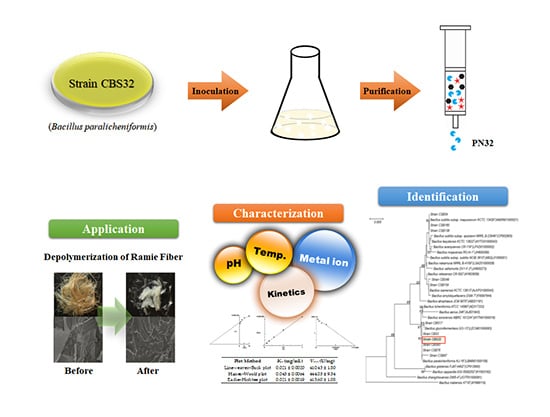
Production of Novel Polygalacturonase from Bacillus paralicheniformis CBS32 and Application to Depolymerization of Ramie Fiber
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Screening, and Identification of the CBS32 Strain
2.3. Protein Measure and Polygalacturonase Activity
2.4. Production and Purification of PN32
2.5. Determination of Molecular Weight
2.6. The N-terminal Sequencing
2.7. Effect of pH and Temperature on Polygalacturonase Activity
2.8. Effects of Metal Ions and Kinetic Parameters
2.9. Enzymatic Depolymerization of Ramie Fiber
3. Results
3.1. Strain Identification, and Enzyme Production
3.2. Purification of PN32
3.3. N-terminal Amino Acid Sequence Analysis
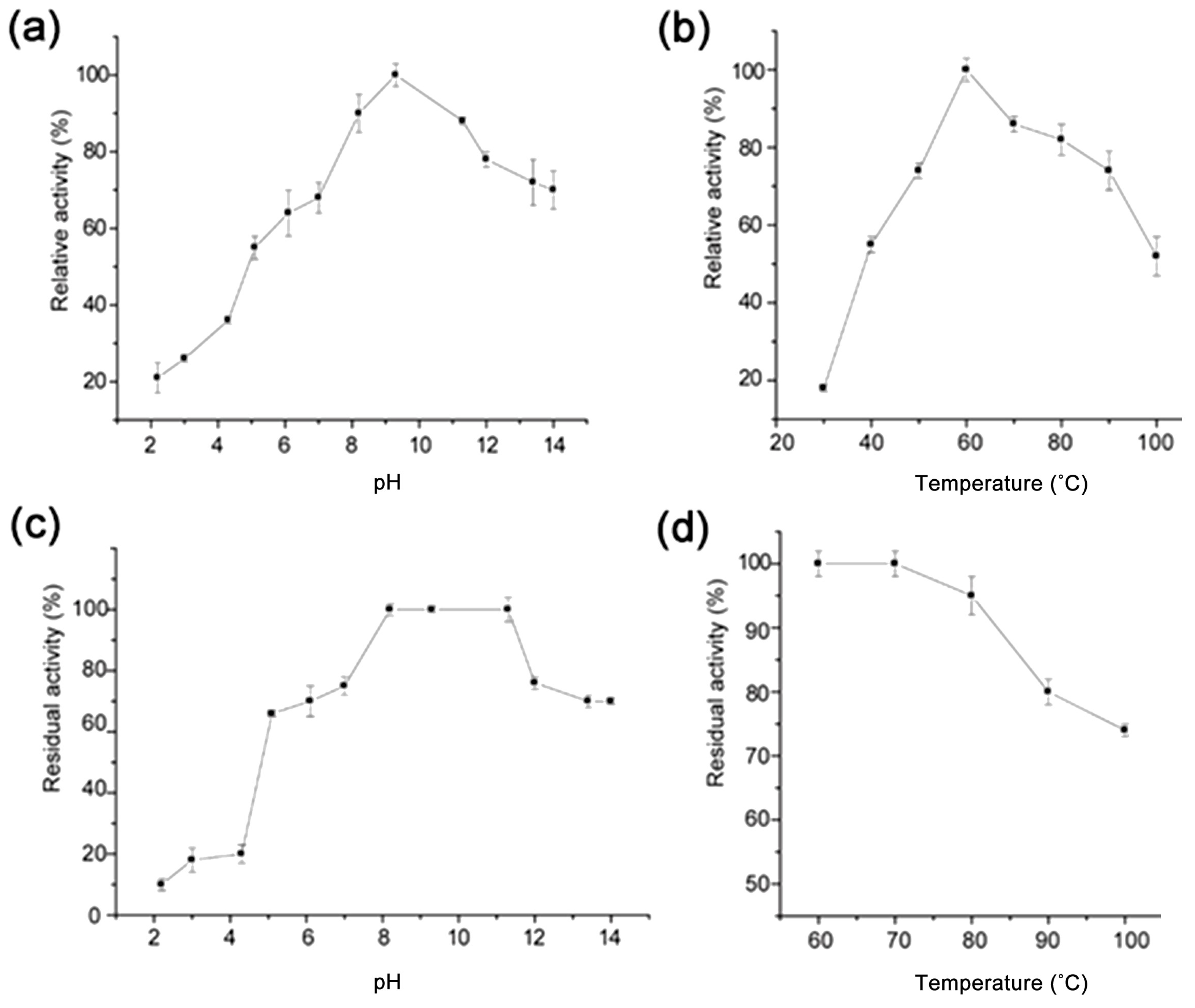
3.4. Effects of pH, Temperature, and Metal Ions on PN32
3.5. Kinetics Parameter of PN32
3.6. Enzymatic Depolymerization of Ramie Fiber by PN32
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Yoo, H.Y.; Lee, J.H.; Kim, D.S.; Lee, J.H.; Lee, S.K.; Lee, S.J.; Park, C.; Kim, S.W. Enhancement of glucose yield from canola agricultural residue by alkali pretreatment based on multi-regression models. Ind. Eng. Chem. Res. 2017, 51, 303–311. [Google Scholar] [CrossRef]
- Siripong, P.; Doungporn, P.; Yoo, H.Y.; Kim, S.W. Improvement of sugar recovery from Sida acuta (Thailand Weed) by NaOH pretreatment and application to bioethanol production. Korean J. Chem. Eng. 2018, 35, 2413–2420. [Google Scholar] [CrossRef]
- Xiao, C.; Charles, T.A. Roles of pectin in biomass yield and processing for biofuels. Front. Plant Sci. 2013, 4, 67. [Google Scholar] [CrossRef] [PubMed]
- Yoo, H.Y.; Yang, X.; Kim, D.S.; Lotrakul, P.; Prasongsuk, S.; Punnapayak, H.; Kim, S.W. Evaluation of the overall process on bioethanol production from miscanthus hydrolysates obtained by dilute acid pretreatment. Biotechnol. Bioprocess Eng. 2016, 21, 733–742. [Google Scholar] [CrossRef]
- Minzanova, S.; Mironov, V.; Arkhipova, D.; Khabibullina, A.; Mironova, L.; Zakirova, Y.; Milyukov, V. Biological activity and pharmacological application of pectic polysaccharides: A review. Polymers 2018, 10, 1407. [Google Scholar] [CrossRef] [PubMed]
- Kashyap, D.R.; Vohra, P.K.; Chopra, S.; Tewari, R. Applications of pectinases in the commercial sector: A review. Bioresour. Technol. 2001, 77, 215–227. [Google Scholar] [CrossRef]
- Chiliveri, S.R.; Koti, S.; Linga, V.R. Retting and degumming of natural fibers by pectinolytic enzymes produced from Bacillus tequilensis SV11-UV37 using solid state fermentation. SpringerPlus 2016, 5, 559. [Google Scholar] [CrossRef] [PubMed]
- Pan, J.; Cao, D.; Ma, X.; Yang, J. Preparation, characterization and in vitro release properties of pectin-based curcumin film. Korean, J. Chem. Eng. 2019, 36, 822–827. [Google Scholar] [CrossRef]
- Kester, H.C.; Visser, J. Purification and characterization of polygalacturonases produced by the hyphal fungus Aspergillus niger. Biotechnol. Appl. Biochem. 1990, 12, 150–160. [Google Scholar] [PubMed]
- Zhao, D.; Pan, C.; Ping, W.; Ge, J. Degumming crude enzyme produced by Bacillus cereus HDYM-02 and its application in flax retting. BioResources 2018, 13, 5213–5224. [Google Scholar]
- Brühlmann, F.; Leupin, M.; Erismann, K.H.; Fiechter, A. Enzymatic degumming of ramie bast fibers. J. Biotechnol. 2001, 76, 43–50. [Google Scholar] [CrossRef]
- Kapoor, M.; Beg, Q.K.; Bhushan, B.; Singh, K.; Dadhich, K.S.; Hoondal, G.S. Application of an alkaline and thermostable polygalacturonase from Bacillus sp. MG-cp-2 in degumming of ramie (Boehmeria nivea) and sunn hemp (Crotalaria juncea) bast fibres. Proc. Biochem. 2001, 36, 803–807. [Google Scholar] [CrossRef]
- Zheng, L.; Du, Y.; Zhang, J. Degumming of ramie fibers by alkalophilic bacteria and their polysaccharide-degrading enzymes. Bioresour. Technol. 2001, 78, 89–94. [Google Scholar] [CrossRef]
- Mohanty, A.K.; Misra, M.A.; Hinrichsen, G.I. Biofibres, biodegradable polymers and biocomposites: An overview. Macromol. Mater. Eng. 2000, 276, 1–24. [Google Scholar] [CrossRef]
- Jiang, W.; Zhou, C.; Han, G.; Via, B.; Swain, T.; Fan, Z.; Liu, S. Classification and identification of plant fibrous material with different species using near infrared technique-A new way to approach determining biomass properties accurately within different species. Front. Plant Sci. 2017, 7, 2000. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Han, G.; Zhang, Y.; Wang, M. Fast compositional analysis of ramie using near-infrared spectroscopy. Carbohydr. Polym. 2010, 81, 937–941. [Google Scholar] [CrossRef]
- He, Y.; Pan, L.; Wang, B. Efficient over-expression and application of high-performance pectin lyase by screening Aspergillus niger pectin lyase gene family. Biotechnol. Bioprocess Eng. 2018, 23, 662–669. [Google Scholar] [CrossRef]
- Johnson, J.; Sudheer, P.D.; Yang, Y.H.; Kim, Y.G.; Choi, K.Y. Hydrolytic activities of hydrolase enzymes from halophilic microorganisms. Biotechnol. Bioprocess Eng. 2017, 22, 450–461. [Google Scholar] [CrossRef]
- Gillespie, A.M.; Keane, D.; Griffin, T.O.; Tuohy, M.G.; Donaghy, J.; Haylock, R.W.; Coughlan, M.P. The application of fungal enzymes in flax retting and the properties of an extracellular polygalacturonase from Penicillium capsulatum. In Biotechnology in Pulp and Paper Manufacture; Butterworth-Heinemann: Stoneham, MA, USA, 1990; pp. 211–219. [Google Scholar]
- Regmi, S.; Yoo, H.Y.; Choi, Y.H.; Choi, Y.S.; Yoo, J.C.; Kim, S.W. Prospects for bio--industrial application of an extremely alkaline mannanase from Bacillus subtilis subsp. inaquosorum CSB31. Biotechnol. J. 2017, 12, 1700113. [Google Scholar] [CrossRef]
- Yoo, H.Y.; Pradeep, G.C.; Lee, S.K.; Park, D.H.; Cho, S.S.; Choi, Y.H.; Yoo, J.C.; Kim, S.W. Understanding β--mannanase from Streptomyces sp. CS147 and its potential application in lignocellulose based biorefining. Biotechnol. J. 2015, 10, 1894–1902. [Google Scholar] [CrossRef]
- Jiang, Z.; Wei, Y.; Li, D.; Li, L.; Chai, P.; Kusakabe, I. High-level production, purification and characterization of a thermostable β-mannanase from the newly isolated Bacillus subtilis WY34. Carbohydr. Polym. 2006, 66, 88–96. [Google Scholar] [CrossRef]
- Khan, M.M.; Choi, Y.S.; Kim, Y.K.; Yoo, J.C. Immobilization of an alkaline endopolygalacturonase purified from Bacillus paralicheniformis exhibits bioscouring of cotton fabrics. Bioproc. Biosyst. Eng. 2018, 41, 1425–1436. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Kim, H.R.; Lee, J.H.; Lee, S.K.; Chun, Y.; Han, S.O.; Yoo, H.Y.; Park, C.; Kim, S.W. Enhanced in-vitro hemozoin polymerization by optimized process using histidine-rich protein II (HRPII). Polymers 2019, 11, 1162. [Google Scholar] [CrossRef] [PubMed]
- Guo, F.; Zou, M.; Li, X.; Zhao, J.; Qu, Y. An effective degumming enzyme from Bacillus sp. Y1 and synergistic action of hydrogen peroxide and protease on enzymatic degumming of ramie fibers. BioMed Res. Int. 2013, 2013, 212315. [Google Scholar] [CrossRef] [PubMed]
- Kohli, P.; Gupta, R. Alkaline pectinases: A review. Biocatal. Agric. Biotechnol. 2015, 4, 279–285. [Google Scholar] [CrossRef]
- Garg, G.; Singh, A.; Kaur, A.; Singh, R.; Kaur, J.; Mahajan, R. Microbial pectinases: An ecofriendly tool of nature for industries. 3 Biotech. 2016, 6, 47. [Google Scholar] [CrossRef] [PubMed]
- Tapre, A.R.; Jain, R.K. Pectinases: Enzymes for fruit processing industry. Int. Food Res. J. 2014, 21, 447–453. [Google Scholar]
- Kobayashi, T.; Higaki, N.; Suzumatsu, A.; Sawada, K.; Hagihara, H.; Kawai, S.; Ito, S. Purification and properties of a high-molecular-weight, alkaline exopolygalacturonase from a strain of Bacillus. Enzym. Microb. Technol. 2001, 29, 70–75. [Google Scholar] [CrossRef]
- Yu, P.; Zhang, Y.; Gu, D. Production optimization of a heat-tolerant alkaline pectinase from Bacillus subtilis ZGL14 and its purification and characterization. Bioengineered 2017, 8, 613–623. [Google Scholar] [CrossRef] [PubMed]
- Jacob, N.; Poorna, C.A.; Prema, P. Purification and partial characterization of polygalacturonase from Streptomyces lydicus. Bioresour. Technol. 2008, 99, 6697–6701. [Google Scholar] [CrossRef] [PubMed]
- Juárez, A.V.; Dreyer, J.; Göpel, P.; Koschke, N.; Frank, D.; Märkl, H.; Müller, R. Characterisation of a new thermoalkaliphilic bacterium for the production of high-quality hemp fibres, Geobacillus thermoglucosidasius strain PB94A. Appl. Microbial. Biotechnol. 2009, 83, 521–527. [Google Scholar] [CrossRef] [PubMed]
- Oumer, O.J.; Abate, D. Characterization of pectinase from Bacillus subtilis strain Btk 27 and its potential application in removal of mucilage from coffee beans. Enzyme Res. 2017, 2017, 7686904. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.; Yao, J.; Zhou, C.; Mao, L.; Zhang, G.; Ma, Y. The alkaline pectate lyase PEL168 of Bacillus subtilis heterologously expressed in Pichia pastoris is more stable and efficient for degumming ramie fiber. BMC Biotechnol. 2013, 13, 26. [Google Scholar] [CrossRef] [PubMed]

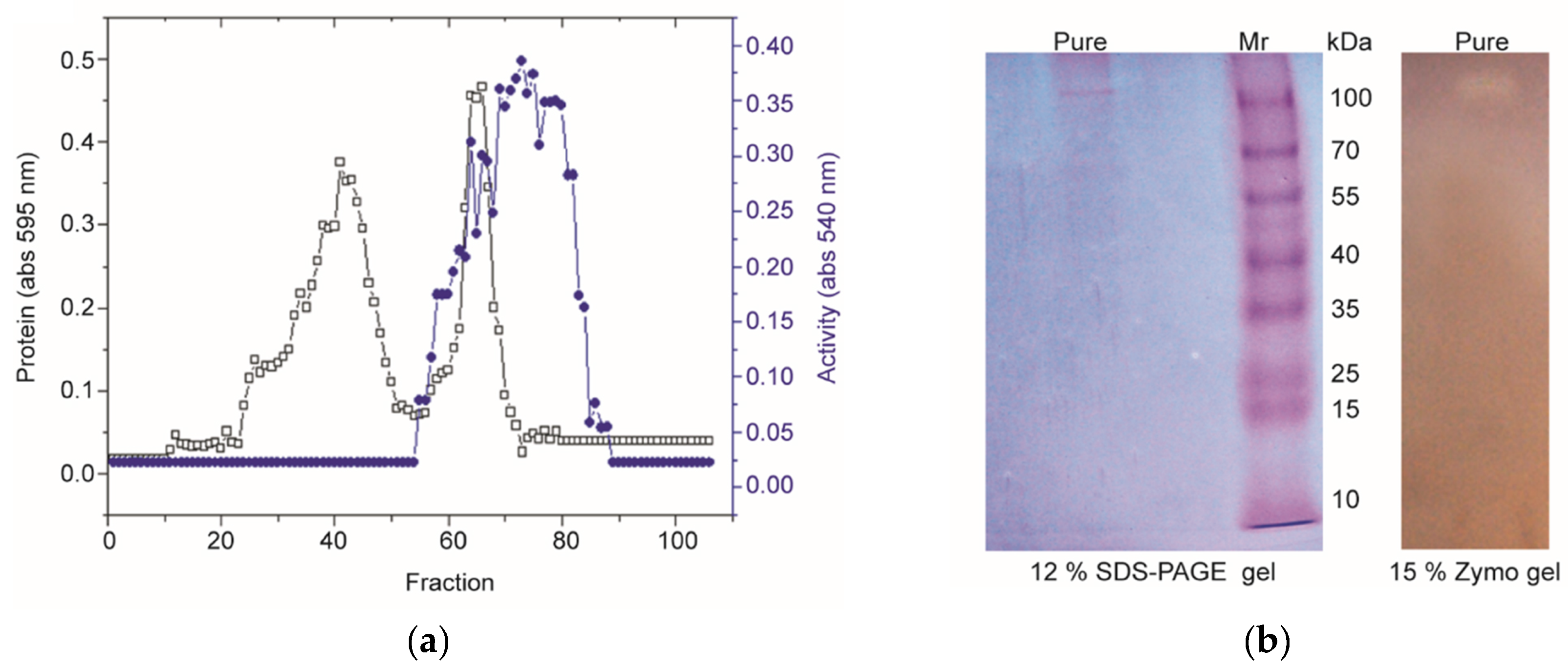


| Purification Step | Total Vol. (mL) | Total Protein (mg) | Total Activity (U) | Specific Activity (U/mg) | Yield (%) | Purity Fold |
|---|---|---|---|---|---|---|
| Crude Broth | 800 | 188 | 23,112 | 123 | 100.0 | 1.0 |
| (NH4)2SO4 | 15 | 21 | 16,254 | 774 | 70.3 | 6.3 |
| Sepharose CL 6B | 18 | 0.96 | 3892 | 4054 | 16.8 | 33.0 |
| Bacillus Strains | Type | pH | Temp. (°C) | Ref. |
|---|---|---|---|---|
| Bacillus sp. MG–cp–2 | Polygalacturonase | 10.0 | 60 | [12] |
| Bacillus paralicheniformis CBS3 | Polygalacturonase | 9.1 | 60 | [23] |
| Bacillus sp. Y1 | Pectate lyase | 8.5 | 50 | [25] |
| Bacillus clausii KSM–K16 | Pectate lyase | 10.5 | 50–55 | [26] |
| Bacillus subtilis PEL168 | Pectate lyase | 9.5 | 50 | [26] |
| Bacillus pumilus DT7 | Polygalacturonase | 8.0 | 45 | [27] |
| Bacillus subtilis | Polygalacturonase | 9.5 | 65 | [27] |
| Bacillus sp. NT–33 | Polygalacturonase | 10.5 | 75 | [28] |
| Paenibacillus polymyxa (Bacillus polymyxa) | Polygalacturonase | 8.4–9.4 | 60 | [28] |
| Bacillus stearothermophilus | Pectate lyase | 9.0 | 70 | [28] |
| Bacillus pumilus | Pectate lyase | 8.0–8.5 | 60 | [28] |
| Bacillus sp. KSM–P576 | Polygalacturonase | 8.0 | 55 | [29] |
| Bacillus subtilis ZGL14 | Polygalacturonase | 8.6 | 50 | [30] |
| Bacillus paralicheniformis CBS32 | Polygalacturonase | 9.0 | 60 | This study |
| Metal Ions | Concentration (mM) | Relative Activity (%) a |
|---|---|---|
| Ca2+ | 5 | 82.7 ± 1.0 |
| Mg2+ | 5 | 105.2 ± 5.0 |
| Cu2+ | 5 | 85.4 ± 1.2 |
| Ni2+ | 5 | 102.4 ± 0.5 |
| Zn2+ | 5 | 101.3 ± 0.9 |
| K+ | 5 | 105.3 ± 4.0 |
| Mn2+ | 5 | 103.6 ± 5.0 |
| Na+ | 5 | 90.1 ± 0.5 |
| Control | - | 100.0 |
| Plot Method | Km (mg/mL) | Vmax (U/mg) |
|---|---|---|
| Lineweaver–Burk plot | 0.021 ± 0.0020 | 410.43 ± 1.80 |
| Hanes–Woolf plot | 0.043 ± 0.0064 | 444.03 ± 9.34 |
| Eadie–Hofstee plot | 0.021 ± 0.0019 | 413.60 ± 1.08 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rahman, M.S.; Choi, Y.S.; Kim, Y.K.; Park, C.; Yoo, J.C. Production of Novel Polygalacturonase from Bacillus paralicheniformis CBS32 and Application to Depolymerization of Ramie Fiber. Polymers 2019, 11, 1525. https://doi.org/10.3390/polym11091525
Rahman MS, Choi YS, Kim YK, Park C, Yoo JC. Production of Novel Polygalacturonase from Bacillus paralicheniformis CBS32 and Application to Depolymerization of Ramie Fiber. Polymers. 2019; 11(9):1525. https://doi.org/10.3390/polym11091525
Chicago/Turabian StyleRahman, Md. Saifur, Yoon Seok Choi, Young Kyun Kim, Chulhwan Park, and Jin Cheol Yoo. 2019. "Production of Novel Polygalacturonase from Bacillus paralicheniformis CBS32 and Application to Depolymerization of Ramie Fiber" Polymers 11, no. 9: 1525. https://doi.org/10.3390/polym11091525