Adsorption and Activity of Lipase on Polyphosphazene-Modified Polypropylene Membrane Surface
Abstract
:1. Introduction
2. Results and Discussion
2.1. Preparation and Characterization of PBAP with Varied Side Group Ratios
2.2. Effect of Side Group Ratios on Polymer Hydrophilicity
2.3. Characterization of the PBAP-Modified PPMM
2.4. Lipase Immobilization and Activity Retention
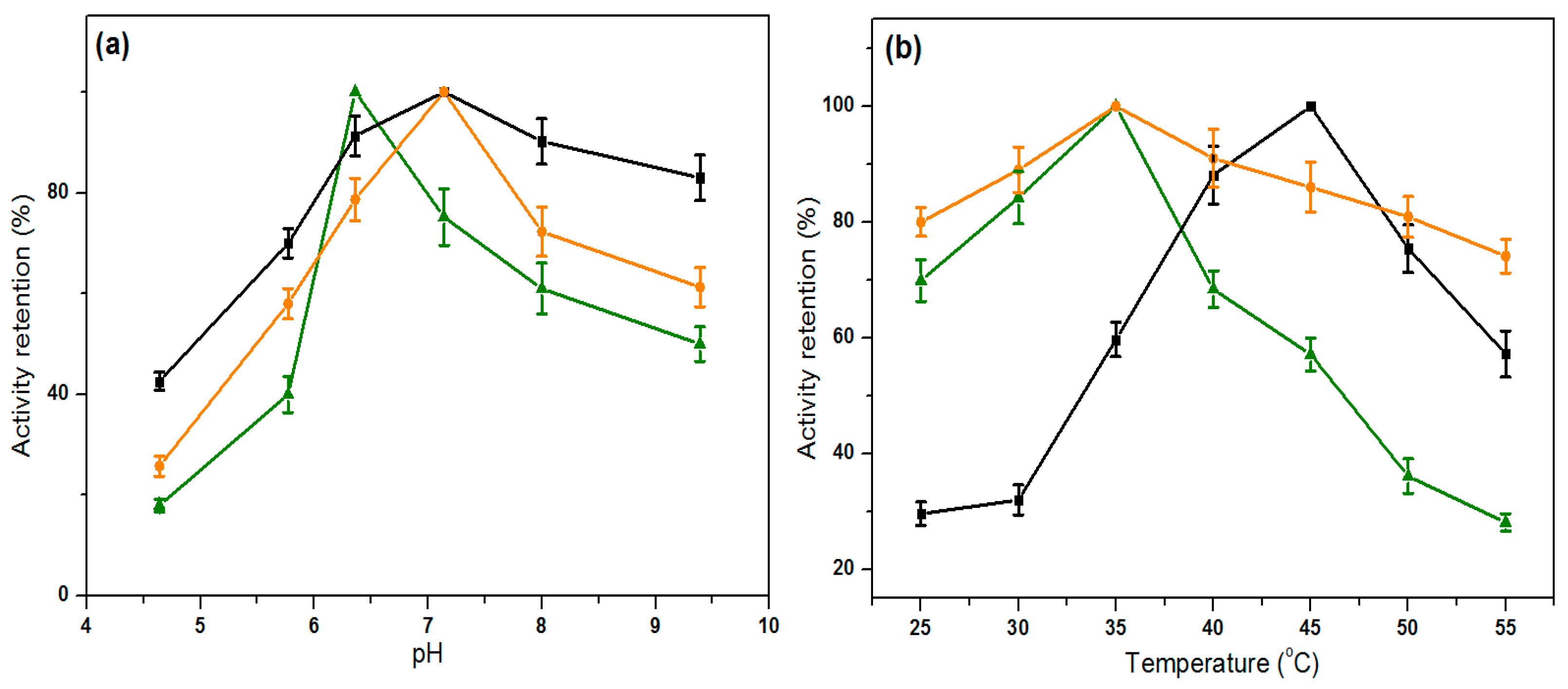
2.5. Influence of pH and Temperature on Lipase Activity
2.6. Thermal Stability of the Immobilized Lipases
3. Materials and Methods
3.1. Materials
3.2. Preparation and Analysis of PBAP
3.3. Surface Modification of PPMM with PBAP
3.4. Immobilization of Lipase by Adsorption
3.5. Assay of Lipase Activity
3.6. Stability Measurements
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Ragupathy, L.; Pluhar, B.; Ziener, U.; Keller, H.; Dyllick-Brenzinger, R.; Landfester, K. Enzymatic aminolysis of lactones in aqueous miniemulsion: Catalysis through a novel pathway. J. Mol. Catal. B Enzym. 2010, 62, 270–276. [Google Scholar] [CrossRef]
- Pan, C.; Luan, Z.Q.; Wang, X.J.; Ma, L. In situ immobilization of horseradish peroxidase on electropsun poly(styrene-co-methacrylic acid) nanofiberous membrane for catalytic treatment of polyphenol. Acta Polym. Sin. 2013, 12, 1508–1513. [Google Scholar]
- Ye, R.; Hayes, D.G.; Burton, R.; Liu, A.J.; Harte, F.M.; Wang, Y.M. Solvent-free lipase-catalyzed synthesis of technical-grade sugar esters and evaluation of their physicochemical and bioactive properties. Catalysts 2016, 6. [Google Scholar] [CrossRef]
- Chen, P.C.; Huang, X.J.; Xu, Z.K. Kinetics-bolstered catalytic study of a high performance lipase-immobilized nanofiber membrane reactor. RSC Adv. 2014, 4, 6151–6158. [Google Scholar] [CrossRef]
- Vaghari, H.; Jafarizadeh-Malmiri, H.; Mohammadlou, M.; Berenjian, A.; Anarjan, N.; Jafari, N.; Nasiri, S. Application of magnetic nanoparticles in smart enzyme immobilization. Biotechnol. Lett. 2016, 38, 223–233. [Google Scholar] [CrossRef]
- Barbosa, O.; Ortiz, C.; Berenguer-Murcia, Á.; Torres, R.; Rodrigues, R.C.; Fernandez-Lafuente, R. Strategies for the one-step immobilization-purification of enzymes as industrial biocatalysts. Biotechnol. Adv. 2015, 33, 435–456. [Google Scholar] [CrossRef]
- Min, K.; Yoo, Y.J. Recent progress in nanobiocatalysis for enzyme immobilization and its application. Biotechnol. Bioprocess Eng. 2014, 19, 553–567. [Google Scholar] [CrossRef]
- Barbosa, O.; Torres, R.; Ortiz, C.; Berenguer-Murcia, A.; Rodrigues, R.C.; Fernandez-Lafuente, R. Heterofunctional supports in enzyme immobilization: From traditional immobilization protocols to opportunities in tuning enzyme properties. Biomacromolecules 2013, 14, 2433–2462. [Google Scholar] [CrossRef] [Green Version]
- Rodrigues, R.C.; Ortiz, C.; Berenguer-Murcia, A.; Torres, R.; Fernández-Lafuente, R. Modifying enzyme activity and selectivity by immobilization. Chem. Soc. Rev. 2013, 42, 6290–6307. [Google Scholar] [CrossRef]
- Verger, R. Interfacial activation of lipase: Facts and artifacts. Trends Biotechnol. 1997, 15, 32–38. [Google Scholar] [CrossRef]
- Brzozowski, A.M.; Derewenda, U.; Derewenda, Z.S.; Dodson, G.G.; Lawson, D.M.; Turkenburg, J.P.; Bjorking, F.; Huge-Jensen, B.; Patrk, S.A.; Thim, L. A model for interfacial activation in lipases from the structure of a fungal lipase-inhibitor complex. Nature 1991, 351, 491–494. [Google Scholar] [CrossRef]
- Manoel, E.A.; dos Santos, J.C.S.; Freire, D.M.G.; Rueda, N.; Fernandez-Lafuente, R. Immobilization of lipases on hydrophobic supports involves the open form of the enzyme. Enzyme Microb. Tech. 2015, 71, 53–57. [Google Scholar] [CrossRef]
- Mateo, C.; Palomo, J.M.; Fernandez-Lorente, G.; Guisan, J.M.; Fernandez-Lafuente, R. Improvement of enzyme activity, stability and selectivity via immobilization techniques. Enzyme Microb. Tech. 2007, 40, 1451–1463. [Google Scholar] [CrossRef]
- Adlercreutz, P. Immobilisation and application of lipase in organic media. Chem. Soc. Rev. 2013, 42, 6406–6436. [Google Scholar] [CrossRef]
- Santos, J.C.; Barbosa, O.; Ortiz, C.; Berenguer-Murcia, A.; Rodrigues, R.C.; Fernandez-Lafuente, R. Importance of the support properties for immobilization or purification of enzymes. ChemCatChem 2015, 7, 2413–2432. [Google Scholar] [CrossRef]
- Chen, P.C.; Huang, X.J.; Xu, Z.K. Activation and deformation of immobilized lipase on self-assembled monolayers with tailored wettability. Phys. Chem. Chem. Phys. 2015, 17, 13457–13465. [Google Scholar] [CrossRef]
- Zhao, K.Y.; Lin, B.B.; Cui, W.K.; Feng, L.Z.; Chen, T.; Wei, J.F. Preparation and adsorption of bovine serum albumin-imprinted polyacrylamide hydrogel membrane grafted on non-woven polypropylene. Talanta 2014, 121, 256–262. [Google Scholar] [CrossRef]
- Makhloufi, C.; Asseuguette, E.; Remigy, J.C.; Belaissaoui, B.; Roizard, D.; Favre, E. Ammonia based CO2 capture process using hollow fiber membrane contactors. J. Membr. Sci. 2013, 455, 236–246. [Google Scholar] [CrossRef]
- Secundo, F. Conformational changes of enzymes upon immobilisation. Chem. Soc. Rev. 2013, 42, 6250–6261. [Google Scholar] [CrossRef]
- Cantone, S.; Ferrario, V.; Corici, L.; Ebert, C.; Fattor, D.; Spizzo, P.; Gardossi, L. Efficient immobilisation of industrial biocatalysts: Criteria and constraints for the selection of organic polymeric carriers and immobilisation methods. Chem. Soc. Rev. 2013, 42, 6262–6276. [Google Scholar] [CrossRef] [Green Version]
- Brady, D.; Jordaan, J. Advances in enzyme immobilisation. Biotechnol. Lett. 2009, 31, 1639–1650. [Google Scholar] [CrossRef]
- Sheldon, R.A.; Van Pelt, S. Enzyme immobilisation in biocatalysis: Why, what and how. Chem. Soc. Rev. 2013, 42, 6223–6235. [Google Scholar] [CrossRef]
- Allcock, H.R.; Phelps, M.V.B.; Barrett, E.W. Ultraviolet photolithographic development of polyphosphazene hydrogel microstructures for potential use in microarray biosensors. Chem. Mater. 2006, 18, 609–613. [Google Scholar] [CrossRef]
- Qian, Y.C.; Huang, X.J.; Chen, C.; Ren, N.; Huang, X.; Xu, Z.K. A versatile approach to the synthesis of polyphosphazene derivatives via the thiol-ene reaction. J. Polym. Sci. Pol. Chem. 2012, 50, 5170–5176. [Google Scholar] [CrossRef]
- Potta, T.; Chun, C.J.; Song, S.C. Dual cross-linking systems of functionally photo-cross-linkable and thermoresponsive polyphosphazene hydrogels for biomedical applications. Biomacromolecules 2010, 11, 1741–1753. [Google Scholar] [CrossRef]
- Qian, Y.C.; Ren, N.; Huang, X.J.; Chen, C.; Yu, A.G.; Xu, Z.K. Glycosylation of polyphosphazene nanofibrous membrane by click chemistry for protein recognition. Macromol. Chem. Phys. 2013, 214, 1852–1858. [Google Scholar] [CrossRef]
- Xue, L.W.; Mao, L.X.; Cai, Q.; Yang, X.P.; Jin, R.G. Preparation of amino acid ester substituted polyphosphazene microparticles via electrohydrodynamic atomization. Polym. Adv. Technol. 2011, 22, 2009–2016. [Google Scholar] [CrossRef]
- Cuetos, A.; Valenzuela, M.L.; Gotor, V.; Carriedo, G.A. Polyphosphazenes as tunable and recyclable supports to immobilize alcohol dehydrogenases and lipases: Synthesis, catalytic activity, and recycling efficiency. Biomacromolecules 2010, 11, 1291–1297. [Google Scholar] [CrossRef]
- Qian, Y.C.; Chen, P.C.; Huang, X.J. Click synthesis of ionic strength-responsive polyphosphazene hydrogel for reversible binding of enzymes. RSC Adv. 2015, 5, 44031–44040. [Google Scholar] [CrossRef]
- Li, X.D.; Wu, J.; Jia, D.C.; Wan, Y.H.; Yang, N.; Qiao, M. Preparation of cross-linked glucoamylase aggregates immobilization by using dextrin and xanthan gum as protecting agents. Catalysts 2016, 6. [Google Scholar] [CrossRef]
- Apetrei, C.; de Saja, J.A.; Zurro, J.; Rodríguez-Méndez, M.L. Advantages of the biomimetic nanostructured films as an immobilization method vs. the carbon paste classical method. Catalysts 2012, 2, 517–531. [Google Scholar] [CrossRef]
- Huang, X.J.; Yu, A.G.; Jiang, J.; Pan, C.; Qian, J.W.; Xu, Z.K. Surface modification of nanofibrous poly(acrylonitrile-co-acrylic acid) membrane with biomacromolecules for lipase immobilization. J. Mol. Catal. B Enzym. 2009, 57, 250–256. [Google Scholar] [CrossRef]
- Bradford, M. A rapid and sensitive method for the quantition of microgram quantities of protein utilizing the principle of dyebinding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Zhang, P.; Wang, Q.Q.; Zhang, J.N.; Li, G.H.; Wei, Q.F. Preparation of amidoxime-modified polyacrylonitrile nanofibers immobilized with laccase for dye degradation. Fibers Polym. 2014, 15, 30–34. [Google Scholar] [CrossRef]
- Huang, X.J.; Chen, P.C.; Huang, F.; Ou, Y.; Chen, M.R.; Xu, Z.K. Immobilization of Candida rugosa lipase on electrospun cellulose nanofiber membrane. J. Mol. Catal. B Enzym. 2011, 70, 95–100. [Google Scholar] [CrossRef]






© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, P.-C.; Qian, Y.-C.; Fang, F.; Zhu, X.-Y.; Huang, X.-J. Adsorption and Activity of Lipase on Polyphosphazene-Modified Polypropylene Membrane Surface. Catalysts 2016, 6, 174. https://doi.org/10.3390/catal6110174
Chen P-C, Qian Y-C, Fang F, Zhu X-Y, Huang X-J. Adsorption and Activity of Lipase on Polyphosphazene-Modified Polypropylene Membrane Surface. Catalysts. 2016; 6(11):174. https://doi.org/10.3390/catal6110174
Chicago/Turabian StyleChen, Peng-Cheng, Yue-Cheng Qian, Fei Fang, Xue-Yan Zhu, and Xiao-Jun Huang. 2016. "Adsorption and Activity of Lipase on Polyphosphazene-Modified Polypropylene Membrane Surface" Catalysts 6, no. 11: 174. https://doi.org/10.3390/catal6110174





