Synthesis of Fluorescent, Small, Stable and Non-Toxic Epitope-Imprinted Polymer Nanoparticles in Water
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Synthesis of the Photoiniferter 2-(N,N-diethyldithiocarbamyl) Acetic Acid (DCAA)
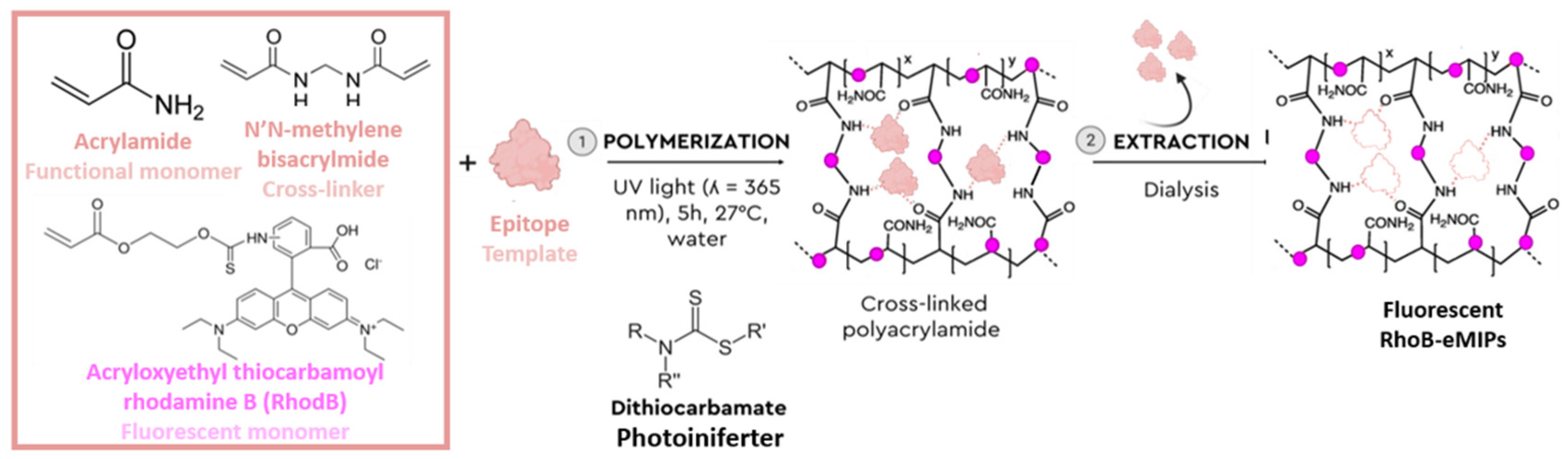
2.3. Synthesis of MIP and NIP
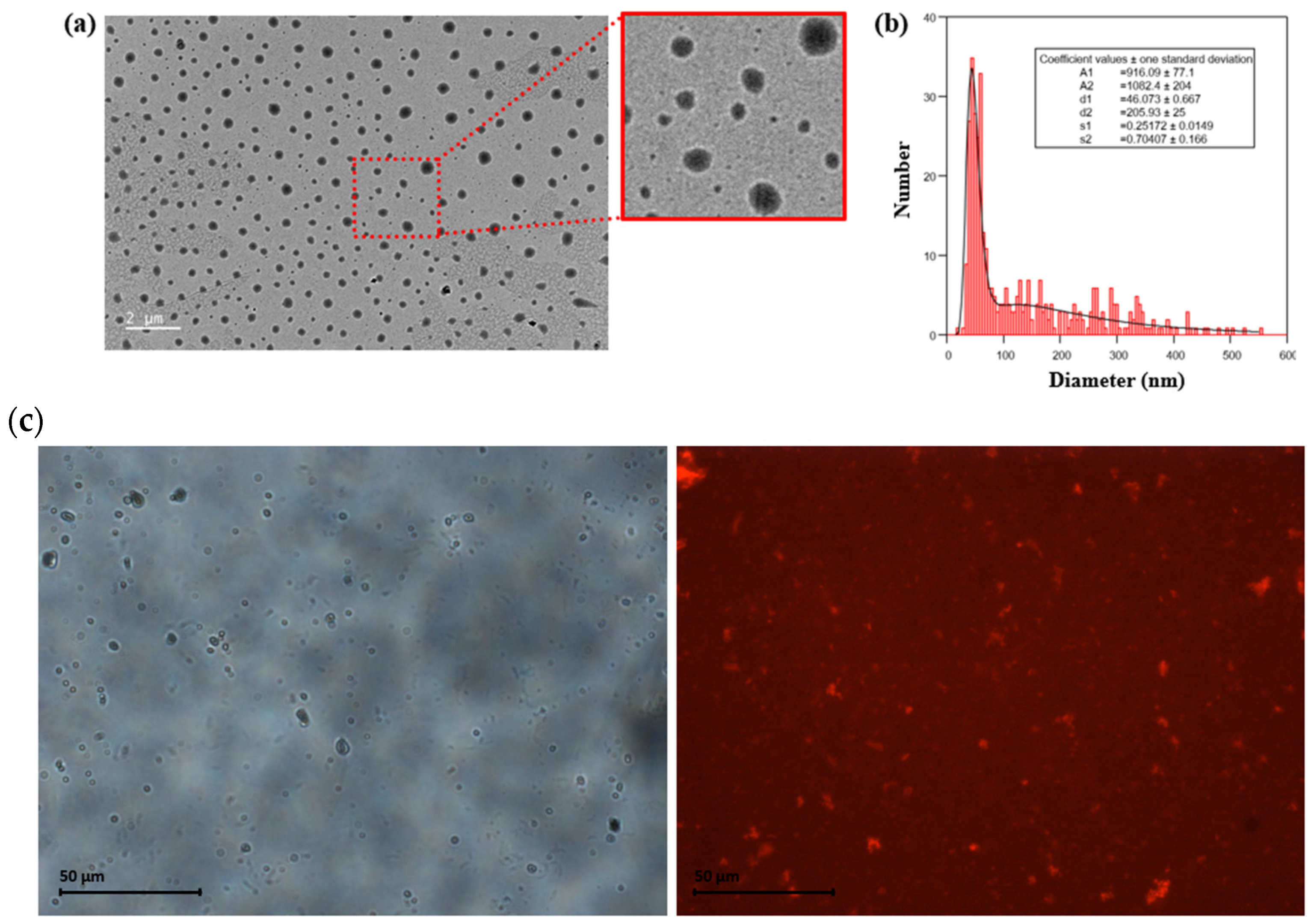
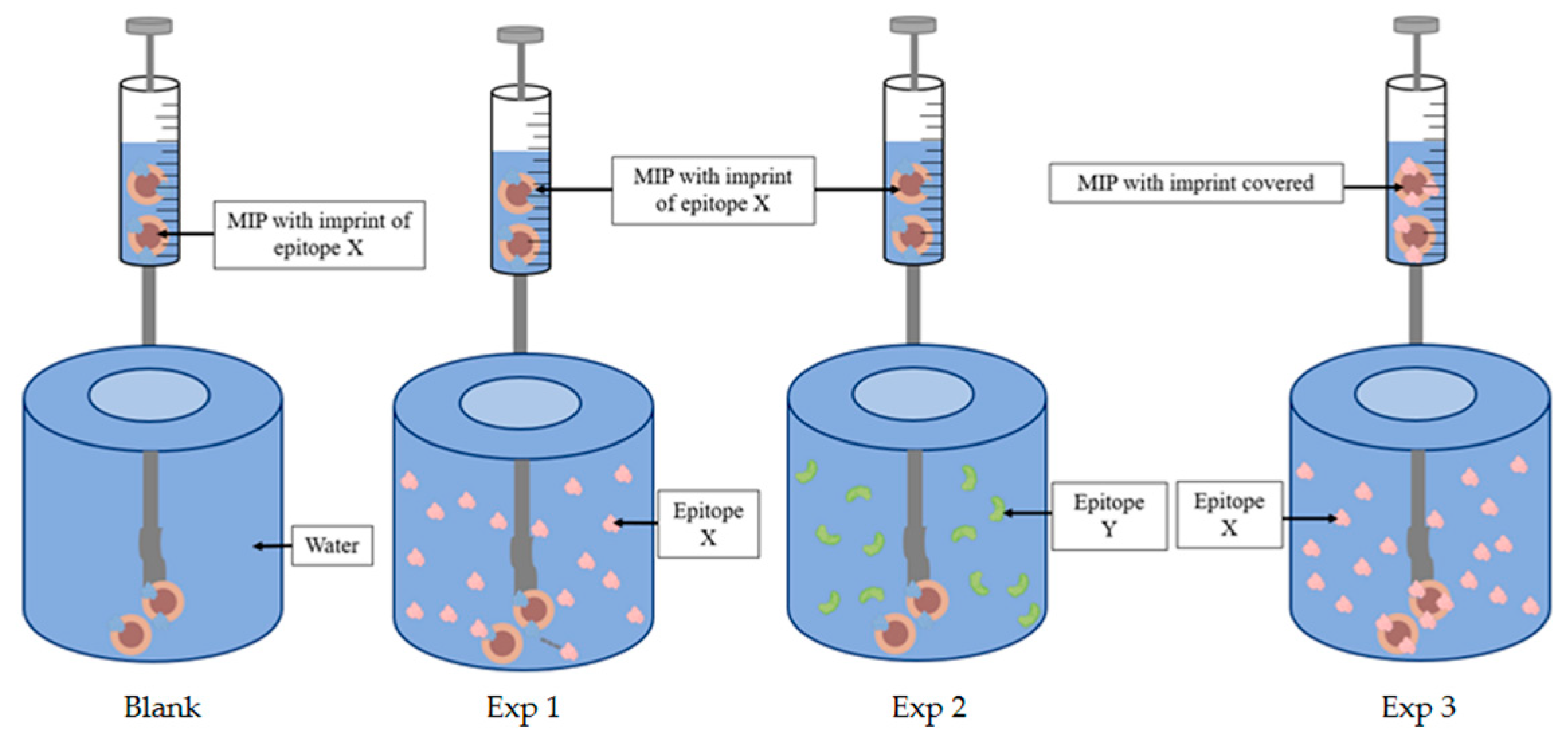
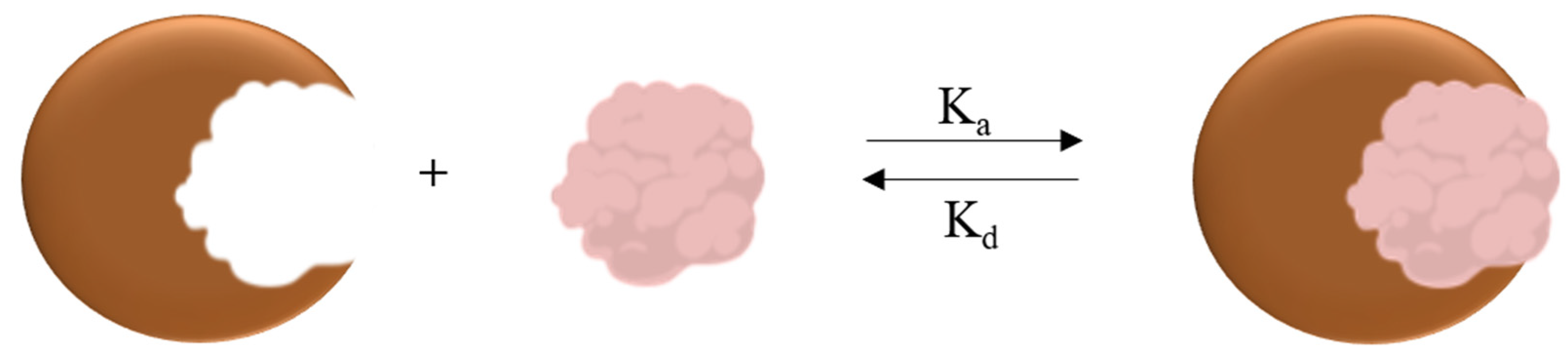
2.4. nanoITC, FTIR, DLS, TEM, Fluorescence
3. Results and Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Mi, Y.; Smith, C.C.; Yang, F.; Qi, Y.; Roche, K.C.; Serody, J.S.; Vincent, B.G.; Wang, A.Z. A Dual Immunotherapy Nanoparticle Improves T-Cell Activation and Cancer Immunotherapy. Adv. Mater. 2018, 30, 1706098. [Google Scholar] [CrossRef] [PubMed]
- Tan, K.X.; Danquah, M.K.; Pan, S.; Yon, L.S. Binding Characterization of Aptamer-Drug Layered Microformulations and In Vitro Release Assessment. J. Pharm. Sci. 2019, 108, 2934. [Google Scholar] [CrossRef] [PubMed]
- Meir, R.; Shamalov, K.; Sadan, T.; Motiei, M.; Yaari, G.; Cohen, C.J.; Popovtzer, R. Fast Image-Guided Stratification Using Anti-Programmed Death Ligand 1 Gold Nanoparticles for Cancer Immunotherapy. ACS Nano 2017, 11, 11127. [Google Scholar] [CrossRef] [PubMed]
- Haupt, K.; Medina Rangel, P.X.; Bui, B.T.S. Molecularly Imprinted Polymers: Antibody Mimics for Bioimaging and Therapy. Chem. Rev. 2020, 120, 9554. [Google Scholar] [CrossRef]
- BelBruno, J.J. Molecularly Imprinted Polymers. Chem. Rev. 2019, 1, 94. [Google Scholar] [CrossRef]
- Vasapollo, G.; Sole, R.D.; Mergola, L.; Lazzoi, M.R.; Scardino, A.; Scorrano, S.; Mele, G. Molecularly Imprinted Polymers: Present and Future Prospective. Int. J. Mol. Sci. 2011, 12, 5908. [Google Scholar] [CrossRef] [Green Version]
- Xu, S.; Wang, L.; Liu, Z. Molecularly Imprinted Polymer Nanoparticles: An Emerging Versatile Platform for Cancer Therapy. Angew. Chem. Int. Ed. 2021, 60, 3858. [Google Scholar] [CrossRef]
- Griffete, N.; Fresnais, J.; Espinosa, A.; Wilhelm, C.; Bée, A.; Ménager, C. Design of magnetic molecularly imprinted polymer nanoparticles for controlled release of doxorubicin under an alternative magnetic field in athermal conditions. Nanoscale 2015, 7, 18891. [Google Scholar] [CrossRef] [Green Version]
- Garnier, M.; Sabbah, M.; Ménager, C.; Griffete, N. Hybrid Molecularly Imprinted Polymers: The Future of Nanomedicine? Nanomaterials 2021, 11, 3091. [Google Scholar] [CrossRef]
- Haupt, K.; Mosbach, K. Molecularly imprinted polymers and their use in biomimetic sensors. Chem. Rev. 2000, 100, 2495–2504. [Google Scholar] [CrossRef]
- Ye, L.; Haupt, K. Molecularly imprinted polymers as antibody and receptor mimics for assays, sensors and drug discovery. Anal. Bioanal. Chem. 2004, 378, 1887. [Google Scholar] [CrossRef] [PubMed]
- Canfarotta, F.; Waters, A.; Sadler, R.; McGill, P.; Guerreiro, A.; Papkovsky, D.; Haupt, K.; Piletsky, S. Biocompatibility and internalization of molecularly imprinted nanoparticles. Nano Res. 2016, 9, 3463. [Google Scholar] [CrossRef]
- Cecchini, A.; Raffa, V.; Canfarotta, F.; Signore, G.; Piletsky, S.; MacDonald, M.P.; Cuschieri, A. In Vivo Recognition of Human Vascular Endothelial Growth Factor by Molecularly Imprinted Polymers. Nano Lett. 2017, 17, 2307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koide, H.; Yoshimatsu, K.; Hoshino, Y.; Lee, S.H.; Okajima, A.; Ariizumi, S.; Narita, Y.; Yonamine, Y.; Weisman, A.C.; Nishimura, Y.; et al. A polymer nanoparticle with engineered affinity for a vascular endothelial growth factor (VEGF165). Nat. Chem. 2017, 9, 715. [Google Scholar] [CrossRef]
- Canfarotta, F.; Lezina, L.; Guerreiro, A.; Czulak, J.; Petukhov, A.; Daks, A.; Smolinska-Kempisty, K.; Poma, A.; Piletsky, S.; Barlev, N.A. Specific Drug Delivery to Cancer Cells with Double-Imprinted Nanoparticles against Epidermal Growth Factor Receptor. Nano Lett. 2018, 18, 4641. [Google Scholar] [CrossRef] [PubMed]
- Canfarotta, F.; Poma, A.; Guerreiro, A.; Piletsky, S. Solid-phase synthesis of molecularly imprinted nanoparticles. Nat. Protoc. 2016, 11, 443. [Google Scholar] [CrossRef] [PubMed]
- Xu, J.; Ambrosini, S.; Tamahkar, E.; Rossi, C.; Haupt, K.; Tse Sum Bui, B. Toward a Universal Method for Preparing Molecularly Imprinted Polymer Nanoparticles with Antibody-like Affinity for Proteins. Biomacromolecules 2016, 11, 345. [Google Scholar] [CrossRef]
- Ambrosini, S.; Beyazit, S.; Haupt, K.; Tse Sum Bui, B. Solid-phase synthesis of molecularly imprinted nanoparticles for protein recognition. Chem. Commun. 2013, 49, 6746. [Google Scholar] [CrossRef]
- Cáceres, C.; Moczko, E.; Basozabal, I.; Guerreiro, A.; Piletsky, S. Molecularly Imprinted Nanoparticles (NanoMIPs) Selective for Proteins: Optimization of a Protocol for Solid-Phase Synthesis Using Automatic Chemical Reactor. Polymers 2021, 13, 314. [Google Scholar] [CrossRef]
- Sadat, S.M.A.; Jahan, S.T.; Azita, H. Effects of Size and Surface Charge of Polymeric Nanoparticles on in Vitro and in Vivo Applications. J. Biomater. Nanobiotechnol. 2016, 7, 91. [Google Scholar] [CrossRef] [Green Version]
- Weber, A.; Dettling, M.; Brunner, H.; Tovar, G. Isothermal Titration Calorimetry of Molecularly Imprinted Polymer Nanospheres. Macromol. Rapid Commun. 2002, 23, 824. [Google Scholar] [CrossRef]
- Huang, R.; Lau, B.L.T. Biomolecule–nanoparticle interactions: Elucidation of the thermodynamics by isothermal titration calorimetry. Biochim. Biophys. Acta Gen. Subj. 2016, 1860, 945. [Google Scholar] [CrossRef]
- Sunayama, H.; Ooya, T.; Takeuchi, T. Fluorescent protein recognition polymer thin films capable of selective signal transduction of target binding events prepared by molecular imprinting with a post-imprinting treatment. Biosens. Bioelectron. 2010, 26, 458–462. [Google Scholar] [CrossRef]




| Sequence | Peptide Structure |
|---|---|
| YRVRFLAKENVTQDAEDNC Labelled X | 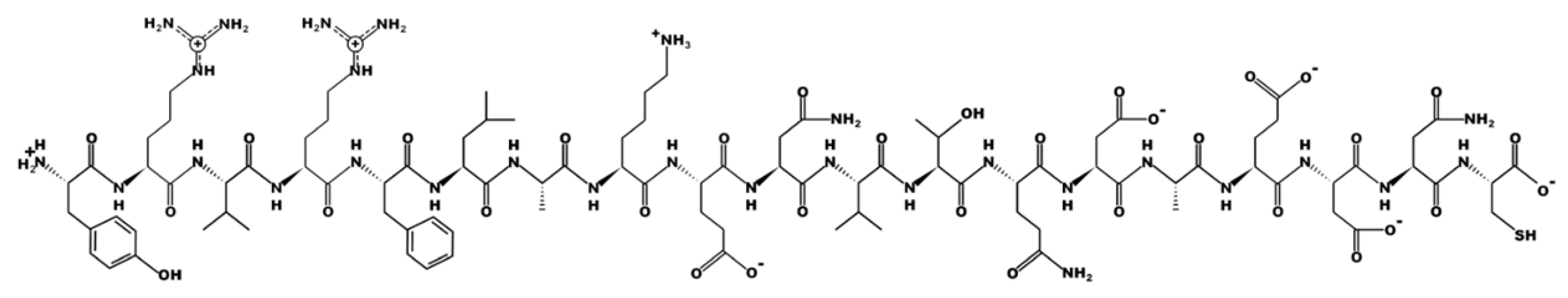 |
| CNLAVAAASHIYQNQFVQ Labelled Y | 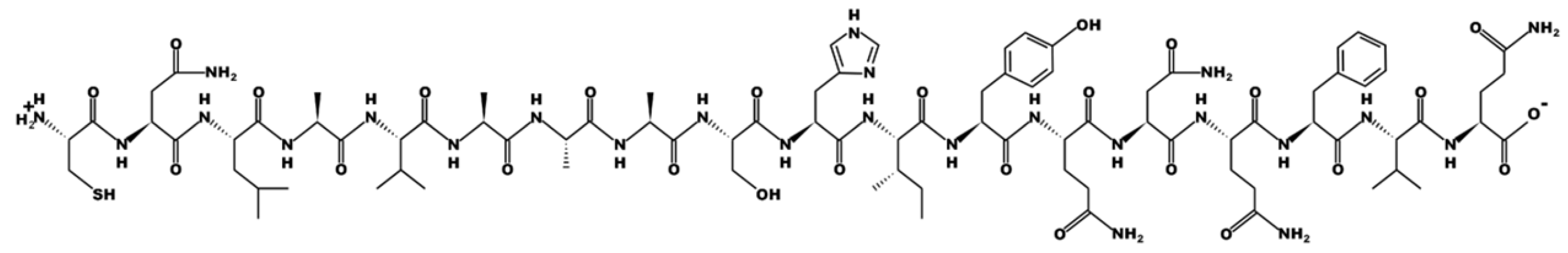 |
| CQMILNSLINKSKSSMFQV Labelled Z | 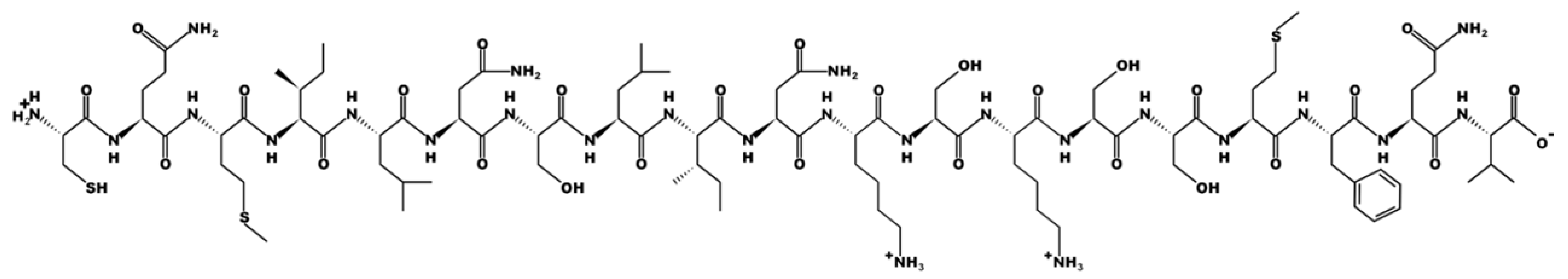 |
| Template | ITC Probe | ΔH (kJ/mol) | ΔG (kJ/mol) | KD | n |
|---|---|---|---|---|---|
| X | X | −26.63 ± 0.036 | −35.51 | 6.003 × 10−7 ± 5.8 × | 1 ± 0.043 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Benghouzi, P.; Louadj, L.; Pagani, A.; Garnier, M.; Fresnais, J.; Gonzato, C.; Sabbah, M.; Griffete, N. Synthesis of Fluorescent, Small, Stable and Non-Toxic Epitope-Imprinted Polymer Nanoparticles in Water. Polymers 2023, 15, 1112. https://doi.org/10.3390/polym15051112
Benghouzi P, Louadj L, Pagani A, Garnier M, Fresnais J, Gonzato C, Sabbah M, Griffete N. Synthesis of Fluorescent, Small, Stable and Non-Toxic Epitope-Imprinted Polymer Nanoparticles in Water. Polymers. 2023; 15(5):1112. https://doi.org/10.3390/polym15051112
Chicago/Turabian StyleBenghouzi, Perla, Lila Louadj, Aurélia Pagani, Maylis Garnier, Jérôme Fresnais, Carlo Gonzato, Michèle Sabbah, and Nébéwia Griffete. 2023. "Synthesis of Fluorescent, Small, Stable and Non-Toxic Epitope-Imprinted Polymer Nanoparticles in Water" Polymers 15, no. 5: 1112. https://doi.org/10.3390/polym15051112








