Poly (Lactic-co-Glycolic Acid) Nanoparticles and Nanoliposomes for Protein Delivery in Targeted Therapy: A Comparative In Vitro Study
Abstract
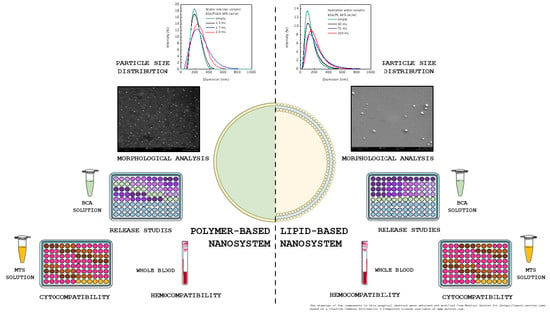
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Preparation of Polymeric Nanoparticles
2.3. Preparation of Nanoliposomes
2.4. Particles Size
2.5. Scanning Electron Microscopy
2.6. Entrapment Efficiency
2.7. In Vitro Release Studies
2.8. Hemolysis
- : is the absorbance of the sample;
- : is the absorbance of the negative control;
- : is the absorbance of the positive control.
2.9. Cell Viability
2.10. Statistical Analysis
3. Results
3.1. Production and Characterization of BSA-Loaded Nanocarriers
3.2. In Vitro Release Studies
3.3. Hemolysis Assay
3.4. Cell Viability
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Maheshwari, R.; Tekade, M.; Sharma, P.; Tekade, R. Nanocarriers assisted siRNA gene therapy for the management of cardiovascular disorders. Curr. Pharm. Des. 2015, 21, 4427–4440. [Google Scholar] [CrossRef] [PubMed]
- Auwal, S.M.; Zarei, M.; Tan, C.; Basri, M.; Saari, N. Improved in vivo efficacy of anti-hypertensive biopeptides encapsulated in chitosan nanoparticles fabricated by ionotropic gelation on spontaneously hypertensive rats. Nanomaterials 2017, 7, 421. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oduk, Y.; Zhu, W.; Kannappan, R.; Zhao, M.; Borovjagin, A.V.; Oparil, S.; Zhang, J. VEGF nanoparticles repair the heart after myocardial infarction. Am. J. Physiol. Heart Circ. Physiol. 2018, 314, H278–H284. [Google Scholar] [CrossRef] [PubMed]
- Theoharis, S.; Krueger, U.; Tan, P.H.; Haskard, D.O.; Weber, M.; George, A.J.T. Targeting gene delivery to activated vascular endothelium using anti E/P-Selectin antibody linked to PAMAM dendrimers. J. Immunol. Methods 2009, 343, 79–90. [Google Scholar] [CrossRef] [PubMed]
- Chan, J.M.; Rhee, J.-W.; Drum, C.L.; Bronson, R.T.; Golomb, G.; Langer, R.; Farokhzad, O.C. In vivo prevention of arterial restenosis with paclitaxel-encapsulated targeted lipid-polymeric nanoparticles. Proc. Natl. Acad. Sci. USA 2011, 108, 19347–19352. [Google Scholar] [CrossRef] [Green Version]
- Olofsson, P.S.; Söderström, L.Å.; Wågsäter, D.; Sheikine, Y.; Ocaya, P.; Lang, F.; Rabu, C.; Chen, L.; Rudling, M.; Aukrust, P.; et al. CD137 is expressed in human atherosclerosis and promotes development of plaque inflammation in hypercholesterolemic mice. Circulation 2008, 117, 1292–1301. [Google Scholar] [CrossRef] [PubMed]
- Fava, C.; Montagnana, M. Atherosclerosis is an inflammatory disease which lacks a common anti-inflammatory therapy: How human genetics can help to this issue. A narrative review. Front. Pharmacol. 2018, 9, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Putney, S.D. Encapsulation of proteins for improved delivery. Curr. Opin. Chem. Biol. 1998, 2, 548–552. [Google Scholar] [CrossRef]
- Perry, S.L.; McClements, D.J. Recent advances in encapsulation, protection, and oral delivery of bioactive proteins and peptides using colloidal systems. Molecules 2020, 25, 1161. [Google Scholar] [CrossRef] [Green Version]
- Giannouli, M.; Karagkiozaki, V.; Pappa, F.; Moutsios, I.; Gravalidis, C.; Logothetidis, S. Fabrication of quercetin-loaded PLGA nanoparticles via electrohydrodynamic atomization for cardiovascular disease. Mater. Today-Proc. 2018, 5, 15998–16005. [Google Scholar] [CrossRef]
- Ferrari, P.F.; Aliakbarian, B.; Zattera, E.; Pastorino, L.; Palombo, D.; Perego, P. Engineered CaCO3 nanoparticles with targeting activity: A simple approach for a vascular intended drug delivery system. Can. J. Chem. Eng. 2017, 95, 1683–1689. [Google Scholar] [CrossRef]
- Lovelyn, C.; Attama, A.A. Current state of nanoemulsions in drug delivery. J. Biomater. Nanobiotechnol. 2011, 2, 626–639. [Google Scholar] [CrossRef] [Green Version]
- Oumzil, K.; Ramin, M.A.; Lorenzato, C.; Hémadou, A.; Laroche, J.; Jacobin-Valat, M.J.; Mornet, S.; Roy, C.-E.; Kauss, T.; Gaudin, K.; et al. Solid lipid nanoparticles for image-guided therapy of atherosclerosis. Bioconj. Chem. 2016, 27, 569–575. [Google Scholar] [CrossRef]
- Liu, J.; Gu, C.; Cabigas, E.B.; Pendergrass, K.D.; Brown, M.E.; Luo, Y.; Davis, M.E. Functionalized dendrimer-based delivery of angiotensin type 1 receptor siRNA for preserving cardiac function following infarction. Biomaterials 2013, 34, 3729–3736. [Google Scholar] [CrossRef] [Green Version]
- Panda, V.S.; Naik, S.R. Cardioprotective activity of Ginkgo biloba phytosomes in isoproterenol-induced myocardial necrosis in rats: A biochemical and histoarchitectural evaluation. Exp. Toxicol. Pathol. 2008, 60, 397–404. [Google Scholar] [CrossRef]
- Peters, D.; Kastantin, M.; Kotamraju, V.R.; Karmali, P.P.; Gujraty, K.; Tirrell, M.; Ruoslahti, E. Targeting atherosclerosis by using modular, multifunctional micelles. Proc. Natl. Acad. Sci. USA 2009, 106, 9815–9819. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Almer, G.; Frascione, D.; Pali-Schöll, I.; Vonach, C.; Lukschal, A.; Stremnitzer, C.; Diesner, S.C.; Jensen-Jarolim, E.; Prassl, R.; Mangge, H. Interleukin-10: An anti-inflammatory marker to target atherosclerotic lesions via PEGylated liposomes. Mol. Pharm. 2013, 10, 175–186. [Google Scholar] [CrossRef] [PubMed]
- Zachman, A.L.; Wang, X.; Tucker-Schwartz, J.M.; Fitzpatrick, S.T.; Lee, S.H.; Guelcher, S.A.; Skala, M.C.; Sung, H.J. Uncoupling angiogenesis and inflammation in peripheral artery disease with therapeutic peptide-loaded microgels. Biomaterials 2014, 35, 9635–9648. [Google Scholar] [CrossRef] [Green Version]
- Cheng, G.; Zhang, X.; Chen, Y.; Lee, R.J.; Wang, J.; Yao, J.; Zhang, Y.; Zhang, C.; Wang, K.; Yu, B. Anticancer activity of polymeric nanoparticles containing linoleic acid-SN38 (LA-SN38) conjugate in a murine model of colorectal cancer. Colloid. Surf. B 2019, 181, 822–829. [Google Scholar] [CrossRef]
- Jana, U.; Mohanty, A.K.; Pal, S.L.; Manna, P.K.; Mohanta, G.P. Felodipine loaded PLGA nanoparticles: Preparation, physicochemical characterization and in vivo toxicity study. Nano Converg. 2014, 1, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Primard, C.; Poecheim, J.; Heuking, S.; Sublet, E.; Esmaeili, F.; Borchard, G. Multifunctional PLGA-based nanoparticles encapsulating simultaneously hydrophilic antigen and hydrophobic immunomodulator for mucosal immunization. Mol. Pharm. 2013, 10, 2996–3004. [Google Scholar] [CrossRef] [PubMed]
- Gentile, P.; Nandagiri, V.; Pabari, R.; Daly, J.; Tonda-Turo, C.; Ciardelli, G.; Ramtoola, Z. Influence of parathyroid hormone-loaded PLGA nanoparticles in porous scaffolds for bone regeneration. Int. J. Mol. Sci. 2015, 16, 20492–20510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chereddy, K.K.; Her, C.-H.; Comune, M.; Moia, C.; Lopes, A.; Porporato, P.E.; Vanacker, J.; Lam, M.C.; Steinstraesser, L.; Sonveaux, P.; et al. PLGA nanoparticles loaded with host defense peptide LL37 promote wound healing. J. Control. Release 2014, 194, 138–147. [Google Scholar] [CrossRef] [PubMed]
- Feczkó, T.; Tóth, J.; Dósa, G.; Gyenis, J. Optimization of protein encapsulation in PLGA nanoparticles. Chem. Eng. Process. 2011, 50, 757–765. [Google Scholar] [CrossRef]
- Liang, G.; Zhu, Y.; Sun, B.; Hu, F.; Tian, T.; Li, S.; Xiao, Z. PLGA-based gene delivering nanoparticle enhance suppression effect of miRNA in HePG2 cells. Nanoscale Res. Lett. 2011, 6, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martins, C.; Sousa, F.; Araújo, F.; Sarmento, B. Functionalizing PLGA and PLGA derivatives for drug delivery and tissue regeneration applications. Adv. Healthc. Mater. 2018, 7. [Google Scholar] [CrossRef]
- Kocbek, P.; Obermajer, N.; Cegnar, M.; Kos, J.; Kristl, J. Targeting cancer cells using PLGA nanoparticles surface modified with monoclonal antibody. J. Control. Release 2007, 120, 18–26. [Google Scholar] [CrossRef]
- Huang, W.; Zhang, C. Tuning the size of poly(lactic-co-glycolic acid) (PLGA) nanoparticles fabricated by nanoprecipitation. Biotechnol. J. 2018, 13. [Google Scholar] [CrossRef]
- Arpagaus, C. PLA/PLGA nanoparticles prepared by nano spray drying. J. Pharm. Investig. 2019, 49, 405–426. [Google Scholar] [CrossRef] [Green Version]
- Cohen-Sela, E.; Chorny, M.; Koroukhov, N.; Danenberg, H.D.; Golomb, G. A new double emulsion solvent diffusion technique for encapsulating hydrophilic molecules in PLGA nanoparticles. J. Control. Release 2009, 133, 90–95. [Google Scholar] [CrossRef]
- Streck, S.; Neumann, H.; Nielsen, H.M.; Rades, T.; McDowell, A. Comparison of bulk and microfluidics methods for the formulation of poly-lactic-co-glycolic acid (PLGA) nanoparticles modified with cell-penetrating peptides of different architectures. Int. J. Pharm. 2019, 1. [Google Scholar] [CrossRef] [PubMed]
- Alshamsan, A. Nanoprecipitation is more efficient than emulsion solvent evaporation method to encapsulate cucurbitacin I in PLGA nanoparticles. Saudi Pharm. J. 2014, 22, 219–222. [Google Scholar] [CrossRef] [PubMed]
- Campardelli, R.; Espirito Santo, I.; Albuquerque, E.C.; de Melo, S.V.; Della Porta, G.; Reverchon, E. Efficient encapsulation of proteins in submicro liposomes using a supercritical fluid assisted continuous process. J. Supercrit. Fluids 2016, 107, 163–169. [Google Scholar] [CrossRef]
- Petersen, A.L.; Hansen, A.E.; Gabizon, A.; Andresen, T.L. Liposome imaging agents in personalized medicine. Adv. Drug Deliv. Rev. 2012, 64, 1417–1435. [Google Scholar] [CrossRef] [PubMed]
- Swaminathan, J.; Ehrhardt, C. Liposomal delivery of proteins and peptides. Expert Opin. Drug Deliv. 2012, 9, 1489–1503. [Google Scholar] [CrossRef]
- Maherani, B.; Arab-Tehrany, E.; Kheirolomoom, A.; Geny, D.; Linder, M. Calcein release behavior from liposomal bilayer; influence of physicochemical/mechanical/structural properties of lipids. Biochimie 2013, 95, 2018–2033. [Google Scholar] [CrossRef]
- Movahedi, F.; Hu, R.G.; Becker, D.L.; Xu, C. Stimuli-responsive liposomes for the delivery of nucleic acid therapeutics. Nanomed Nanotechnol. 2015, 11, 1575–1584. [Google Scholar] [CrossRef]
- Patil, Y.P.; Jadhav, S. Novel methods for liposome preparation. Chem. Phys. Lipids 2014, 177, 8–18. [Google Scholar] [CrossRef]
- Akbarzadeh, A.; Rezaei-Sadabady, R.; Davaran, S.; Joo, S.W.; Zarghami, N.; Hanifehpour, Y.; Samiei, M.; Kouhi, M.; Nejati-Koshki, K. Liposome: Classification, preparation, and applications. Nanoscale Res. Lett. 2013, 8, 102. [Google Scholar] [CrossRef] [Green Version]
- Khosravi-Darani, K.; Mozafari, M.R. Nanoliposome potentials in nanotherapy: A concise overview. J. Nanosci. Nanotechnol. 2010, 6, 3–13. [Google Scholar]
- Fornaguera, C.; Calderó, G.; Mitjans, M.; Vinardell, M.P.; Solans, C.; Vauthier, C. Interactions of PLGA nanoparticles with blood components: Protein adsorption, coagulation, activation of the complement system and hemolysis studies. Nanoscale 2015, 7, 6045–6058. [Google Scholar] [CrossRef] [PubMed]
- Michanetzis, G.P.; Markoutsa, E.; Mourtas, S.; Missirlis, Y.F.; Antimisiaris, S.G. Hemocompatibility of amyloid and/or brain targeted liposomes. Future Med. Chem. 2019, 11, 693–705. [Google Scholar] [CrossRef] [PubMed]
- Freytag, T.; Dashevsky, A.; Tillman, L.; Hardee, G.E.; Bodmeier, R. Improvement of the encapsulation efficiency of oligonucleotide-containing biodegradable microspheres. J. Control. Release 2000, 69, 197–207. [Google Scholar] [CrossRef]
- Alex, R.; Bodmeier, R. Encapsulation of water-soluble drugs by a modified solvent evaporation method. I. Effect of process and formulation variables on drug entrapment. J. Microencapsul. 1990, 7, 347–355. [Google Scholar] [CrossRef]
- Herrmann, J.; Bodmeier, R. Somatostatin containing biodegradable microspheres prepared by a modified solvent evaporation method based on W/O/W-multiple emulsions. Int. J. Pharm. 1995, 126, 129–138. [Google Scholar] [CrossRef]
- Chen, L.Q.; Fang, L.; Ling, J.; Ding, C.Z.; Kang, B.; Huang, C.Z. Nanotoxicity of silver nanoparticles to red blood cells: Size dependent adsorption, uptake, and hemolytic activity. Chem. Res. Toxicol. 2015, 28, 501–509. [Google Scholar] [CrossRef]


 PNPBSA (30 mg/mL),
PNPBSA (30 mg/mL),  PNPBSA (35 mg/mL),
PNPBSA (35 mg/mL),  PNPBSA (40 mg/mL) and from (B) NLPs:
PNPBSA (40 mg/mL) and from (B) NLPs:  NLPBSA (3 mg/mL),
NLPBSA (3 mg/mL),  NLPBSA (4 mg/mL),
NLPBSA (4 mg/mL),  NLPBSA (6 mg/mL). Data are expressed as mean of three measurements. Error bars indicate SD. BSA: bovine serum albumin, PNPs: polymeric nanoparticles, NLPs: nanoliposomes.
NLPBSA (6 mg/mL). Data are expressed as mean of three measurements. Error bars indicate SD. BSA: bovine serum albumin, PNPs: polymeric nanoparticles, NLPs: nanoliposomes.
 PNPBSA (30 mg/mL),
PNPBSA (30 mg/mL),  PNPBSA (35 mg/mL),
PNPBSA (35 mg/mL),  PNPBSA (40 mg/mL) and from (B) NLPs:
PNPBSA (40 mg/mL) and from (B) NLPs:  NLPBSA (3 mg/mL),
NLPBSA (3 mg/mL),  NLPBSA (4 mg/mL),
NLPBSA (4 mg/mL),  NLPBSA (6 mg/mL). Data are expressed as mean of three measurements. Error bars indicate SD. BSA: bovine serum albumin, PNPs: polymeric nanoparticles, NLPs: nanoliposomes.
NLPBSA (6 mg/mL). Data are expressed as mean of three measurements. Error bars indicate SD. BSA: bovine serum albumin, PNPs: polymeric nanoparticles, NLPs: nanoliposomes.
 PNPE,
PNPE,  PNPBSA (30 mg/mL),
PNPBSA (30 mg/mL),  PNPBSA (35 mg/mL),
PNPBSA (35 mg/mL),  PNPBSA (40 mg/mL) and of (B) NLPs:
PNPBSA (40 mg/mL) and of (B) NLPs:  NLPE,
NLPE,  NLPBSA (3 mg/mL),
NLPBSA (3 mg/mL),  NLPBSA (4 mg/mL),
NLPBSA (4 mg/mL),  NLPBSA (6 mg/mL). RBCs: red blood cells, PNPE: empty polymeric nanoparticles, PNPBSA: BSA-loaded polymeric nanoparticles, NLPE: empty nanoliposomes, NLPBSA: BSA-loaded nanoliposomes. Data are expressed as mean of three measurements. Error bars indicate SD. Different symbols refer to statistically significant differences among results (p < 0.05, ANOVA with Tukey’s HSD post hoc multiple comparison test). a: statistically different to empty nanocarriers (PNPE or NLPE), aa: statistically different to PNPBSA (30 mg/mL) or NLPBSA (3 mg/mL), aaa: statistically different to PNPBSA (35 mg/mL) or NLPBSA (4 mg/mL). The dotted line refers to 5% of hemolysis.
NLPBSA (6 mg/mL). RBCs: red blood cells, PNPE: empty polymeric nanoparticles, PNPBSA: BSA-loaded polymeric nanoparticles, NLPE: empty nanoliposomes, NLPBSA: BSA-loaded nanoliposomes. Data are expressed as mean of three measurements. Error bars indicate SD. Different symbols refer to statistically significant differences among results (p < 0.05, ANOVA with Tukey’s HSD post hoc multiple comparison test). a: statistically different to empty nanocarriers (PNPE or NLPE), aa: statistically different to PNPBSA (30 mg/mL) or NLPBSA (3 mg/mL), aaa: statistically different to PNPBSA (35 mg/mL) or NLPBSA (4 mg/mL). The dotted line refers to 5% of hemolysis.
 PNPE,
PNPE,  PNPBSA (30 mg/mL),
PNPBSA (30 mg/mL),  PNPBSA (35 mg/mL),
PNPBSA (35 mg/mL),  PNPBSA (40 mg/mL) and of (B) NLPs:
PNPBSA (40 mg/mL) and of (B) NLPs:  NLPE,
NLPE,  NLPBSA (3 mg/mL),
NLPBSA (3 mg/mL),  NLPBSA (4 mg/mL),
NLPBSA (4 mg/mL),  NLPBSA (6 mg/mL). RBCs: red blood cells, PNPE: empty polymeric nanoparticles, PNPBSA: BSA-loaded polymeric nanoparticles, NLPE: empty nanoliposomes, NLPBSA: BSA-loaded nanoliposomes. Data are expressed as mean of three measurements. Error bars indicate SD. Different symbols refer to statistically significant differences among results (p < 0.05, ANOVA with Tukey’s HSD post hoc multiple comparison test). a: statistically different to empty nanocarriers (PNPE or NLPE), aa: statistically different to PNPBSA (30 mg/mL) or NLPBSA (3 mg/mL), aaa: statistically different to PNPBSA (35 mg/mL) or NLPBSA (4 mg/mL). The dotted line refers to 5% of hemolysis.
NLPBSA (6 mg/mL). RBCs: red blood cells, PNPE: empty polymeric nanoparticles, PNPBSA: BSA-loaded polymeric nanoparticles, NLPE: empty nanoliposomes, NLPBSA: BSA-loaded nanoliposomes. Data are expressed as mean of three measurements. Error bars indicate SD. Different symbols refer to statistically significant differences among results (p < 0.05, ANOVA with Tukey’s HSD post hoc multiple comparison test). a: statistically different to empty nanocarriers (PNPE or NLPE), aa: statistically different to PNPBSA (30 mg/mL) or NLPBSA (3 mg/mL), aaa: statistically different to PNPBSA (35 mg/mL) or NLPBSA (4 mg/mL). The dotted line refers to 5% of hemolysis.

 control (without particles),
control (without particles),  PNPE (A,C,E),
PNPE (A,C,E),  PNPBSA (40mg/mL) (A,C,E),
PNPBSA (40mg/mL) (A,C,E),  NLPE (B,D,F),
NLPE (B,D,F),  NLPBSA (3 mg/mL) (B,D,F). PNPE: empty polymeric nanoparticles, PNPBSA: BSA-loaded polymeric nanoparticles, NLPE: empty nanoliposomes, NLPBSA: BSA-loaded nanoliposomes. Data are expressed as mean of three measurements. Error bars indicate SD. Different symbols refer to statistically significant differences among results (p < 0.05, ANOVA with Tukey’s HSD post hoc multiple comparison test). a: statistically different to control, aa: statistically different to empty carrier.
NLPBSA (3 mg/mL) (B,D,F). PNPE: empty polymeric nanoparticles, PNPBSA: BSA-loaded polymeric nanoparticles, NLPE: empty nanoliposomes, NLPBSA: BSA-loaded nanoliposomes. Data are expressed as mean of three measurements. Error bars indicate SD. Different symbols refer to statistically significant differences among results (p < 0.05, ANOVA with Tukey’s HSD post hoc multiple comparison test). a: statistically different to control, aa: statistically different to empty carrier.
 control (without particles),
control (without particles),  PNPE (A,C,E),
PNPE (A,C,E),  PNPBSA (40mg/mL) (A,C,E),
PNPBSA (40mg/mL) (A,C,E),  NLPE (B,D,F),
NLPE (B,D,F),  NLPBSA (3 mg/mL) (B,D,F). PNPE: empty polymeric nanoparticles, PNPBSA: BSA-loaded polymeric nanoparticles, NLPE: empty nanoliposomes, NLPBSA: BSA-loaded nanoliposomes. Data are expressed as mean of three measurements. Error bars indicate SD. Different symbols refer to statistically significant differences among results (p < 0.05, ANOVA with Tukey’s HSD post hoc multiple comparison test). a: statistically different to control, aa: statistically different to empty carrier.
NLPBSA (3 mg/mL) (B,D,F). PNPE: empty polymeric nanoparticles, PNPBSA: BSA-loaded polymeric nanoparticles, NLPE: empty nanoliposomes, NLPBSA: BSA-loaded nanoliposomes. Data are expressed as mean of three measurements. Error bars indicate SD. Different symbols refer to statistically significant differences among results (p < 0.05, ANOVA with Tukey’s HSD post hoc multiple comparison test). a: statistically different to control, aa: statistically different to empty carrier.
| BSA Concentration (mg/mL) | Water Internal Phase Volume (mL) | MD ± SD (nm) | EE ± SD (%) | |
|---|---|---|---|---|
| PNPs | 0 | 1.7 | 204 ± 20 | - |
| 30 | 2.0 | 195 ± 11 | 97.15 ± 0.07 a | |
| 35 | 1.7 | 185 ± 10 | 97.82 ± 0.07 b | |
| 40 | 1.5 | 170 ± 12 | 98.01 ± 0.05 c | |
| NLPs | 0 | 75 | 130 ± 51 | - |
| 3 | 100 | 175 ± 62 | 46.14 ± 14.17 d | |
| 4 | 75 | 152 ± 68 | 49.49 ± 2.18 d | |
| 6 | 50 | 144 ± 60 | 80.16 ± 7.46 e |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Negri Atanasio, G.; Ferrari, P.F.; Campardelli, R.; Perego, P.; Palombo, D. Poly (Lactic-co-Glycolic Acid) Nanoparticles and Nanoliposomes for Protein Delivery in Targeted Therapy: A Comparative In Vitro Study. Polymers 2020, 12, 2566. https://doi.org/10.3390/polym12112566
De Negri Atanasio G, Ferrari PF, Campardelli R, Perego P, Palombo D. Poly (Lactic-co-Glycolic Acid) Nanoparticles and Nanoliposomes for Protein Delivery in Targeted Therapy: A Comparative In Vitro Study. Polymers. 2020; 12(11):2566. https://doi.org/10.3390/polym12112566
Chicago/Turabian StyleDe Negri Atanasio, Giulia, Pier Francesco Ferrari, Roberta Campardelli, Patrizia Perego, and Domenico Palombo. 2020. "Poly (Lactic-co-Glycolic Acid) Nanoparticles and Nanoliposomes for Protein Delivery in Targeted Therapy: A Comparative In Vitro Study" Polymers 12, no. 11: 2566. https://doi.org/10.3390/polym12112566