Fluorescent Molecularly Imprinted Polymers Loaded with Avenanthramides for Inhibition of Advanced Glycation End Products
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Apparatus
2.2. Design and Synthesis of CDs
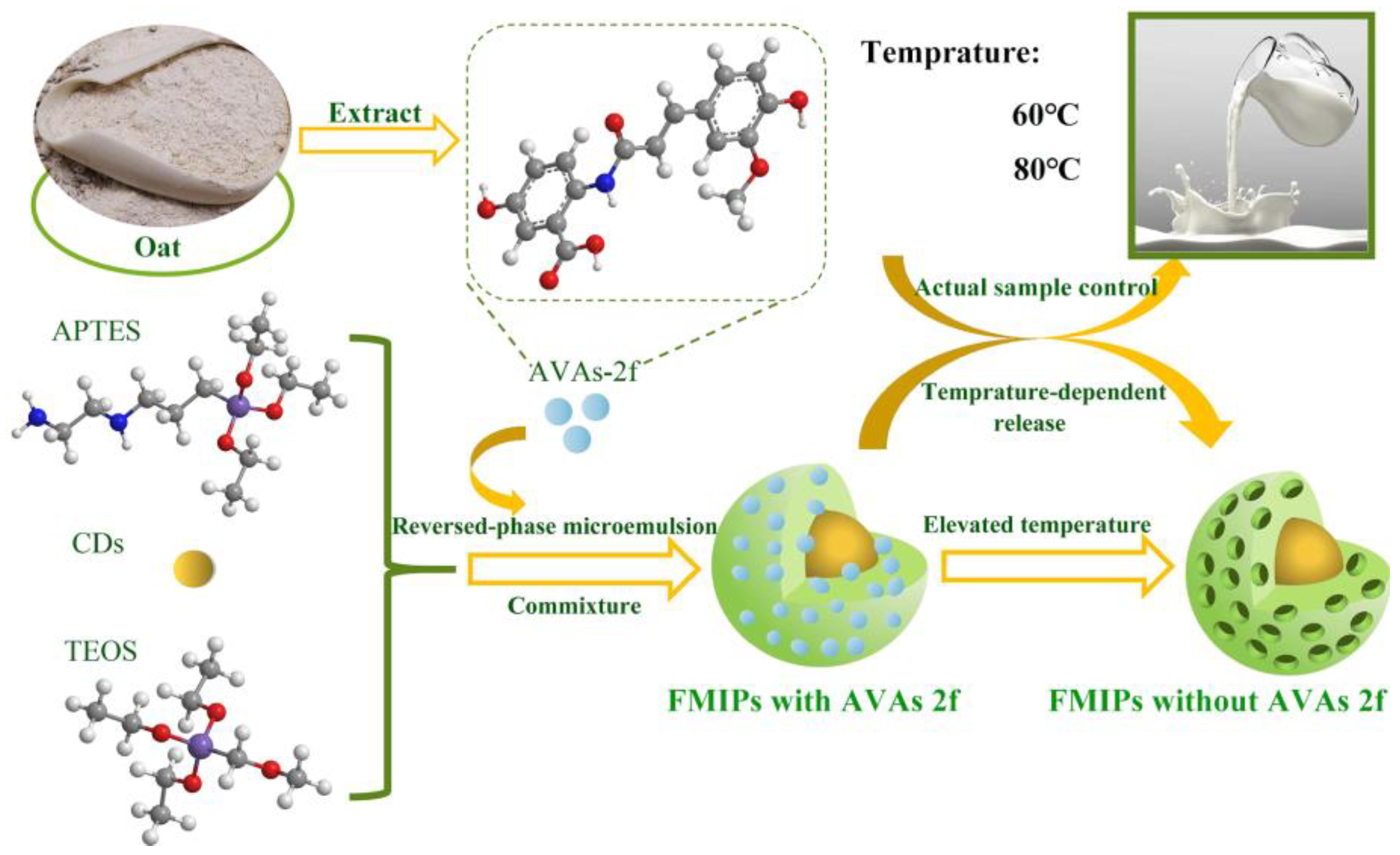
2.3. Preparation of FMIPs
2.4. Efficiency of AVAs-2f Encapsulation into FMIPs
2.5. Determination of DPPH Radical Scavenging Activity
2.6. Fluorescence Stability Performance
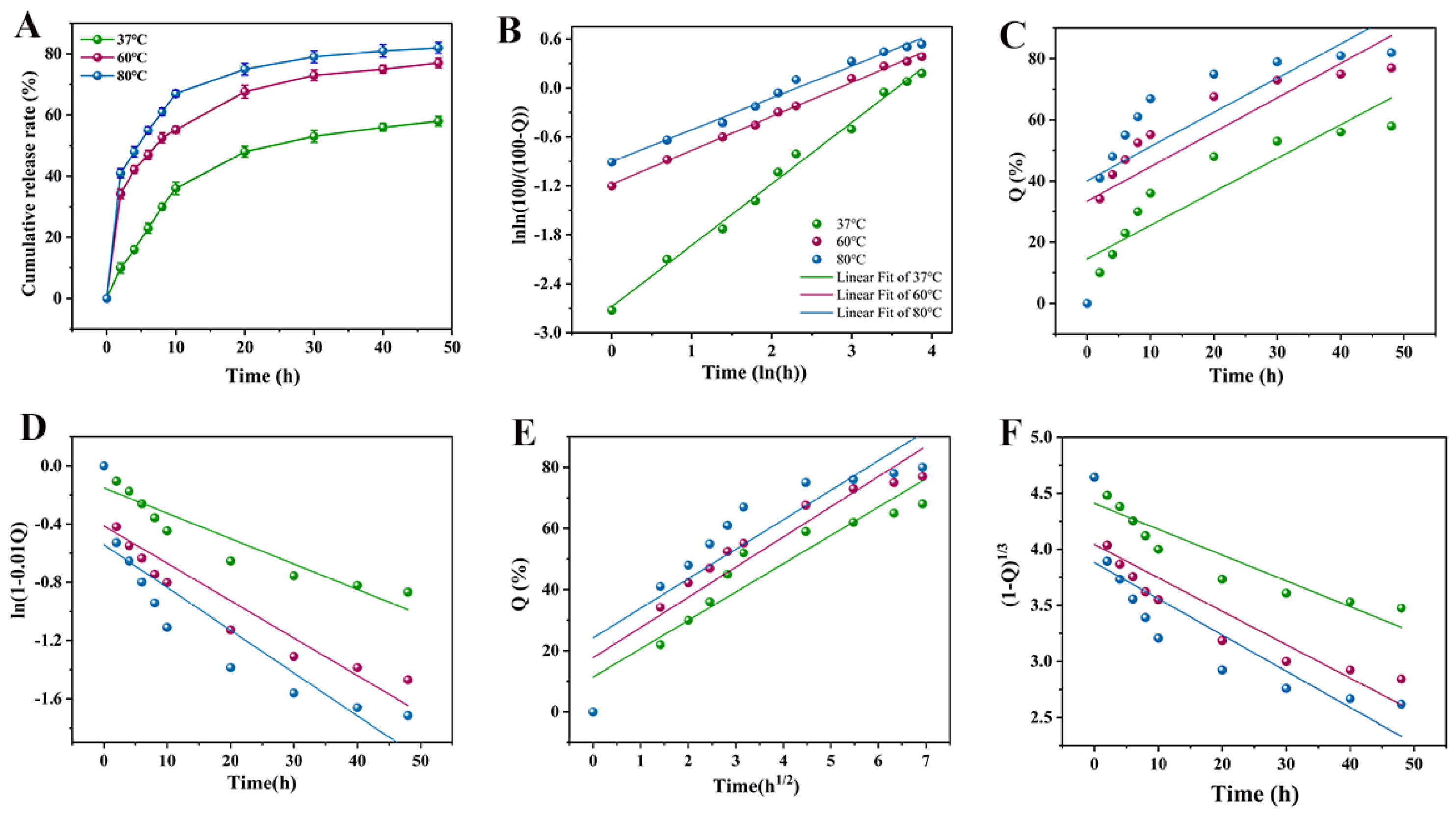
2.7. In Vitro Release of AVAs 2f
2.8. Preparation of Casein-Ribose Simulation System
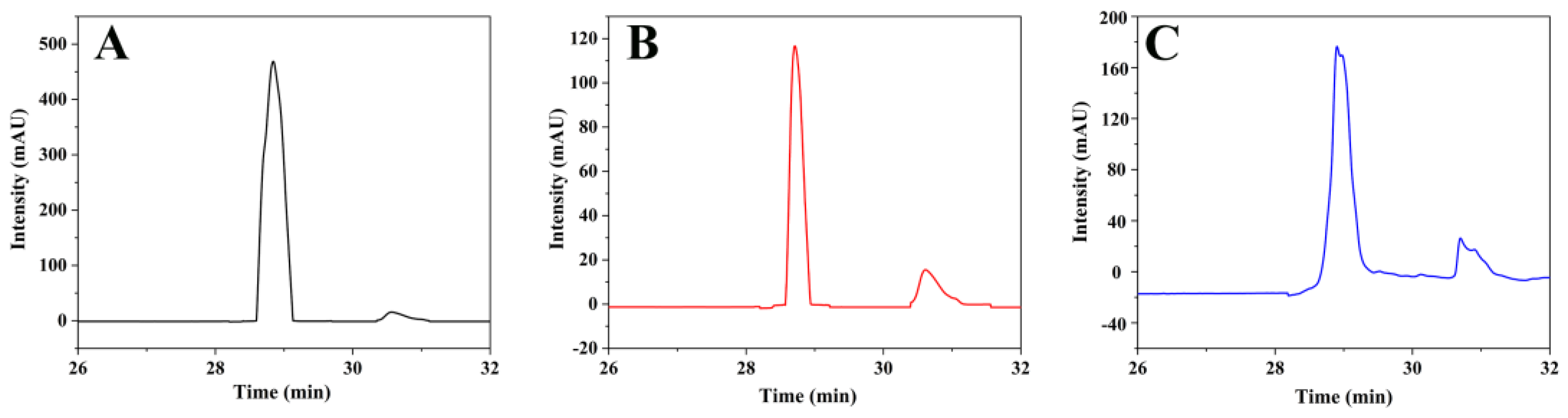
2.9. Detection of PRL by HPLC
2.10. Testing of Milk Samples
2.11. Statistical Analysis
3. Results
3.1. Morphological and Structural Characterization of FMIPs
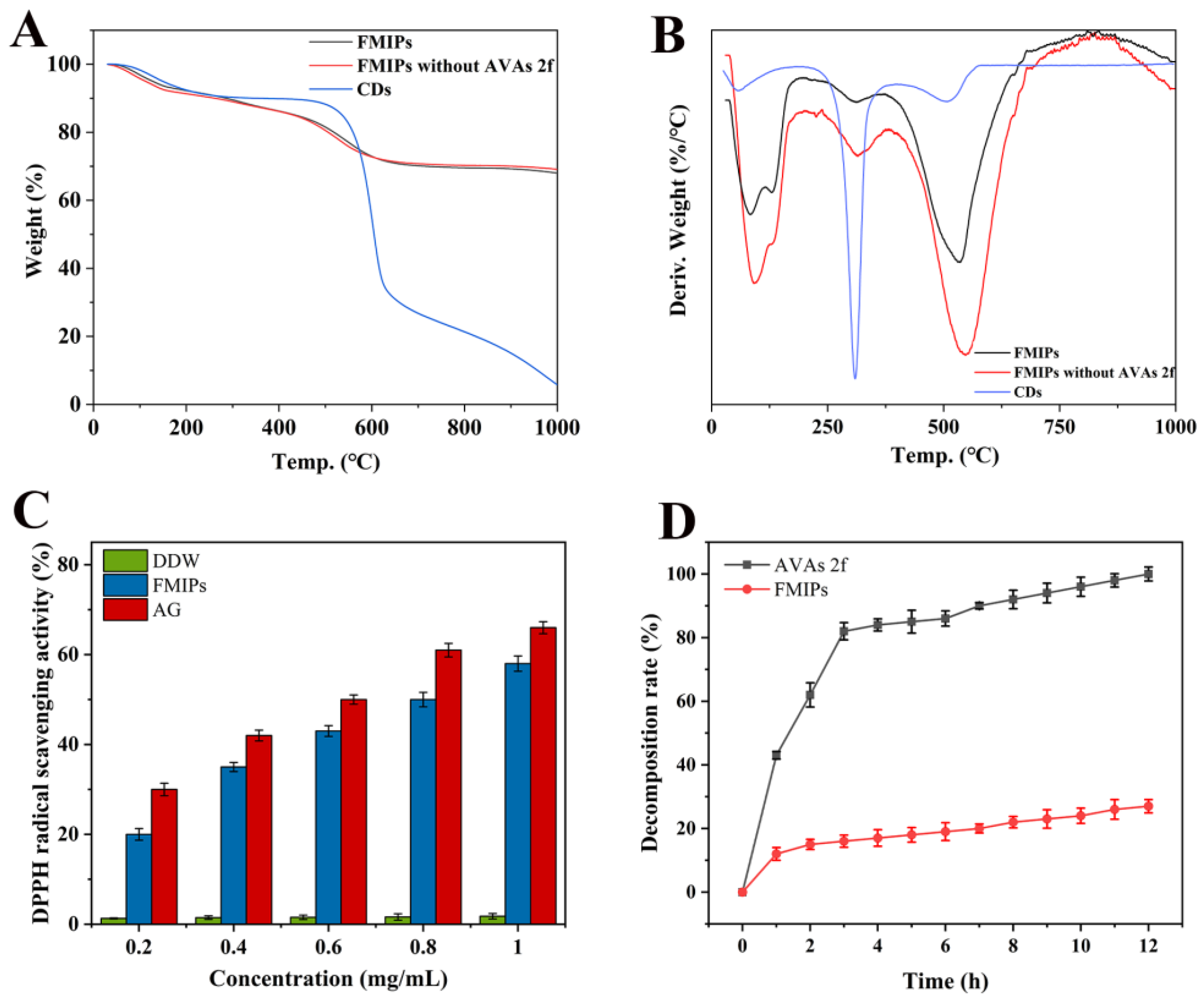
3.2. Thermogravimetric Analysis of FMIPs
3.3. DPPH Radical Scavenging Analysis
3.4. Fluorescence Stability
3.5. AVAs-2f In Vitro Release
3.6. Inhibition of PRL by FMIPs in Simulation System
3.7. Inhibition of PRL by FMIPs in Milk Samples
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Nowotny, K.; Schröter, D.; Schreiner, M.; Grune, T. Dietary advanced glycation end products and their relevance for human health. Ageing Res. Rev. 2018, 47, 55–66. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.; Liu, H.; Wang, J.; Sun, B. Inhibitory effect of phenolic compounds and plant extracts on the formation of advance glycation end products: A comprehensive review. Food Res. Int. 2020, 130, 108933. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Zheng, T.; Sang, S.; Lv, L. Quercetin Inhibits Advanced Glycation End Product Formation by Trapping Methylglyoxal and Glyoxal. J. Agric. Food Chem. 2014, 62, 12152–12158. [Google Scholar] [CrossRef]
- Zhang, D.; Zhu, P.; Han, L.; Chen, X.; Liu, H.; Sun, B. Highland Barley and Its By-Products Enriched with Phenolic Compounds for Inhibition of Pyrraline Formation by Scavenging α-Dicarbonyl Compounds. Foods 2021, 10, 1109. [Google Scholar] [CrossRef]
- Peterson, D.M.; Dimberg, L.H. Avenanthramide concentrations and hydroxycinnamoyl-CoA:hydroxyanthranilate N-hydroxycinnamoyltransferase activities in developing oats. J. Cereal Sci. 2008, 47, 101–108. [Google Scholar] [CrossRef]
- Perrelli, A.; Goitre, L.; Salzano, A.M.; Moglia, A.; Scaloni, A.; Retta, S.F. Biological Activities, Health Benefits, and Therapeutic Properties of Avenanthramides: From Skin Protection to Prevention and Treatment of Cerebrovascular Diseases. Oxidative Med. Cell. Longev. 2018, 2018, 6015351. [Google Scholar] [CrossRef] [PubMed]
- Ding, J.; Johnson, J.; Chu, Y.F.; Feng, H. Enhancement of γ-aminobutyric acid, avenanthramides, and other health-promoting metabolites in germinating oats (Avena sativa L.) treated with and without power ultrasound. Food Chem. 2019, 283, 239–247. [Google Scholar] [CrossRef]
- de Bruijn, W.J.; Van Dinteren, S.; Gruppen, H.; Vincken, J.-P. Mass spectrometric characterisation of avenanthramides and enhancing their production by germination of oat (Avena sativa). Food Chem. 2019, 277, 682–690. [Google Scholar] [CrossRef]
- Zhu, P.; Zhang, Y.; Zhang, D.; Han, L.; Liu, H.; Sun, B. Inhibitory Mechanism of Advanced Glycation End-Product Formation by Avenanthramides Derived from Oats through Scavenging the Intermediates. Foods 2022, 11, 1813. [Google Scholar] [CrossRef]
- Liang, C.; Zhang, Z.; Zhang, H.; Ye, L.; He, J.; Ou, J.; Wu, Q. Ordered macroporous molecularly imprinted polymers prepared by a surface imprinting method and their applications to the direct extraction of flavonoids from Gingko leaves. Food Chem. 2020, 309, 125680. [Google Scholar] [CrossRef]
- Chang, P.; Zhang, Z.; Yang, C. Molecularly imprinted polymer-based chemiluminescence array sensor for the detection of proline. Anal. Chim. Acta. 2010, 666, 70–75. [Google Scholar] [CrossRef] [PubMed]
- Hu, Q.; Bu, Y.; Zhen, X.; Xu, K.; Ke, R.; Xie, X.; Wang, S. Magnetic carbon nanotubes camouflaged with cell membrane as a drug discovery platform for selective extraction of bioactive compounds from natural products. Chem. Eng. J. 2019, 364, 269–279. [Google Scholar] [CrossRef]
- Wang, C.; Javadi, A.; Ghaffari, M.; Gong, S. A pH-sensitive molecularly imprinted nanospheres/hydrogel composite as a coating for implantable biosensors. Biomaterials 2010, 31, 4944–4951. [Google Scholar] [CrossRef] [PubMed]
- Whitcombe, M.J.; Chianella, I.; Larcombe, L.; Piletsky, S.A.; Noble, J.; Porter, R.; Horgan, A. The rational development of molecularly imprinted polymer-based sensors for protein detection. Chem. Soc. Rev. 2010, 40, 1547–1571. [Google Scholar] [CrossRef] [Green Version]
- Ali, M.; Horikawa, S.; Venkatesh, S.; Saha, J.; Hong, J.W.; Byrne, M.E. Zero-order therapeutic release from imprinted hydrogel contact lenses within in vitro physiological ocular tear flow. J. Control. Release 2008, 124, 154–162. [Google Scholar] [CrossRef]
- Adali-Kaya, Z.; Bui, B.T.S.; Falcimaigne-Cordin, A.; Haupt, K. Molecularly Imprinted Polymer Nanomaterials and Nanocomposites: Atom-Transfer Radical Polymerization with Acidic Monomers. Angew. Chem. Int. Ed. Engl. 2015, 127, 5281–5284. [Google Scholar] [CrossRef]
- Zhao, M.; Chen, X.; Zhang, H.; Yan, H.; Zhang, H. Well-Defined Hydrophilic Molecularly Imprinted Polymer Microspheres for Efficient Molecular Recognition in Real Biological Samples by Facile RAFT Coupling Chemistry. Biomacromolecules 2014, 15, 1663–1675. [Google Scholar] [CrossRef]
- Zhu, X.; Han, L.; Liu, H.; Sun, B. A smartphone-based ratiometric fluorescent sensing system for on-site detection of pyrethroids by using blue-green dual-emission carbon dots. Food Chem. 2022, 379, 132154. [Google Scholar] [CrossRef]
- Wen, J.; Anderson, S.M.; Du, J.; Yan, M.; Wang, J.; Shen, M.; Lu, Y.; Segura, T. Controlled Protein Delivery Based on Enzyme-Responsive Nanocapsules. Adv. Mater. 2011, 23, 4549–4553. [Google Scholar] [CrossRef] [Green Version]
- Piletska, E.V.; Abd, B.H.; Krakowiak, A.S.; Parmar, A.; Pink, D.L.; Wall, K.S.; Wharton, L.; Moczko, E.; Whitcombe, M.J.; Karim, K.; et al. Magnetic high throughput screening system for the development of nano-sized molecularly imprinted polymers for controlled delivery of curcumin. Analyst 2015, 140, 3113–3120. [Google Scholar] [CrossRef]
- Nematollahzadeh, A.; Sun, W.; Aureliano, C.S.A.; Lütkemeyer, D.; Stute, J.; Abdekhodaie, M.J.; Shojaei, A.; Sellergren, B. High-Capacity Hierarchically Imprinted Polymer Beads for Protein Recognition and Capture. Angew. Chem. 2011, 50, 495–498. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, Y.; Koide, H.; Urakami, T.; Kanazawa, H.; Kodama, T.; Oku, N.; Shea, K.J. Recognition, Neutralization, and Clearance of Target Peptides in the Bloodstream of Living Mice by Molecularly Imprinted Polymer Nanoparticles: A Plastic Antibody. J. Am. Chem. Soc. 2010, 132, 6644–6645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takeuchi, T.; Mukawa, T.; Shinmori, H. Signaling molecularly imprinted polymers: Molecular recognition-based sensing materials. Chem. Rec. 2010, 5, 263–275. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Zeng, S.; El-Aty, A.M.A.; Hacımüftüoğlu, A.; Yohannes, W.K.; Khan, M.; She, Y. Development of Water-Compatible Molecularly Imprinted Polymers Based on Functionalized β-Cyclodextrin for Controlled Release of Atropine. Polymers 2020, 12, 130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Song, R.; Xie, J.; Yu, X.; Ge, J.; Liu, M.; Guo, L. Preparation of Molecularly Imprinted Polymer Microspheres for Selective Solid-Phase Extraction of Capecitabine in Urine Samples. Polymers 2022, 14, 3968. [Google Scholar] [CrossRef] [PubMed]
- Zamruddin, N.M.; Herman, H.; Rijai, L.; Hasanah, A.N. Factors Affecting the Analytical Performance of Magnetic Molecularly Imprinted Polymers. Polymers 2022, 14, 3008. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Zhang, Y.; Han, L.; Liu, H.; Sun, B. Quantum confined peptide assemblies in a visual photoluminescent hydrogel platform and smartphone-assisted sample-to-answer analyzer for detecting trace pyrethroids. Biosens. Bioelectron. 2022, 210, 114265. [Google Scholar] [CrossRef]
- Das, R.; Bandyopadhyay, R.; Pramanik, P. Carbon quantum dots from natural resource: A review. Mater. Today Chem. 2018, 8, 96–109. [Google Scholar] [CrossRef]
- Mathew, S.; Thara, C.R.; John, N.; Mathew, B. Carbon dots from green sources as efficient sensor and as anticancer agent. J. Photochem. Photobiol. A Chem. 2023, 434, 114237. [Google Scholar] [CrossRef]
- Yu, C.; Xuan, T.; Chen, Y.; Zhao, Z.; Sun, Z.; Li, H. A facile, green synthesis of highly fluorescent carbon nanoparticles from oatmeal for cell imaging. J. Mater. Chem. C 2015, 3, 9514–9518. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhu, X.; Li, M.; Liu, H.; Sun, B. Temperature-Responsive Covalent Organic Framework-Encapsulated Carbon Dot-Based Sensing Platform for Pyrethroid Detection via Fluorescence Response and Smartphone Readout. J. Agric. Food Chem. 2022, 70, 6059–6071. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Xu, Y.; Liu, H.; Sun, B. Ultrahigh sensitivity nitrogen-doping carbon nanotubes-based metamaterial-free flexible terahertz sensing platform for insecticides detection. Food Chem. 2022, 394, 133467. [Google Scholar] [CrossRef] [PubMed]
- Espín, J.C.; Soler-Rivas, C.; Wichers, H.J. Characterization of the Total Free Radical Scavenger Capacity of Vegetable Oils and Oil Fractions Using 2,2-Diphenyl-1-picrylhydrazyl Radical. J. Agric. Food Chem. 2000, 48, 648–656. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.-L.; Huang, S.; Guo, H.-H.; Han, Y.-X.; Zheng, W.-S.; Jiang, J.-D. In situ delivery of thermosensitive gel-mediated 5-fluorouracil microemulsion for the treatment of colorectal cancer. Drug Des. Dev. Ther. 2016, 10, 2855–2867. [Google Scholar] [CrossRef] [Green Version]
- Portero-Otin, M.; Nagaraj, R.H.; Monnier, V.M. Chromatographic evidence for pyrraline formation during protein glycation in vitro and in vivo. Biochim. Biophys. Acta (BBA)-Protein Struct. Mol. Enzym. 1995, 1247, 74–80. [Google Scholar] [CrossRef]
- Gao, Y.; Xiao, Y.; Mao, K.; Qin, X.; Zhang, Y.; Li, D.; Zhang, Y.; Li, J.; Wan, H.; He, S. Thermoresponsive polymer-encapsulated hollow mesoporous silica nanoparticles and their application in insecticide delivery. Chem. Eng. J. 2020, 383, 123169. [Google Scholar] [CrossRef]
- Jain, A.; Jain, S.K. In vitro release kinetics model fitting of liposomes: An insight. Chem. Phys. Lipids 2016, 201, 28–40. [Google Scholar] [CrossRef]


| Sample | Temperature | Inhibitor | Detection (μg/L) | RSD | Inhibition Rate (%) |
|---|---|---|---|---|---|
| Simulation system | 60 °C | Blank | 115.3 ± 0.11 | 0.1 | 50.6 |
| FMIPs | 56.96 ± 0.06 | 0.11 | |||
| 80 °C | Blank | 115.3 ± 0.11 | 0.1 | 43.1 | |
| FMIPs | 69.61 ± 0.09 | 0.13 | |||
| Milk 1 | 60 °C | Blank | 4.62 ± 0.08 | 1.73 | 36.3 |
| FMIPs | 2.94 ± 0.09 | 3.06 | |||
| AG | 3.17 ± 0.01 | 0.31 | |||
| 80 °C | Blank | 4.49 ± 0.16 | 3.56 | 31.6 | |
| MIPs | 3.07 ± 0.02 | 0.65 | |||
| AG | 3.22 ± 0.03 | 0.93 | |||
| Milk 2 | 60 °C | Blank | 4.03 ± 0.02 | 0.49 | 44.91 |
| FMIPs | 2.22 ± 0.04 | 1.8 | |||
| AG | 2.55 ± 0.02 | 0.78 | |||
| 80 °C | Blank | 4.53 ± 0.06 | 1.32 | 32.9 | |
| FMIPs | 3.04 ± 0.10 | 3.29 | |||
| AG | 3.19 ± 0.03 | 0.94 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhu, P.; Zhang, Y.; Zhang, D.; Liu, H.; Sun, B. Fluorescent Molecularly Imprinted Polymers Loaded with Avenanthramides for Inhibition of Advanced Glycation End Products. Polymers 2023, 15, 538. https://doi.org/10.3390/polym15030538
Zhu P, Zhang Y, Zhang D, Liu H, Sun B. Fluorescent Molecularly Imprinted Polymers Loaded with Avenanthramides for Inhibition of Advanced Glycation End Products. Polymers. 2023; 15(3):538. https://doi.org/10.3390/polym15030538
Chicago/Turabian StyleZhu, Pei, Ying Zhang, Dianwei Zhang, Huilin Liu, and Baoguo Sun. 2023. "Fluorescent Molecularly Imprinted Polymers Loaded with Avenanthramides for Inhibition of Advanced Glycation End Products" Polymers 15, no. 3: 538. https://doi.org/10.3390/polym15030538







