Biotransformation of Ergostane Triterpenoid Antcin K from Antrodia cinnamomea by Soil-Isolated Psychrobacillus sp. AK 1817
Abstract
:1. Introduction
2. Results
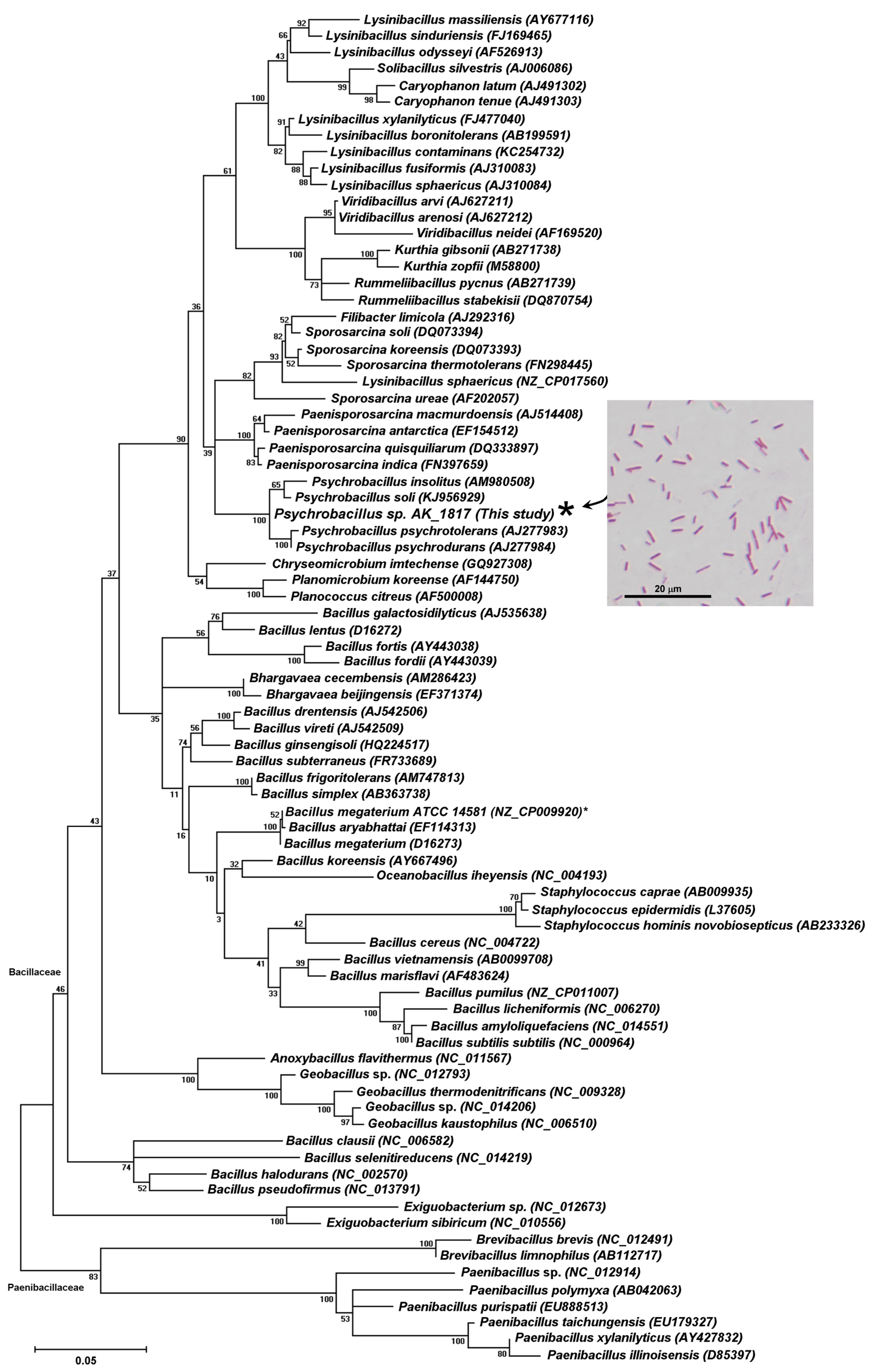
2.1. Screening and Identification of Soil Bacteria with Biotransformation Activity
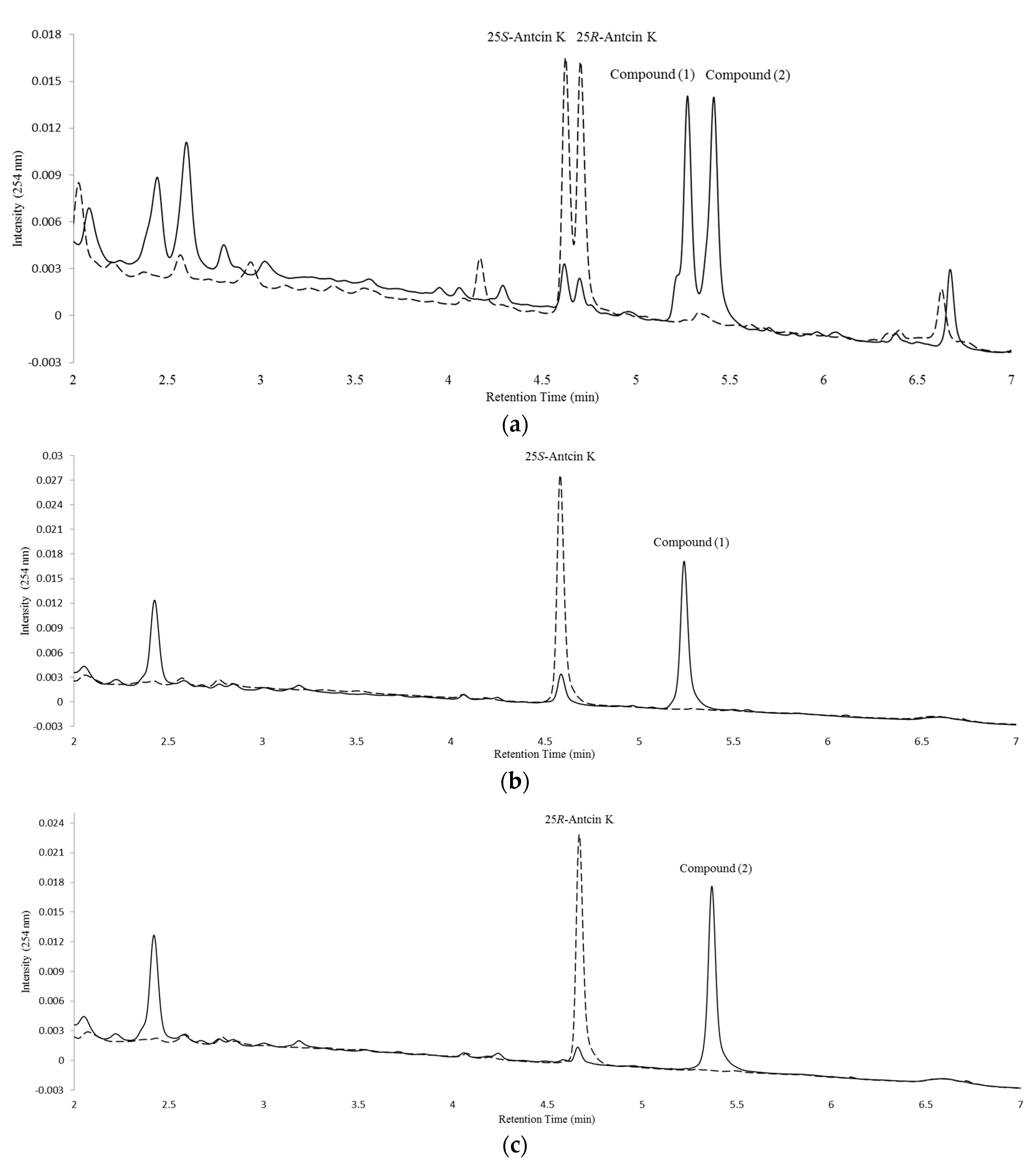
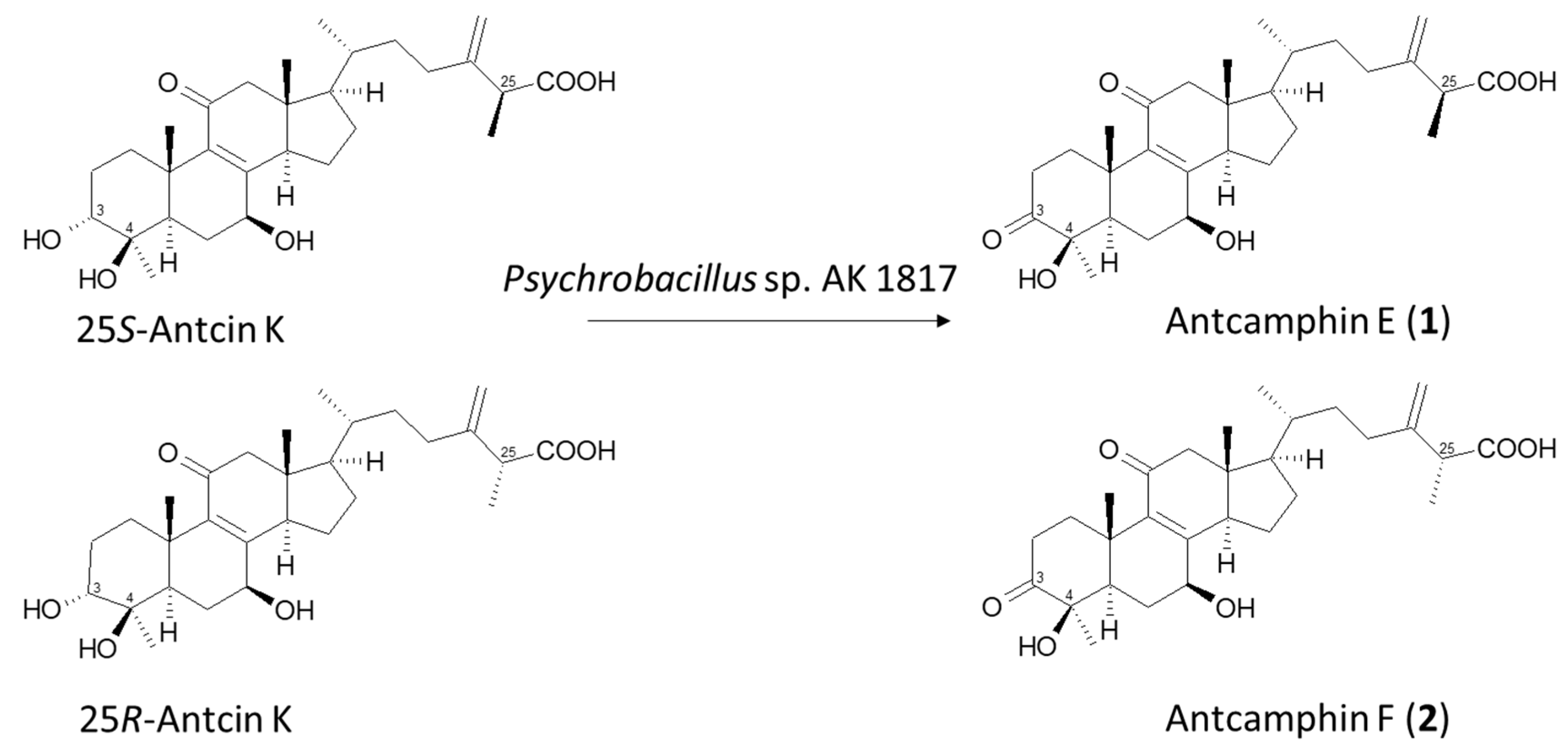
2.2. Isolation and Identification of Biotransformation Metabolites
3. Discussion
4. Materials and Methods
4.1. Microorganism and Chemicals
4.2. Preparation of Antcin K
4.3. Screening and Identification of Soil Bacteria with Biotransformation Activity
4.4. UPLC Analysis
4.5. Candidate Strain Classification via 16S rRNA Gene Analysis
4.6. Scale-Up Fermentation, Isolation, and Identification of the Biotransformation Products
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Sultana, N.; Saify, Z.S. Enzymatic biotransformation of terpenes as bioactive agents. J. Enzym. Inhib. Med. Chem. 2013, 28, 1113–1128. [Google Scholar] [CrossRef] [PubMed]
- Lu, M.C.; El-Shazly, M.; Wu, T.Y.; Du, Y.C.; Chang, T.T.; Chen, C.F.; Hsu, Y.M.; Lai, C.P.; Chang, F.R.; Wu, Y.C. Recent research and development of Antrodia cinnamomea. Pharmacol. Ther. 2013, 139, 124–156. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.C.; Wang, Y.H.; Chou, Y.C.; Chen, C.F.; Lin, L.X.; Chang, T.T.; Tien, J.H.; Chou, C.J. Evaluation of the anti-inflammatory activity of zhankuic acids isolated from the fruiting bodies of Antrodia camphorate. Planta Med. 2004, 70, 310–314. [Google Scholar] [PubMed]
- Geethangili, M.; Fang, S.H.; Lai, C.H.; Rao, Y.K.; Lien, H.M.; Tzeng, Y.M. Inhibitory effect of Antrodia camphorata constituents on the Helicobacter pylori-associated gastric inflammation. Food Chem. 2010, 119, 149–153. [Google Scholar] [CrossRef]
- Lai, C.I.; Chu, Y.L.; Ho, C.T.; Su, Y.C.; Kuo, Y.H.; Sheen, L.Y. Antcin K, an active triterpenoid from the fruiting bodies of basswood cultivated Antrodia cinnamomea, induces mitochondria and endoplasmic reticulum stress-mediated apoptosis in human hepatoma cells. J. Tradit. Complement. Med. 2016, 6, 48–56. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.L.; Chu, Y.L.; Ho, C.T.; Chung, J.G.; Lai, C.I.; Su, Y.C.; Kuo, Y.H.; Sheen, L.Y. Antcin K, an active triterpenoid from the fruiting bodies of basswood cultivated Antrodia cinnamomea, inhibits metastasis via suppression of integrin-mediated adhesion, migration, and invasion in human hepatoma cells. J. Agric. Food Chem. 2015, 63, 4561–4569. [Google Scholar] [CrossRef] [PubMed]
- Tien, A.J.; Chien, C.Y.; Chen, Y.H.; Lin, L.C.; Chien, C.T. Fruiting bodies of Antrodia cinnamomea and its active triterpenoid, antcin K, ameliorates N-nitrosodiethylamine-induced hepatic inflammation, fibrosis and carcinogenesis in rats. Am. J. Chin. Med. 2017, 45, 173. [Google Scholar] [CrossRef] [PubMed]
- Kuo, Y.H.; Lin, C.H.; Shih, C.C.; Yang, C.S. Antcin K, a triterpenoid compound from Antrodia camphorate, displays antidiabetic and antihyperlipidemic effects via glucose transporter 4 and AMP-activated protein kinase phosphorylation in muscles. Evid. Based Complement. Alternat. Med. 2016. [Google Scholar] [CrossRef] [PubMed]
- Muffler, K.; Leipold, D.; Scheller, M.C.; Haas, C.; Steingroewer, J.; Bley, T.; Neuhaus, H.E.; Mirata, M.A.; Schrader, J.; Ulber, R. Biotransformation of triterpenes. Process Biochem. 2011, 46, 1–15. [Google Scholar] [CrossRef]
- Wang, Y.; Sun, Y.; Wang, C.; Huo, X.; Liu, P.; Wang, C.; Zhang, B.; Zhan, L.; Zhang, H.; Deng, S.; et al. Biotransformation of 11-keto-β-boswellic acid by Cunninghamella blakesleana. Phytochemistry 2013, 96, 330–336. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Liu, D.; Xi, R.G.; Wang, X.; Wang, Y.; Hou, J.; Zhang, B.; Wang, X.; Liu, K.; Ma, X. Microbial transformation of acetyl-11-keto-β-boswellic acid and their inhibitory activity on LPS-induced NO production. Bioorg. Med. Chem. Lett. 2013, 23, 1338–1342. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.W.; Xu, S.H.; Zhao, Y.Z.; Zhang, C.; Zhang, Y.Y.; Yu, B.Y.; Zhang, J. Microbial hydroxylation and glycosylation of pentacyclic triterpenes as inhibitors on tissue factor procoagulant activity. Bioorg. Med. Chem. Lett. 2017, 27, 1026–1030. [Google Scholar] [CrossRef] [PubMed]
- Martinez, A.; Rivas, F.; Perojil, A.; Parra, A.; Garcia-Granados, A.; Fernandez-Vivas, A. Biotransformation of oleanolic and maslinic acids by Rhizomucor miehei. Phytochemistry 2013, 94, 229–237. [Google Scholar] [CrossRef] [PubMed]
- Qian, L.W.; Zhang, J.; Liu, J.H.; Yu, B.Y. Direct microbial-catalyzed asymmetric α-hydroxylation of betulinic acid by Nocardia sp. NRRL 5646. Tetrahedron Lett. 2009, 50, 2193–2195. [Google Scholar] [CrossRef]
- Feng, X.; Luan, J.; Guo, F.F.; Li, D.P.; Chu, Z.Y. Microbial transformation of maslinic acid by Cunninghamella blakesleana. J. Mol. Catal. B Enzym. 2012, 82, 127–130. [Google Scholar] [CrossRef]
- Huang, F.X.; Yang, W.Z.; Ye, F.; Tian, J.Y.; Hu, H.B.; Feng, L.M.; Guo, D.A.; Ye, M. Microbial transformation of ursolic acid by Syncephalastrum racemosum (Cohn) Schroter AS 3.264. Phytochemistry 2012, 82, 56–60. [Google Scholar] [CrossRef] [PubMed]
- Fu, S.B.; Yang, J.S.; Cui, J.L.; Sun, D.A. Biotransformation of ursolic acid by Syncephalastrum racemosum CGMCC 3.2500 and anti-HCV activity. Fitoterapia 2013, 86, 123–128. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Xu, S.H.; Ma, B.L.; Wang, W.W.; Yu, B.Y.; Zhang, J. New derivatives of ursolic acid through the biotransformation by Bacillus megaterium CGMCC 1.1741 as inhibitors on nitric oxide production. Bioorg. Med. Chem. Lett. 2017, 27, 2575–2578. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Qiao, L.; Xie, D.; Zhang, Y.; Zou, J.; Chen, X.; Dai, J. Microbial transformation of ginsenoside-Rg1 by Absidia coerulea and the reversal activity of the metabolites towards multi-drug resistant tumor cells. Fitoterapia 2011, 82, 1313–1317. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Yang, M.; Nong, S.; Yang, X.; Ling, Y.; Wang, D.; Wang, X.; Zhang, W. Microbial transformation of 20(S)-protopanaxadiol by Absidia corymbifera. Cytotoxic activity of the metabolites against human prostate cancer cells. Fitoterapia 2013, 84, 6–10. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.; Yang, X.; Li, J.; Ge, H.; Song, Y.; Ren, J. Biotransformation of 20(S)-protopanaxadiol by Aspergillus niger AS 3.1858. Fitoterapia 2013, 91, 256–260. [Google Scholar] [CrossRef] [PubMed]
- Schmitz, D.; Zapp, J.; Bernhardt, R. Hydroxylation of the triterpenoid dipterocarpol with CYP106A2 from Bacillus megaterium. FEBS J. 2012, 279, 1663–1674. [Google Scholar] [CrossRef] [PubMed]
- Nei, M.; Kumar, S. Molecular Evolution and Phylogenetics; Oxford University Press: New York, NY, USA, 2000; ISBN 100-1-95-13585-7. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.; Lin, X.; Qiao, X.; Ji, S.; Liu, K.; Yeh, C.; Tzeng, Y.M.; Guo, D.; Ye, M. Antcamphins A-L, ergostanoids from Antrodia camphorate. J. Nat. Prod. 2014, 77, 118–124. [Google Scholar] [CrossRef] [PubMed]
- Kouzi, S.A.; Chatterjee, P.; Pezzuto, J.M.; Hamann, M.T. Microbial transformations of the antimelanoma agent betulinic acid. J. Nat. Prod. 2000, 63, 1653–1657. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.C.; Wang, P.H.; Lee, S.S. Microbial transformation of protopanaxadiol and protopanaxatriol derivatives with Mycobacterium sp. (NRRL B-3805). J. Nat. Prod. 1997, 60, 1236–1241. [Google Scholar] [CrossRef]
- Feng, X.; Zou, Z.; Fu, S.; Sun, L.; Su, Z.; Sun, D.A. Microbial oxidation and glucosidation of echinocystic acid by Nocardia corallina. J. Mol. Catal. B Enzym. 2010, 66, 219–223. [Google Scholar] [CrossRef]
- Grishko, V.V.; Tarasova, E.V.; Ivshina, I.B. Biotransformation of botulin to betulone by growing and resting cells of the actinobacterium Rhodococcus rhodochrous IEGM 66. Process Biochem. 2013, 48, 1640–1644. [Google Scholar] [CrossRef]
- Qiao, X.; Song, W.; Wang, Q.; Liu, K.D.; Zhang, Z.X.; Bo, T.; Li, R.Y.; Liang, L.N.; Tzeng, Y.M.; Guo, D.A.; et al. Comprehensive chemical analysis of triterpenoids and polysaccharides in the medicinal mushroom Antrodia cinnamomea. RSC Adv. 2015, 5, 47040–47052. [Google Scholar] [CrossRef]
- Li, Z.W.; Kuang, Y.; Tang, S.N.; Li, K.; Huang, Y.; Qiao, X.; Yu, S.W.; Tzeng, Y.M.; Lo, J.Y.; Ye, M. Hepatoprotective activities of Antrodia camphorata and its triterpenoid compounds against CCl4-induced liver injury in mice. J. Ethnopharm. 2017, 206, 31–39. [Google Scholar] [CrossRef] [PubMed]
- Bertani, G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J. Bacteriol. 1951, 62, 293–300. [Google Scholar] [PubMed]
- Hugenholtz, P.; Goebel, B.M. Environmental Molecular Microbiology: Protocols and Applications; Horizon Scientific Press: Norfolk, UK, 2001; pp. 31–42. ISBN 101-8-98-486-29-8. [Google Scholar]
- Thompson, J.D.; Gibson, T.J.; Higgins, D.G. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinform. 2002. [Google Scholar] [CrossRef]
- Felsenstein, J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985, 39, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Posada, D. Model test server: A web-based tool for the statistical selection of models of nucleotide substitution online. Nucleic Acids Res. 2006, 34, W700–W703. [Google Scholar] [CrossRef] [PubMed]
- Chiang, C.M.; Chang, Y.J.; Wu, J.Y.; Chang, T.S. Production and anti-melanoma activity of methoxyisoflavones from biotransformation of genistein by two recombinant Escherichia coli. Molecules 2017, 22, 87. [Google Scholar] [CrossRef] [PubMed]
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chiang, C.-M.; Wang, T.-Y.; Ke, A.-N.; Chang, T.-S.; Wu, J.-Y. Biotransformation of Ergostane Triterpenoid Antcin K from Antrodia cinnamomea by Soil-Isolated Psychrobacillus sp. AK 1817. Catalysts 2017, 7, 299. https://doi.org/10.3390/catal7100299
Chiang C-M, Wang T-Y, Ke A-N, Chang T-S, Wu J-Y. Biotransformation of Ergostane Triterpenoid Antcin K from Antrodia cinnamomea by Soil-Isolated Psychrobacillus sp. AK 1817. Catalysts. 2017; 7(10):299. https://doi.org/10.3390/catal7100299
Chicago/Turabian StyleChiang, Chien-Min, Tzi-Yuan Wang, An-Ni Ke, Te-Sheng Chang, and Jiumn-Yih Wu. 2017. "Biotransformation of Ergostane Triterpenoid Antcin K from Antrodia cinnamomea by Soil-Isolated Psychrobacillus sp. AK 1817" Catalysts 7, no. 10: 299. https://doi.org/10.3390/catal7100299





