S.; Chemlali, W.; Schnitzler, A.; Morel, C.; Dubot, C.; Jeannot, E.; Meseure, D.; Klijanienko, J.; et al. Immune gene expression in head and neck squamous cell carcinoma patients. Eur. J. Cancer 2019, 121, 210–223. [Google Scholar] [CrossRef]
- Bie, L.; Zhao, G.; Wang, Y.P.; Zhang, B. Kinesin family member 2C (KIF2C/MCAK) is a novel marker for prognosis in human gliomas. Clin. Neurol. Neurosurg. 2012, 114, 356–360. [Google Scholar] [CrossRef] [PubMed]
- Ishikawa, K.; Kamohara, Y.; Tanaka, F.; Haraguchi, N.; Mimori, K.; Inoue, H.; Mori, M. Mitotic centromere-associated kinesin is a novel marker for prognosis and lymph node metastasis in colorectal cancer. Br. J. Cancer 2008, 98, 1824–1829. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y.; Tanaka, F.; Haraguchi, N.; Mimori, K.; Matsumoto, T.; Inoue, H.; Yanaga, K.; Mori, M. Clinicopathological and biological significance of mitotic centromere-associated kinesin overexpression in human gastric cancer. Br. J. Cancer 2007, 97, 543–549. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Yang, L.; Qiao, F.; Hu, X.; Li, S.; Yao, L.; Yang, X.L.; Shao, Z.M. High Levels of Nucleolar Spindle-Associated Protein and Reduced Levels of BRCA1 Expression Predict Poor Prognosis in Triple-Negative Breast Cancer. PLoS ONE 2015, 10, e0140572. [Google Scholar] [CrossRef]
- Su, R.; Cai, L.; Xiong, P.; Liu, Z.; Chen, S.; Liu, X.; Lin, R.; Lei, Z.; Tian, D.; Su, M. TLR3 Expression is a Potential Prognosis Biomarker and Shapes the Immune-Active Tumor Microenvironment in Esophageal Squamous Cell Carcinoma. J. Inflamm. Res. 2022, 15, 1437–1456. [Google Scholar] [CrossRef] [PubMed]
- Zeng, L.; Sun, X. Correlation of INHBA Overexpression with Pathological Features, Antitumor Immune Response and Clinical Prognosis in Cervical Cancer. Medicina 2023, 59, 495. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhou, Z.; Jiang, Y.; Lin, Y.; Yang, Y.; Tian, C.; Liu, J.; Lin, H.; Huang, B. EPHA3 Could Be a Novel Prognosis Biomarker and Correlates with Immune Infiltrates in Bladder Cancer. Cancers 2023, 15, 621. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, A.; Chaudhary, A.; Ghosh, A.; Arun, P.; Mukherjee, G.; Arun, I.; Maitra, A.; Biswas, N.; Majumder, P.P. Overexpression of CD73 is associated with recurrence and poor prognosis of gingivobuccal oral cancer as revealed by transcriptome and deep immune profiling of paired tumor and margin tissues. Cancer Med. 2023, 12, 16774–16787. [Google Scholar] [CrossRef] [PubMed]
- Hu, X.; Zhu, H.; Feng, S.; Wang, C.; Ye, Y.; Xiong, X. Transmembrane and coiled-coil domains 3 is a diagnostic biomarker for predicting immune checkpoint blockade efficacy in hepatocellular carcinoma. Front. Genet. 2022, 13, 1006357. [Google Scholar] [CrossRef] [PubMed]
- He, X.; Li, W.; Liang, X.; Zhu, X.; Zhang, L.; Huang, Y.; Yu, T.; Li, S.; Chen, Z. IGF2BP2 Overexpression Indicates Poor Survival in Patients with Acute Myelocytic Leukemia. Cell. Physiol. Biochem. Int. J. Exp. Cell. Physiol. Biochem. Pharmacol. 2018, 51, 1945–1956. [Google Scholar] [CrossRef]
- Fan, T.; Xue, L.; Dong, B.; He, H.; Zhang, W.; Hao, L.; Ma, W.; Zang, G.; Han, C.; Dong, Y. CDH1 overexpression predicts bladder cancer from early stage and inversely correlates with immune infiltration. BMC Urol. 2022, 22, 156. [Google Scholar] [CrossRef] [PubMed]
- Ohtaki, S.; Wanibuchi, M.; Kataoka-Sasaki, Y.; Sasaki, M.; Oka, S.; Noshiro, S.; Akiyama, Y.; Mikami, T.; Mikuni, N.; Kocsis, J.D.; et al. ACTC1 as an invasion and prognosis marker in glioma. J. Neurosurg. 2017, 126, 467–475. [Google Scholar] [CrossRef] [PubMed]
- Rajamani, D.; Bhasin, M.K. Identification of key regulators of pancreatic cancer progression through multidimensional systems-level analysis. Genome Med. 2016, 8, 38. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.; Shu, B.; Chen, L.; Qian, K.; Wang, Y.; Qian, G.; Zhu, Y.; Cao, X.; Xie, C.; Xiao, Y.; et al. Overexpression of COL3A1 confers a poor prognosis in human bladder cancer identified by co-expression analysis. Oncotarget 2017, 8, 70508–70520. [Google Scholar] [CrossRef] [PubMed]
- Chan, Y.X.; Alfonso, H.; Paul Chubb, S.A.; Ho, K.K.Y.; Gerard Fegan, P.; Hankey, G.J.; Golledge, J.; Flicker, L.; Yeap, B.B. Higher IGFBP3 is associated with increased incidence of colorectal cancer in older men independently of IGF1. Clin. Endocrinol. 2018, 88, 333–340. [Google Scholar] [CrossRef] [PubMed]
- Brunner, A.; Prelog, M.; Verdorfer, I.; Tzankov, A.; Mikuz, G.; Ensinger, C. EpCAM is predominantly expressed in high grade and advanced stage urothelial carcinoma of the bladder. J. Clin. Pathol. 2008, 61, 307–310. [Google Scholar] [CrossRef] [PubMed]
- Baeuerle, P.A.; Gires, O. EpCAM (CD326) finding its role in cancer. Br. J. Cancer 2007, 96, 417–423. [Google Scholar] [CrossRef] [PubMed]
- Harada, K.; Masuda, S.; Hirano, M.; Nakanuma, Y. Reduced expression of syndecan-1 correlates with histologic dedifferentiation, lymph node metastasis, and poor prognosis in intrahepatic cholangiocarcinoma. Hum. Pathol. 2003, 34, 857–863. [Google Scholar] [CrossRef]
- Yang, N.; Mosher, R.; Seo, S.; Beebe, D.; Friedl, A. Syndecan-1 in breast cancer stroma fibroblasts regulates extracellular matrix fiber organization and carcinoma cell motility. Am. J. Pathol. 2011, 178, 325–335. [Google Scholar] [CrossRef]
- Bauer, M.; Eickhoff, J.C.; Gould, M.N.; Mundhenke, C.; Maass, N.; Friedl, A. Neutrophil gelatinase-associated lipocalin (NGAL) is a predictor of poor prognosis in human primary breast cancer. Breast Cancer Res. Treat. 2008, 108, 389–397. [Google Scholar] [CrossRef]
- Zhao, Y.Y.; Xiang, Q.M.; Chen, J.L.; Zhang, L.; Zheng, W.L.; Ke, D.; Shi, R.S.; Yang, K.W. SLC25A25-AS1 over-expression could be predicted the dismal prognosis and was related to the immune microenvironment in prostate cancer. Front. Oncol. 2022, 12, 990247. [Google Scholar] [CrossRef]
- Liu, S.; Song, A.; Wu, Y.; Yao, S.; Wang, M.; Niu, T.; Gao, C.; Li, Z.; Zhou, X.; Huo, Z.; et al. Analysis of genomics and immune infiltration patterns of epithelial-mesenchymal transition related to metastatic breast cancer to bone. Transl. Oncol. 2021, 14, 100993. [Google Scholar] [CrossRef]
- Wang, B.; Shen, Y.; Zou, Y.; Qi, Z.; Huang, G.; Xia, S.; Gao, R.; Li, F.; Huang, Z. TOP2A Promotes Cell Migration, Invasion and Epithelial-Mesenchymal Transition in Cervical Cancer via Activating the PI3K/AKT Signaling. Cancer Manag. Res. 2020, 12, 3807–3814. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Shi, M.; Chen, T.; Zhang, B. Characterization of the Immune Cell Infiltration Landscape in Head and Neck Squamous Cell Carcinoma to Aid Immunotherapy. Mol. Ther.-Nucleic Acids 2020, 22, 298–309. [Google Scholar] [CrossRef] [PubMed]
- Wu, M.-H.; Lee, W.-J.; Hua, K.-T.; Kuo, M.-L.; Lin, M.-T. Macrophage Infiltration Induces Gastric Cancer Invasiveness by Activating the β-Catenin Pathway. PLoS ONE 2015, 10, e0134122. [Google Scholar] [CrossRef] [PubMed]
- Richmond, A.; Yang, J. The role of NF-kB in modulating antitumor immunity. Oncoimmunology 2016, 5, e1005522. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, S.; Verma, A.K.; Dev, K.; Goyal, Y.; Bhatt, D.; Alsahli, M.A.; Rahmani, A.H.; Almatroudi, A.; Almatroodi, S.A.; Alrumaihi, F.; et al. Role of Cytokines and Chemokines in NSCLC Immune Navigation and Proliferation. Oxidative Med. Cell. Longev. 2021, 2021, 5563746. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.S.; Mellman, I. Elements of cancer immunity and the cancer-immune set point. Nature 2017, 541, 321–330. [Google Scholar] [CrossRef] [PubMed]
- Şenbabaoğlu, Y.; Gejman, R.S.; Winer, A.G.; Liu, M.; Van Allen, E.M.; de Velasco, G.; Miao, D.; Ostrovnaya, I.; Drill, E.; Luna, A.; et al. Tumor immune microenvironment characterization in clear cell renal cell carcinoma identifies prognostic and immunotherapeutically relevant messenger RNA signatures. Genome Biol. 2016, 17, 231. [Google Scholar] [CrossRef] [PubMed]
- Dunn, G.P.; Old, L.J.; Schreiber, R.D. The immunobiology of cancer immunosurveillance and immunoediting. Immunity 2004, 21, 137–148. [Google Scholar] [CrossRef] [PubMed]
- Ferrone, C.; Dranoff, G. Dual roles for immunity in gastrointestinal cancers. J. Clin. Oncol. Off. J. Am. Soc. Clin. Oncol. 2010, 28, 4045–4051. [Google Scholar] [CrossRef] [PubMed]
- Funada, Y.; Noguchi, T.; Kikuchi, R.; Takeno, S.; Uchida, Y.; Gabbert, H.E. Prognostic significance of CD8+ T cell and macrophage peritumoral infiltration in colorectal cancer. Oncol. Rep. 2003, 10, 309–313. [Google Scholar] [CrossRef]
- Correale, P.; Rotundo, M.S.; Botta, C.; Del Vecchio, M.T.; Ginanneschi, C.; Licchetta, A.; Conca, R.; Apollinari, S.; De Luca, F.; Tassone, P.; et al. Tumor infiltration by T lymphocytes expressing chemokine receptor 7 (CCR7) is predictive of favorable outcome in patients with advanced colorectal carcinoma. Clin. Cancer Res. Off. J. Am. Assoc. Cancer Res. 2012, 18, 850–857. [Google Scholar] [CrossRef] [PubMed]
- Sarkar, T.; Dhar, S.; Sa, G. Tumor-infiltrating T-regulatory cells adapt to altered metabolism to promote tumor-immune escape. Curr. Res. Immunol. 2021, 2, 132–141. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhang, N.; Li, Q.; Zhang, W.; Ke, F.; Leng, Q.; Wang, H.; Chen, J.; Wang, H. Tumor-Associated Macrophages Recruit CCR6+ Regulatory T Cells and Promote the Development of Colorectal Cancer via Enhancing CCL20 Production in Mice. PLoS ONE 2011, 6, e19495. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, Z.; Zhao, G. Recent advances in the study of regulatory T cells in gastric cancer. Int. Immunopharmacol. 2019, 73, 560–567. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhang, N.; Li, Q.; Zhang, W.; Ke, F.; Leng, Q.; Wang, H.; Chen, J.; Wang, H. CCL1 is a major regulatory T cell attracting factor in human breast cancer. BMC Cancer 2018, 18, 1278. [Google Scholar]
- Koyama, S.; Nishikawa, H. Mechanisms of regulatory T cell infiltration in tumors: Implications for innovative immune precision therapies. J. Immunother. Cancer 2021, 9, e002591. [Google Scholar] [CrossRef] [PubMed]
- Redjimi, N.; Raffin, C.; Raimbaud, I.; Pignon, P.; Matsuzaki, J.; Odunsi, K.; Valmori, D.; Ayyoub, M. CXCR3+ T regulatory cells selectively accumulate in human ovarian carcinomas to limit type I immunity. Cancer Res. 2012, 72, 4351–4360. [Google Scholar] [CrossRef] [PubMed]
- Borregaard, N. Neutrophils, from marrow to microbes. Immunity 2010, 33, 657–670. [Google Scholar] [CrossRef] [PubMed]
- Fioretti, F.; Fradelizi, D.; Stoppacciaro, A.; Ramponi, S.; Ruco, L.; Minty, A.; Sozzani, S.; Garlanda, C.; Vecchi, A.; Mantovani, A. Reduced tumorigenicity and augmented leukocyte infiltration after monocyte chemotactic protein-3 (MCP-3) gene transfer: Perivascular accumulation of dendritic cells in peritumoral tissue and neutrophil recruitment within the tumor. J. Immunol. 1998, 161, 342–346. [Google Scholar] [CrossRef]
- Fridlender, Z.G.; Albelda, S.M. Tumor-associated neutrophils: Friend or foe? Carcinogenesis 2012, 33, 949–955. [Google Scholar] [CrossRef]
- Fridlender, Z.G.; Sun, J.; Kim, S.; Kapoor, V.; Cheng, G.; Ling, L.; Worthen, G.S.; Albelda, S.M. Polarization of tumor-associated neutrophil phenotype by TGF-beta: “N1” versus “N2”, T.A.N. Cancer Cell 2009, 16, 183–194. [Google Scholar] [CrossRef] [PubMed]
- Shen, M.; Hu, P.; Donskov, F.; Wang, G.; Liu, Q.; Du, J. Tumor-associated neutrophils as a new prognostic factor in cancer: A systematic review and meta-analysis. PLoS ONE 2014, 9, e98259. [Google Scholar] [CrossRef] [PubMed]
- Uribe-Querol, E.; Rosales, C. Neutrophils in Cancer: Two Sides of the Same Coin. J. Immunol. Res. 2015, 2015, 983698. [Google Scholar] [CrossRef] [PubMed]
- Swierczak, A.; Mouchemore, K.A.; Hamilton, J.A.; Anderson, R.L. Neutrophils: Important contributors to tumor progression and metastasis. Cancer Metastasis Rev. 2015, 34, 735–751. [Google Scholar] [CrossRef] [PubMed]
- Mantovani, A.; Cassatella, M.A.; Costantini, C.; Jaillon, S. Neutrophils in the activation and regulation of innate and adaptive immunity. Nat. Rev. Immunol. 2011, 11, 519–531. [Google Scholar] [CrossRef] [PubMed]
- Masucci, M.T.; Minopoli, M.; Carriero, M.V. Tumor Associated Neutrophils. Their Role in Tumorigenesis, Metastasis, Prognosis and Therapy. Front. Oncol. 2019, 9, 1146. [Google Scholar] [CrossRef] [PubMed]
- Piccard, H.; Muschel, R.J.; Opdenakker, G. On the dual roles and polarized phenotypes of neutrophils in tumor development and progression. Crit. Rev. Oncol. Hematol. 2012, 82, 296–309. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zhang, W.; Yuan, X.; Fu, M.; Qian, H.; Xu, W. Neutrophils in cancer development and progression: Roles, mechanisms, and implications (Review). Int. J. Oncol. 2016, 49, 857–867. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhao, J.; Zhao, H.; A, S.; Liu, Z.; Zhang, Y.; Liu, X.; Wang, F. Infiltrating CD4/CD8 high T cells shows good prognostic impact in pancreatic cancer. Int. J. Clin. Exp. Pathol. 2017, 10, 8820–8828. [Google Scholar] [PubMed]
- Ino, Y.; Yamazaki-Itoh, R.; Shimada, K.; Iwasaki, M.; Kosuge, T.; Kanai, Y.; Hiraoka, N. Immune cell infiltration as an indicator of the immune microenvironment of pancreatic cancer. Br. J. Cancer 2013, 108, 914–923. [Google Scholar] [CrossRef] [PubMed]
- Fukunaga, A.; Miyamoto, M.; Cho, Y.; Murakami, S.; Kawarada, Y.; Oshikiri, T.; Kato, K.; Kurokawa, T.; Suzuoki, M.; Nakakubo, Y.; et al. CD8+ tumor-infiltrating lymphocytes together with CD4+ tumor-infiltrating lymphocytes and dendritic cells improve the prognosis of patients with pancreatic adenocarcinoma. Pancreas 2004, 28, e26–e31. [Google Scholar] [CrossRef]


| S. No. | Immune Biomarker (Official Gene Symbol) | High Expression— Unfavorable Prognosis | High Expression— Favorable Prognosis | Reference |
|---|---|---|---|---|
| 1 | PSMB8 | LAML, LAUD, PDAC | BLCA, BRCA, MESO | [133] |
| 2 | LCN2 | BLCA, KIRC, GBM | BRCA, ESCA, LGG, THCA | [134] |
| 3 | TIGIT | KIRC, KIRP, LGG, UVM | BRCA, CECS, HNSC, SKCM | [135] |
| 4 | HMGBs | BLCA, PRAD | GC | [136] [143] [137] [144] |
| 5 | STC2 | BLCA, COAD, ESCA HNSC, KIRP, LIHC LUAD, MESO, SARC THYM | HNSCC, BRCA, LGG | [145] |
| 6 | FN1 | BRCA | [146] | |
| 7 | PD-1 and PD-L1 | RCC, BRCA | HNSCC | [147] [148] |
| 8 | KIF2C | GBM, CRC, GC | [149] [150] [151] | |
| 9 | NUSAP1 | TNBC | [152] | |
| 10 | TLR3 | ESCC | [153] | |
| 11 | INHBA | CESC | [154] | |
| 12 | EPHA3 | BLCA | [155] | |
| 13 | CD73 | GBOV | [156] | |
| 14 | HSPB11 | HCC | [138] | |
| 15 | JAM2 | BRCA | [141] | |
| 16 | OX40L | HNSCC | [148] | |
| 17 | PDGFRB | HNSCC | [148] | |
| 18 | TMCO3 | LIHC | [157] | |
| 19 | IGF2BP2 | AML | [158] | |
| 20 | ACTA2 | BLCA | [159] | |
| 21 | TPM1 | BLCA | [159] | |
| 22 | ACTC1 | BLCA, GBM | PDAC | [159] [160] [161] |
| 23 | ACTN1 | BLCA | [159] | |
| 24 | PPARG | BLCA | [159] | |
| 25 | COL3A1 | BLCA | [159] [162] | |
| 26 | IGFBP3 | CRC | [163] | |
| 27 | EPCAM | BLCA | LAUD BRCA BLCA ESCC | [164] [165] |
| 28 | TPM1 | GC | [142] | |
| 29 | SDC1 | BRCA | ICC, CRC | [166] [167] [139] |
| 30 | NGAL | BRCA | [168] | |
| 31 | DZIP1 | GC | [140] | |
| 32 | SLC25A25-AS1 | PRAD | [169] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dakal, T.C.; George, N.; Xu, C.; Suravajhala, P.; Kumar, A. Predictive and Prognostic Relevance of Tumor-Infiltrating Immune Cells: Tailoring Personalized Treatments against Different Cancer Types. Cancers 2024, 16, 1626. https://doi.org/10.3390/cancers16091626
Dakal TC, George N, Xu C, Suravajhala P, Kumar A. Predictive and Prognostic Relevance of Tumor-Infiltrating Immune Cells: Tailoring Personalized Treatments against Different Cancer Types. Cancers. 2024; 16(9):1626. https://doi.org/10.3390/cancers16091626
Chicago/Turabian StyleDakal, Tikam Chand, Nancy George, Caiming Xu, Prashanth Suravajhala, and Abhishek Kumar. 2024. "Predictive and Prognostic Relevance of Tumor-Infiltrating Immune Cells: Tailoring Personalized Treatments against Different Cancer Types" Cancers 16, no. 9: 1626. https://doi.org/10.3390/cancers16091626





