High Resolution Melting and Insertion Site-Based Polymorphism Markers for Wheat Variability Analysis and Candidate Genes Selection at Drought and Heat MQTL Loci
Abstract
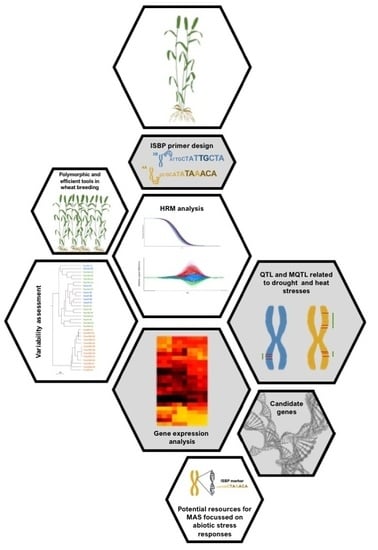
:1. Introduction
2. Material and methods
2.1. Plant Material and DNA Isolation
2.2. Insertion Site-Based Polymorphism Markers Development
2.3. Candidate Genes and Gene Expression Analyses
2.4. Wheat Variability Assessment
3. Results
3.1. Markers Sequence Validation and HRM Pattern Types Assignment
3.2. Candidate Gene Analysis
3.3. Wheat Variability Assessment by High Resolution Melting Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lawlor, D.W. Wheat and Wheat Improvement. Soil Sci. 1988, 146, 292–293. [Google Scholar] [CrossRef]
- Rampino, P.; Pataleo, S.; Gerardi, C.; Mita, G.; Perrotta, C. Drought stress response in wheat: Physiological and molecular analysis of resistant and sensitive genotypes. Plant Cell Environ. 2006, 29, 2143–2152. [Google Scholar] [CrossRef] [PubMed]
- Nezhadahmadi, A.; Prodhan, Z.H.; Faruq, G. Drought tolerance in wheat. Sci. World J. 2013, 2013, 610721. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Farooq, M.; Hussain, M.; Siddique, K.H.M. Drought stress in wheat during flowering and grain-filling periods. Crit. Rev. Plant Sci. 2014, 33, 331–349. [Google Scholar] [CrossRef]
- Sallam, A.; Alqudah, A.M.; Dawood, M.F.A.; Baenziger, P.S.; Börner, A. Drought stress tolerance in wheat and barley: Advances in physiology, breeding and genetics research. Int. J. Mol. Sci. 2019, 20, 3137. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Vinocur, B.; Altman, A. Plant responses to drought, salinity and extreme temperatures: Towards genetic engineering for stress tolerance. Planta 2003, 218, 1–14. [Google Scholar] [CrossRef]
- Wang, W.; Vinocur, B.; Shoseyov, O.; Altman, A. Role of plant heat-shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci. 2004, 9, 244–252. [Google Scholar] [CrossRef]
- Gooding, M.J.; Ellis, R.H.; Shewry, P.R.; Schofield, J.D. Effects of restricted water availability and increased temperature on the grain filling, drying and quality of winter wheat. J. Cereal Sci. 2003, 37, 295–309. [Google Scholar] [CrossRef]
- IPCC Assessment Report. Published Online. 2020. Available online: https//www.ipccch/srccl/ (accessed on 3 March 2020).
- Lesk, C.; Rowhani, P.; Ramankutty, N. Influence of extreme weather disasters on global crop production. Nature 2016, 529, 84–87. [Google Scholar] [CrossRef]
- Fathi, A.; Tari, D.B. Effect of drought stress and its mechanism in plants. Int. J. Life Sci. 2016, 10, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Gupta, A.; Balyan, H.; Gahlaut, V. QTL analysis for drought tolerance in wheat: Present status and future possibilities. Agronomy 2017, 7, 5. [Google Scholar] [CrossRef]
- Reynolds, M.P.; Ortiz, R. Adapting crops to climate change: A summary. In Climate Change and Crop Production; CABI: Oxfordshire, UK, 2010; pp. 1–8. [Google Scholar] [CrossRef]
- Kantar, M.; Lucas, S.J.; Budak, H. Drought Stress: Molecular Genetics and Genomics Approaches, 1st ed.; Academic Press: Cambridge, MA, USA, 2011; Volume 57, ISBN 9780123876928. [Google Scholar]
- Li, J.; Zhou, R.; Endo, T.R.; Stein, N. High-throughput development of SSR marker candidates and their chromosomal assignment in rye (Secale cereale L.). Plant Breed. 2018, 137, 561–572. [Google Scholar] [CrossRef] [Green Version]
- Farooq, M.; Bramley, H.; Palta, J.A.; Siddique, K.H.M. Heat stress in wheat during reproductive and grain-filling phases. Crit. Rev. Plant Sci. 2011, 30, 491–507. [Google Scholar] [CrossRef]
- Cattivelli, L.; Rizza, F.; Badeck, F.W.; Mazzucotelli, E.; Mastrangelo, A.M.; Francia, E.; Marè, C.; Tondelli, A.; Stanca, A.M. Drought tolerance improvement in crop plants: An integrated view from breeding to genomics. Field Crop. Res. 2008, 105, 1–14. [Google Scholar] [CrossRef]
- McWilliam, J.R. The Dimensions of Drought. In Drought Resistant Cereal; Bak, F., Ed.; CAB Int.: Wallingford, UK, 1989; pp. 1–11. [Google Scholar]
- Blum, A. Drought resistance is it really a complex trait? Funct. Plant Biol. 2011, 38, 753–757. [Google Scholar] [CrossRef]
- Ingram, J.; Bartels, D. The molecular basis of dehydration tolerance in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996, 47, 377–403. [Google Scholar] [CrossRef] [Green Version]
- Chaves, M.M.; Maroco, J.P.; Pereira, J.S. Understanding plant responses to drought—From genes to the whole plant. Funct. Plant Biol. 2003, 30, 239–264. [Google Scholar] [CrossRef]
- Liu, Z.; Xin, M.; Qin, J.; Peng, H.; Ni, Z.; Yao, Y.; Sun, Q. Temporal transcriptome profiling reveals expression partitioning of homeologous genes contributing to heat and drought acclimation in wheat (Triticum aestivum L.). BMC Plant Biol. 2015, 15, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Li, M.; Wei, X.; Zhang, X.; Xue, R.; Zhao, Y.; Zhao, H. Transcriptome analysis of drought-responsive genes regulated by hydrogen sulfide in wheat (Triticum aestivum L.) leaves. Mol. Genet. Genom. 2017, 292, 1091–1110. [Google Scholar] [CrossRef]
- Chaichi, M.; Sanjarian, F.; Razavi, K.; Gonzalez-Hernandez, J.L. Analysis of transcriptional responses in root tissue of bread wheat landrace (Triticum aestivum L.) reveals drought avoidance mechanisms under water scarcity. PLoS ONE 2019, 14, e0212671. [Google Scholar] [CrossRef]
- Iquebal, M.A.; Sharma, P.; Jasrotia, R.S.; Jaiswal, S.; Kaur, A.; Saroha, M.; Angadi, U.B.; Sheoran, S.; Singh, R.; Singh, G.P.; et al. RNAseq analysis reveals drought-responsive molecular pathways with candidate genes and putative molecular markers in root tissue of wheat. Sci. Rep. 2019, 9, 1–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, J.; Li, R.; Wang, H.; Li, D.; Wang, X.; Zhang, Y.; Zhen, W.; Duan, H.; Yan, G.; Li, Y. Transcriptomics analyses reveal wheat responses to drought stress during reproductive stages under field conditions. Front. Plant Sci. 2017, 8, 592. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gálvez, S.; Mérida-García, R.; Camino, C.; Borrill, P.; Abrouk, M.; Ramírez-González, R.H.; Biyiklioglu, S.; Amil-Ruiz, F.; Dorado, G.; Budak, H.; et al. Hotspots in the genomic architecture of field drought responses in wheat as breeding targets. Funct. Integr. Genom. 2019, 19, 295–309. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wahid, A.; Gelani, S.; Ashraf, M.; Foolad, M.R. Heat tolerance in plants: An overview. Environ. Exp. Bot. 2007, 61, 199–223. [Google Scholar] [CrossRef]
- Schöffl, F.; Prandl, R.; Reindl, A. Molecular Responses to Heat Stress. In Molecular Responses to Cold, Drought, Heat Salt Stress in Higher Plants; Shinozaki, K., Yamaguchi-Shinozaki, K., Eds.; R.G. Landes Co.: Austin, TX, USA, 1999; pp. 81–98. [Google Scholar]
- Paux, E.; Faure, S.; Choulet, F.; Roger, D.; Gauthier, V.; Martinant, J.P.; Sourdille, P.; Balfourier, F.; Le Paslier, M.C.; Chauveau, A.; et al. Insertion site-based polymorphism markers open new perspectives for genome saturation and marker-assisted selection in wheat. Plant Biotechnol. J. 2010, 8, 196–210. [Google Scholar] [CrossRef]
- Mourad, A.M.I.; Alomari, D.Z.; Alqudah, A.M.; Sallam, A.; Salem, K.F.M. Recent Advances in Wheat (Triticum spp.) Breeding; Springer: Cham, Switzerland, 2019; Volume 5, ISBN 9783030231088. [Google Scholar]
- Landjeva, S.; Korzun, V.; Börner, A. Molecular markers: Actual and potential contributions to wheat genome characterization and breeding. Euphytica 2007, 156, 271–296. [Google Scholar] [CrossRef]
- IWGSC. Shifting the limits in wheat research and breeding using a fully annotated reference genome. Science 2018, 361, eaar7191. [Google Scholar] [CrossRef] [Green Version]
- Paux, E.; Roger, D.; Badaeva, E.; Gay, G.; Bernard, M.; Sourdille, P.; Feuillet, C. Characterizing the composition and evolution of homoeologous genomes in hexaploid wheat through BAC-end sequencing on chromosome 3B. Plant J. 2006, 48, 463–474. [Google Scholar] [CrossRef]
- Paux, E.; Gao, L.; Faure, S.; Choulet, F.; Roger, D.; Chevalier, K.; Saintenac, C.; Balfourier, F.; Paux, K.; Cakir, M.; et al. Insertion site-based polymorphism: A Swiss army knife for wheat genomics. In Proceedings of the 11th International Wheat Genetics Symposium, Brisbane, QLT, Australia, 24–19 August 2008; pp. 4–6. [Google Scholar]
- Li, G.; Fang, T.; Zhang, H.; Xie, C.; Li, H.; Yang, T.; Nevo, E.; Fahima, T.; Sun, Q.; Liu, Z. Molecular identification of a new powdery mildew resistance gene Pm41 on chromosome 3BL derived from wild emmer (Triticum turgidum var. dicoccoides). Theor. Appl. Genet. 2009, 119, 531–539. [Google Scholar] [CrossRef]
- Cubizolles, N.; Rey, E.; Choulet, F.; Rimbert, H.; Laugier, C.; Balfourier, F.; Bordes, J.; Poncet, C.; Jack, P.; James, C.; et al. Exploiting the Repetitive Fraction of the Wheat Genome for High-Throughput Single-Nucleotide Polymorphism Discovery and Genotyping. Plant Genom. 2016, 9, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Rimbert, H.; Darrier, B.; Navarro, J.; Kitt, J.; Choulet, F.; Leveugle, M.; Duarte, J.; Rivière, N.; Eversole, K.; Le Gouis, J.; et al. High throughput SNP discovery and genotyping in hexaploid wheat. PLoS ONE 2018, 13, e0186329. [Google Scholar] [CrossRef] [Green Version]
- Oliveira, H.R.; Campana, M.G.; Jones, H.; Hunt, H.V.; Leigh, F.; Redhouse, D.I.; Lister, D.L.; Jones, M.K. Tetraploid wheat landraces in the Mediterranean basin: Taxonomy, evolution and genetic diversity. PLoS ONE 2012, 7, e37063. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lucas, S.J.; Budak, H. Sorting the wheat from the chaff: Identifying miRNAs in genomic survey sequences of Triticum aestivum chromosome 1AL. PLoS ONE 2012, 7, e40859. [Google Scholar] [CrossRef] [PubMed]
- Sehgal, S.K.; Li, W.; Rabinowicz, P.D.; Chan, A.; Šimková, H.; Doležel, J.; Gill, B.S. Chromosome arm-specific BAC end sequences permit comparative analysis of homoeologous chromosomes and genomes of polyploid wheat. BMC Plant Biol. 2012, 12, 64. [Google Scholar] [CrossRef] [Green Version]
- Vaissayre, L.; Ardisson, M.; Borries, C.; Santoni, S.; David, J.; Roumet, P. Elite durum wheat genetic map and recombination rate variation in a multiparental connected design. Euphytica 2012, 185, 61–75. [Google Scholar] [CrossRef]
- Salina, E.A.; Nesterov, M.A.; Frenkel, Z.; Kiseleva, A.A.; Timonova, E.M.; Magni, F.; Vrána, J.; Šafár, J.; Šimková, H.; Doležel, J.; et al. Features of the organization of bread wheat chromosome 5BS based on physical mapping. BMC Genom. 2018, 19, 129–141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McNeil, M.D.; Kota, R.; Paux, E.; Dunn, D.; McLean, R.; Feuillet, C.; Li, D.; Kong, X.; Lagudah, E.; Zhang, J.C.; et al. BAC-derived markers for assaying the stem rust resistance gene, Sr2, in wheat breeding programs. Mol. Breed. 2008, 22, 15–24. [Google Scholar] [CrossRef]
- Vossen, R.H.A.M.; Aten, E.; Roos, A.; Den Dunnen, J.T. High-resolution melting analysis (HRMA)—More than just sequence variant screening. Hum. Mutat. 2009, 30, 860–866. [Google Scholar] [CrossRef]
- Montgomery, J.L.; Sanford, L.N.; Wittwer, C.T. High-resolution DNA melting analysis in clinical research and diagnostics. Expert Rev. Mol. Diagn. 2010, 10, 219–240. [Google Scholar] [CrossRef]
- Arias Aguirre, A.; Studer, B.; Do Canto, J.; Frei, U.; Lübberstedt, T.; Rognli, O.A. Validation of two models for self-incompatibility in autotetraploid perennial ryegrass using high resolution melting-based markers. Plant Breed. 2014, 133, 765–770. [Google Scholar] [CrossRef]
- Reed, G.H.; Wittwer, C.T. Sensitivity and specificity of single-nucleotide polymorphism scanning by high-resolution melting analysis. Clin. Chem. 2004, 50, 1748–1754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shatalina, M.; Messmer, M.; Feuillet, C.; Mascher, F.; Paux, E.; Choulet, F.; Wicker, T.; Keller, B. High-resolution analysis of a QTL for resistance to Stagonospora nodorum glume blotch in wheat reveals presence of two distinct resistance loci in the target interval. Theor. Appl. Genet. 2014, 127, 573–586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mondini, L.; Nachit, M.M.; Porceddu, E.; Pagnotta, M.A. HRM technology for the identification and characterization of INDEL and SNPs mutations in genes involved in drought and salt tolerance of durum wheat. Plant Genet. Resour. Charact. Util. 2011, 9, 166–169. [Google Scholar] [CrossRef] [Green Version]
- Iehisa, J.C.M.; Shimizu, A.; Sato, K.; Nasuda, S.; Takumi, S. Discovery of high-confidence single nucleotide polymorphisms from large-scale de novo analysis of leaf transcripts of aegilops tauschii, a wild wheat progenitor. DNA Res. 2012, 19, 487–497. [Google Scholar] [CrossRef] [Green Version]
- Matsuda, R.; Iehisa, J.C.M.; Takumi, S. Application of real-time PCR-based SNP detection for mapping of Net2, a causal D-genome gene for hybrid necrosis in interspecific crosses between tetraploidwheat and Aegilops tauschii. Genes Genet. Syst. 2012, 87, 137–143. [Google Scholar] [CrossRef] [Green Version]
- Dong, C.; Vincent, K.; Sharp, P. Simultaneous mutation detection of three homoeologous genes in wheat by high resolution melting analysis and mutation Surveyor®. BMC Plant. Biol. 2009, 9, 143. [Google Scholar] [CrossRef] [Green Version]
- Botticella, E.; Sestili, F.; Hernandez-Lopez, A.; Phillips, A.; Lafiandra, D. High resolution melting analysis for the detection of EMS induced mutations in wheat SbeIIa genes. BMC Plant Biol. 2011, 11, 156. [Google Scholar] [CrossRef] [Green Version]
- Mondini, L.; Nachit, M.; Porceddu, E.; Pagnotta, M.A. Identification of SNP mutations in DREB1, HKT1, and WRKY1 genes involved in drought and salt stress tolerance in durum wheat (Triticum turgidum L. var durum). Omics J. Integr. Biol. 2012, 16, 178–187. [Google Scholar] [CrossRef]
- Sestili, F.; Palombieri, S.; Botticella, E.; Mantovani, P.; Bovina, R.; Lafiandra, D. TILLING mutants of durum wheat result in a high amylose phenotype and provide information on alternative splicing mechanisms. Plant Sci. 2015, 233, 127–133. [Google Scholar] [CrossRef]
- Lehmensiek, A.; Sutherland, M.W.; McNamara, R.B. The use of high resolution melting (HRM) to map single nucleotide polymorphism markers linked to a covered smut resistance gene in barley. Theor. Appl. Genet. 2008, 117, 721–728. [Google Scholar] [CrossRef] [Green Version]
- Terracciano, I.; Maccaferri, M.; Bassi, F.; Mantovani, P.; Sanguineti, M.C.; Salvi, S.; Šimková, H.; Doležel, J.; Massi, A.; Ammar, K.; et al. Development of COS-SNP and HRM markers for high-throughput and reliable haplotype-based detection of Lr14a in durum wheat (Triticum durum Desf.). Theor. Appl. Genet. 2013, 126, 1077–1101. [Google Scholar] [CrossRef] [PubMed]
- Curvers, K.; Pycke, B.; Kyndt, T.; Vanrompay, D.; Haesaert, G.; Gheysen, G. A high-resolution melt (HRM) assay to characterize CYP51 haplotypes of the wheat pathogen Mycosphaerella graminicola. Crop. Prot. 2015, 71, 12–18. [Google Scholar] [CrossRef]
- Li, F.; Wen, W.; Zhang, Y.; Cao, S.; He, Z. Genetic architecture of grain yield in bread wheat based on genome-wide association studies. BMC Plant Biol. 2019, 19, 168. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, P.; Martis, M.; Dorado, G.; Pfeifer, M.; Gálvez, S.; Schaaf, S.; Jouve, N.; Šimková, H.; Valárik, M.; Doležel, J.; et al. Next-generation sequencing and syntenic integration of flow-sorted arms of wheat chromosome 4A exposes the chromosome structure and gene content. Plant J. 2012, 69, 377–386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Araki, E.; Miura, H.; Sawada, S. Identification fo genetic loci affecting amylose content and agronomic traits on chromosome 4A of wheat. Theor. Appl. Genet. 1999, 98, 977–984. [Google Scholar] [CrossRef]
- Borner, A.; Schumann, E.; Furste, A.; Coster, H.; Leithold, B.; Roder, S. Mapping of quantitative trait loci determining agronomic important characters in hexaploid wheat (Triticum aestivum L.). Theor. Appl. Genet. 2002, 105, 921–936. [Google Scholar] [CrossRef]
- McCartney, C.A.; Somers, D.J.; Humphreys, D.G.; Lukow, O.; Ames, N.; Noll, J. Mapping quantitative trait loci controlling agronomic traits in the spring wheat cross RL445x’AC Domain’. Genome 2005, 48, 870–883. [Google Scholar] [CrossRef]
- McDonald, G.K.; Gene, Y.; Nurzhanuly, B.; Trethowan, R.; Reynolds, M.; Yaqub Mujahid, M. Quantifying the value to grain yield of QTL for adaptation and tolerance to abiotic stress in bread wheat. In Proceedings of the 11th International Wheat Genetics Symposium, Brisbane Convention Exhibition Centre, Brisbane, QLD, Australia, 24–29 August 2008. [Google Scholar]
- Liu, L.; Wang, L.; Yao, J.; Zheng, Y.; Zhao, C. Association mapping of six agronomic traits on chromosome 4A of wheat (Triticum aestivum L.). Mol. Plant Breed. 2010, 1. [Google Scholar] [CrossRef]
- Jakobson, I.; Peusha, H.; Timofejeva, L.; Järve, K. Adult plant and seedling resistance to powdery mildew in a Triticum aesivum x Triticum miitinae hybrid line. Theor. Appl. Genet. 2006, 112, 760–766. [Google Scholar] [CrossRef]
- Kirigwi, F.M.; Van Ginkel, M.; Brown-Guedira, G.; Gill, B.S.; Paulsen, G.M.; Fritz, A.K. Markers associated with a QTL for grain yield in wheat under drought. Mol. Breed. 2007, 20, 401–413. [Google Scholar] [CrossRef]
- Rana, R.M.; Rehman, S.U.; Ahmed, J.; Bilal, M. A comprehensive overview of recent advances in drought stress tolerance research in wheat (Triticum aestivum L.). Asian J. Agric. Biol. 2013, 1, 29–37. [Google Scholar]
- Shorinola, O.; Bird, N.; Simmonds, J.; Berry, S.; Henriksson, T.; Jack, P. The wheat Phs-A1 pre-harvest sprouting resistance locus delays the rate of seed dormancy loss and maps 0.3cM to the PM19 genes in UK germplasm. J. Exp. Bot. 2016, 67, 4169–4178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Acuña-Galindo, M.A.; Mason, R.E.; Subramanian, N.K.; Hays, D.B. Meta-analysis of wheat QTL regions associated with adaptation to drought and heat stress. Crop. Sci. 2015, 55, 477–492. [Google Scholar] [CrossRef]
- Zhang, J.; Zhang, S.; Cheng, M.; Jiang, H.; Zhang, X.; Peng, C.; Lu, X.; Zhang, M.; Jin, J. Effect of drought on agronomic traits of rice and wheat: A meta-analysis. Int. J. Environ. Res. Public Health 2018, 15, 839. [Google Scholar] [CrossRef] [Green Version]
- Murray, M.; Thompson, W. Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 1980, 8, 4321–4325. [Google Scholar] [CrossRef] [Green Version]
- Hernández, P.; De la Rosa, R.; Rallo, L.; Martín, A.; Dorado, G. First evidence of a retrotransposon-like element in olive (Olea europaea): Implications in plant variety identification by SCAR-marker development. Theor. Appl. Genet. 2001, 102, 1082–1087. [Google Scholar] [CrossRef]
- Gaujoux, R.; Seoighe, C. A flexible R package for nonnegative matrix factorization. BMC Inform. 2010, 11, 367. [Google Scholar] [CrossRef] [Green Version]
- Liu, K.; Muse, S.V. PowerMaker: An integrated analysis environment for genetic maker analysis. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef] [Green Version]
- Nei, M. Genetic distance between populations. Am. Nat. 1972, 106, 283–292. [Google Scholar] [CrossRef]
- Felsenstein, J. Phylogeny Inference Package; Version 3.6; Department of Genetics, University of Washington: Seattle, WA, USA, 2005. [Google Scholar]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [Green Version]
- Dighe, A.S.; Jangid, K.; González, J.M.; Pidiyar, V.J.; Patole, M.S.; Ranade, D.R.; Shouche, Y.S. Comparison of 16S rRNA gene sequences of genus Methanobrevibacter. BMC Microbiol. 2004, 4, 20. [Google Scholar] [CrossRef] [Green Version]
- Kumar, A.; Hirochika, H. Applications of retrotransposons as genetic tools in plant biology. Trends Plant Sci. 2001, 6, 127–134. [Google Scholar] [CrossRef]
- Schulman, A.H.; Flavell, A.J.; Paux, E.; Noel Ellis, T.H. The Application of LTR Retrotransposons as Molecular Markers in Plants. Methods Mol. Biol. 2012, 859, 115–153. [Google Scholar] [CrossRef] [PubMed]
- Barabaschi, D.; Orru, L.; Lacrima, K.; Faccioli, P.; Colaiacovo, M.; Bagnaresi, P. On the road to a high density genetic linkage map of wheat chromosome 5A. J. Med. Plants Res. 2010, 150, 473. [Google Scholar] [CrossRef]
- Lucas, S.J.; Šimková, H.; Šafář, J.; Jurman, I.; Cattonaro, F.; Vautrin, S.; Bellec, A.; Berges, H.; Doležel, J.; Budak, H. Functional features of a single chromosome arm in wheat (1AL) determined from its structure. Funct. Integr. Genom. 2012, 12, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Druml, B.; Cichna-Markl, M. High resolution melting (HRM) analysis of DNA—Its role and potential in food analysis. Food Chem. 2014, 158, 245–254. [Google Scholar] [CrossRef]
- Hoffmann, M.; Hurlebaus, J.; Weilke, C. High-resolution melting curve analysis on the LightCycler ® 480 PCR system. Nat. Methods Suppl. 2007, S, AN17–AN18. [Google Scholar]
- Wittwer, C.T. High-resolution DNA melting analysis: Advancements and limitations. Hum. Mutat. 2009, 30, 857–859. [Google Scholar] [CrossRef]
- Distefano, G.; La Malfa, S.; Gentile, A.; Wu, S.B. EST-SNP genotyping of citrus species using high-resolution melting curve analysis. Tree Genet. Genom. 2013, 9, 1271–1281. [Google Scholar] [CrossRef]
- Liew, M.; Pryor, R.; Palais, R.; Meadows, C.; Erali, M.; Lyon, E.; Wittwer, C. Genotyping of single-nucleotide polymorphisms by high-resolution melting of small amplicons. Clin. Chem. 2004, 50, 1156–1164. [Google Scholar] [CrossRef]
- Erali, M.; Palais, R.; Wittwer, C. SNP Genotyping by Unlabeled Probe Melting Analysis. In Molecular Beacons: Signalling Nucleic Acid Probes, Methods, and Protocols; Marx, A., Seiz, O., Eds.; Humana Press: Totowa, NJ, USA, 2008; p. 429. [Google Scholar] [CrossRef]
- Tindall, E.A.; Petersen, D.C.; Woodbridge, P.; Schipany, K.; Hayes, V.M. Assessing high-resolution melt curve analysis for accurate detection of gene variants in complex DNA fragments—Tindall—2009—Human Mutation—Wiley Online Library. Hum. Mutat. 2009, 30, 876–883. [Google Scholar] [CrossRef] [PubMed]
- Reed, G.H.; Kent, J.O.; Wittwer, C.T. High-resolution DNA melting analysis for simple and efficient molecular diagnostics. Pharmacogenomics 2007, 8, 597–608. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ganopoulos, I.; Argiriou, A.; Tsaftaris, A. Microsatellite high resolution melting (SSR-HRM) analysis for authenticity testing of protected designation of origin (PDO) sweet cherry products. Food Control 2011, 22, 532–541. [Google Scholar] [CrossRef]
- Ganopoulos, I.; Madesis, P.; Zambounis, A.; Tsaftaris, A. High-resolution melting analysis allowed fast and accurate closed-tube genotyping of Fusarium oxysporum formae speciales complex. FEMS Microbiol. Lett. 2012, 334, 16–21. [Google Scholar] [CrossRef] [PubMed]
- Ganopoulos, I.; Xanthopoulou, A.; Mastrogianni, A.; Drouzas, A.; Kalivas, A.; Bletsos, F.; Krommydas, S.K.; Ralli, P.; Tsaftaris, A.; Madesis, P. High Resolution Melting (HRM) analysis in eggplant (Solanum melongena L.): A tool for microsatellite genotyping and molecular characterization of a Greek Genebank collection. Biochem. Syst. Ecol. 2015, 58, 64–71. [Google Scholar] [CrossRef]
- MacKay, J.F.; Wright, C.D.; Bonfiglioli, R.G. A new approach to varietal identification in plants by microsatellite high resolution melting analysis: Application to the verification of grapevine and olive cultivars. Plant Methods 2008, 4, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Madesis, P.; Ganopoulos, I.; Anagnostis, A.; Tsaftaris, A. The application of Bar-HRM (Barcode DNA-High Resolution Melting) analysis for authenticity testing and quantitative detection of bean crops (Leguminosae) without prior DNA purification. Food Control 2012, 25, 576–582. [Google Scholar] [CrossRef]
- Wu, S.B.; Wirthensohn, M.G.; Hunt, P.; Gibson, J.P.; Sedgley, M. High resolution melting analysis of almond SNPs derived from ESTs. Theor. Appl. Genet. 2008, 118, 1–14. [Google Scholar] [CrossRef]
- Culbertson, A.T.; Ehrlich, J.J.; Choe, J.Y.; Honzatko, R.B.; Zabotina, O.A. Structure of xyloglucan xylosyltransferase 1 reveals simple steric rules that define biological patterns of xyloglucan polymers. Proc. Natl. Acad. Sci. USA 2018, 115, 6064–6069. [Google Scholar] [CrossRef] [Green Version]
- Kaur, K.; Gupta, A.K.; Kaur, N. Effect of water defficit on carbohydrate status and enzymes of carbohydrate metabolism in seedlings of wheat cultivars. Indian J. Biochem. Biophys. 2007, 44, 223–230. [Google Scholar]
- Guo, R.; Shi, L.; Jiao, Y.; Li, M.; Zhong, X.; Gu, F.; Liu, Q.; Xia, X.; Li, H. Metabolic responses to drought stress in the tissues of drought-tolerant and drought-sensitive wheat genotype seedlings. AoB Plants 2018, 10, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abebe, T.; Melmaiee, K.; Berg, V.; Wise, R.P. Drought response in the spikes of barley: Gene expression in the lemma, palea, awn, and seed. Funct. Integr. Genom. 2010, 10, 191–205. [Google Scholar] [CrossRef] [PubMed]
- Mindrebo, J.T.; Nartey, C.M.; Seto, Y.; Burkart, M.D.; Noel, J.P. Unveiling the functional diversity of the alpha/beta hydrolase superfamily in the plant kingdom. Curr. Opin. Struct. Biol. 2016, 41, 233–246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, M.; Pandey, G.K. Expansion and function of repeat domain proteins during stress and development in plants. Front. Plant Sci. 2016, 6, 1218. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hunt, M.R. Biological roles of small RNAs expressed during infection of barley by the obligate fungal biotroph, Blumeria graminis f. sp. hordei. Master’s Thesis, Iowa State University, Ames, IA, USA, 2018. [Google Scholar]
- Shibagaki, N.; Grossman, A.R. The role of the STAS domain in the function and biogenesis of a sulfate transporter as probed by random mutagenesis. J. Biol. Chem. 2006, 281, 22964–22973. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shibagaki, N.; Grossman, A.R. Probing the function of STAS domains of the Arabidopsis sulfate transporters. J. Biol. Chem. 2004, 279, 30791–30799. [Google Scholar] [CrossRef] [Green Version]
- Rouached, H.; Berthomieu, P.; El Kassis, E.; Cathala, N.; Catherinot, V.; Labesse, G. Structural and Functional Analysis of the C-terminal STAS (Sulfate Transporter and Anti-sigma Antagonist) domain of the Arabidopsis thaliana sulfate transporter SULTR1.1. J. Biol. Chem. 2005, 280, 15976–15983. [Google Scholar] [CrossRef] [Green Version]
- Tabe, L.; Hagan, N.; Higgins, T.J.V. Plasticity of seed protein composition in response to nitrogen and sulfur availability. Curr. Opin. Plant Biol. 2002, 5, 212–217. [Google Scholar] [CrossRef]
- Miazek, A.; Zagdańska, B. Involvement of exopeptidases in dehydration tolerance of spring wheat seedlings. Biol. Plant. 2008, 52, 687–694. [Google Scholar] [CrossRef]
- Bray, E.A. Plant responses to water deficit. Trends Plant Sci. 1997, 2, 48–54. [Google Scholar] [CrossRef]
- Vierstra, R.D. Proteolysis in plants: Mechanism and functions. Plant Mol. Biol. 2004, 32, 275–302. [Google Scholar] [CrossRef] [PubMed]
- Simova-Stoilova, L.; Vaseva, I.; Grigorova, B.; Demirevska, K.; Feller, U. Proteolytic activity and cysteine protease expression in wheat leaves under severe soil drought and recovery. Plant Physiol. Biochem. 2010, 48, 200–206. [Google Scholar] [CrossRef] [PubMed]
- Bohlmann, H.; Apel, K. Thionins. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1991, 42, 227–240. [Google Scholar] [CrossRef]
- Begcy, K.; Walia, H. Drought stress delays endosperm development and misregulates genes associated with cytoskeleton organization and grain quality proteins in developing wheat seeds. Plant Sci. 2015, 240, 109–119. [Google Scholar] [CrossRef] [PubMed]
- Ye, Y.; Ding, Y.; Jiang, Q.; Wang, F.; Sun, J.; Zhu, C. The role of receptor-like protein kinases (RLKs) in abiotic stress response in plants. Plant Cell Rep. 2017, 36, 235–242. [Google Scholar] [CrossRef]
- Cabouté, M.-E.; Clément, B.; Sekine, M.; Philipps, G.; Chaubet-Gigot, N. Cell cycle regulation of the tobacco ribonucleotide reductase small subunit gene is mediated by E2F-like elements. Plant Cell 2000, 12, 1987–1999. [Google Scholar] [CrossRef]
- Sauge-Merle, S.; Laulhère, J.P.; Covès, J.; Le Pape, L.; Ménage, S.; Fontecave, M. Ribonucleotide reductase from the higher plant Arabidopsis thaliana: Expression of the R2 component and characterization of its iron-radical center. J. Biol. Inorg. Chem. 1997, 2, 586–594. [Google Scholar] [CrossRef]
- Sparkes, I. Recent advances in understanding plant myosin function: Life in the fast lane. Mol. Plant 2011, 4, 805–812. [Google Scholar] [CrossRef] [Green Version]
- Vogt, T.; Jones, P. Glycosyltransferases in plant-natural product synthesis: Characterization of a supergene family. Trends Plant Sci. 2000, 5, 380–386. [Google Scholar] [CrossRef]
- Akdogan, G.; Tufekci, E.D.; Uranbey, S.; Unver, T. miRNA-based drought regulation in wheat. Funct. Integr. Genom. 2016, 16, 221–233. [Google Scholar] [CrossRef]
- Kang, J.; Park, J.; Choi, H.; Burla, B.; Kretzschmar, T.; Lee, Y.; Martinoia, E. Plant ABC Transporters. Arabidopsis Book 2011, 9, e0153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rampino, P.; Mita, G.; Fasano, P.; Borrelli, G.M.; Aprile, A.; Dalessandro, G.; De Bellis, L.; Perrotta, C. Novel durum wheat genes up-regulated in response to a combination of heat and drought stress. Plant Physiol. Biochem. 2012, 56, 72–78. [Google Scholar] [CrossRef]
- Walter, S.; Kahla, A.; Arunachalam, C.; Perochon, A.; Khan, M.R.; Scofield, S.R.; Doohan, F.M. A wheat ABC transporter contributes to both grain formation and mycotoxin tolerance. J. Exp. Bot. 2015, 66, 2583–2593. [Google Scholar] [CrossRef] [Green Version]
- Kulik, A.; Wawer, I.; Krzywińska, E.; Bucholc, M.; Dobrowolska, G. SnRK2 protein Kinases—Key regulators of plant response to abiotic stresses. Omics J. Integr. Biol. 2011, 15, 859–872. [Google Scholar] [CrossRef] [PubMed]
- Wei, K.; Li, Y. Functional genomics of the protein kinase superfamily from wheat. Mol. Breed. 2019, 39, 141. [Google Scholar] [CrossRef]








| Marker ID | Chr | Pattern | Forward Primer (5′→3′) | Reverse Primer (5′→3′) | Amp. Size |
|---|---|---|---|---|---|
| HRM4A_2791416 * | 4AS | A | TCCTACAAAAACGTCTTATATTTTGG | GATCACTTGCACGTTGCATT | 103 |
| HRM4A_9618320 | 4AS | C | CGTCAGCTCAAAGGAAAACC | GGGAGGAAATTTGCGAGTC | 151 |
| HRM4A_73613394 | 4AS | D | CGGTCCTTGTGATGATGTTG | CTTTGTAGGCCCCATCTGAA | 136 |
| HRM4A_36371442 | 4AS | D | GCATGTGGTCATGTTCTTGG | TCAAAAACGCTTTTATATTATGGGA | 193 |
| HRM4A_38654555 * | 4AS | A | TCTGAAAGAACCTCAGCTTATTACTT | TTTGGTAAAATTGAGGGACCA | 101 |
| HRM4A_47108251 | 4AS | n/s | ACACGTGGGAGATATAGCCG | AAACCCTAGGTCCCACTGCT | 168 |
| HRM4A_60660613 | 4AS | C | CATCCTCTCAGGCCATGAAT | CAAAATTCAAACTTTTCGGTTTC | 141 |
| HRM4A_U-1 | - | n/s | AGAGCGCTTAAGTTTGGCTG | TCACTCATTTCAGTCCGGAAG | 161 |
| HRM4A_56502024 | 4AS | D | CAAACGATGCTCCTCTGTCA | TCCATCTATCTGTATTTCGTATTGAA | 119 |
| HRM4A_67413676 | 4AS | A | CGAGTCCTAGCGAGTTCCTG | AAAAACATTGCATAAATGGATGG | 109 |
| HRM4A_59280888 | 4AS | B | CATCCACATGGATTTTGCAG | CGATTTGGTACGCTAGGAGG | 207 |
| HRM4A_141912346 | 4AS | A | AGCACAAGCATGCAATGAAG | TTATCTCGTGTAGGACCGGG | 143 |
| HRM4A_99034796 | 4AS | n/s | CCTCCGTTCGGAATTACTTG | TTCATGCAGCAGCAGTTACC | 157 |
| HRM4A_109848074 * | 4AS | A | ATGAGACTTTTGACGACCGC | CAAGCTTTTTCAGACGGAGG | 139 |
| HRM4A_141111880 | 4AS | D | AGATCTGCTGGACATAAGCACA | CCCTCCGTCCCAAAATAAGT | 117 |
| HRM4A_162423177 | 4AS | C | AAGCAAGAAGCGAAAACAGC | AATCATCTAGTCGGTTGCGG | 180 |
| HRM4A_176868209 | 4AS | C | TTCCGAATTACTTGTCTCGGAT | CACAGGCTCGGATAGGCTAC | 191 |
| HRM4A_183985221 | 4AS | C | TCCAGAAAATCTGTAGGCACTG | AGATGGACGCGATAAGATGG | 108 |
| HRM4A_149049302 | 4AS | D | CCTCCGTTCGGAATTACTTG | ATGAAAGGCAGGCTAGACCA | 223 |
| HRM4A_617938526 | 4AL | C | CTTCGTCCTTCCTCGCATAG | GTAAGGTAGTGATCTAAACGGTGTT | 113 |
| HRM4A_618105078 | 4AL | B | AGTCATGGCACCAACAACAA | AGAGTTGCCGTGCCACTTAT | 162 |
| HRM4A_U-2 | - | n/s | TTTCTAAGGGGTAAGGGCGT | TAGAGGGTTGTGCTGGTTCC | 100 |
| HRM4A_716986193 * | 4AL | A | CTGCACCATAGATCGAAGCA | ACAGGACAATTGGAGACCCA | 159 |
| HRM4A_U-3 | - | A | TTGAACTGCCAAAAACGTCTCA | CTCCCTCCTATAACCACCATTG | 100 |
| HRM4A_548541053 | 4AL | D | CGGTGCTAGATACATCCGTTT | AACCAAGTAAGCATGTACTAGAGAAAA | 112 |
| HRM4A_714743756 | 4AL | A | CGCTAGTATAGTGTCAAAAACGC | ATGTAGGATGTCCCTGCGTC | 117 |
| HRM4A_U-4 * | - | A | CAGACAATGTGCAAAACAACC | AAAAGAGTTCATGTACAAGGGGA | 116 |
| HRM4A_U-5 | - | n/s | TCCACCTTATAAACACCCGC | TGTATTTCCAGGACGGAAGG | 202 |
| HRM4A_460238681 | 4AL | C | GGTCTGTATTGAAATCTCTAAAAGTGC | CCTTTTGTTCAGCCTGTGGT | 119 |
| HRM4A_583704598 * | 4AL | A | CCCTCTGTAAACTAATATAGAATGCG | CTTGTTCCCTCTGCTCCTTG | 165 |
| HRM4A_317085557 | 4AS | C | ACATGGGTGACCCTATCCAA | CGGACTGGTCCATTAGGGTT | 162 |
| HRM4A_291420130 | 4AS | D | AAGTGGAAAAGGCACAATGC | CAAACACTTTGCCAACATGG | 183 |
| HRM4A_681664894 | 4AL | D | TATGCTTGAGTGCTTGGCTG | CATCCATTTGAGCGACAACT | 121 |
| HRM4A_683608822 | 4AL | C | TCAGTTTTAGGTCCCGTTGC | TGACACTACTCTAAGTTACTCCCCA | 196 |
| HRM4A_U-6 | - | D | TTCCAAGAAAATGTTCGCAA | TCCTTCGTTCAAGACTCGCT | 177 |
| HRM4A_U-7 | - | B | TCCCTAGCTGATGATTTGGG | ATAATAGCTCCATACGCGCC | 107 |
| HRM4A_660524139 * | 4AL | A | GATAAATCTAAGATAAGCTTTTTGGG | GGGACACAATGTGATGGTGA | 116 |
| HRM4A_U-8 | - | C | GGCCCTAGAAATGCAAATGA | TTCCCACCTCAATAACTGGG | 127 |
| HRM4A_U-9 | - | B | TCTTCACTCGTTTCAGTCCG | AGACGAGCACACACGCATAC | 131 |
| HRM4A_702156718 | 4AL | A | CCTTTGGCAACAACACAATG | ATTGGCAGATTCTTCAAGCG | 133 |
| HRM4A_U-10 | - | D | AGCCGAGGAAGGTTCACATA | TCCATTTATAAACAAATATAAGAGCG | 119 |
| HRM4A_U-11 | - | A | CGGTCAATGTATACTACCGTCG | ATCGGGAACCACCAATGTTA | 164 |
| HRM4A_U-12 | - | D | TGTTTGCTGAGGACCAACAG | CCGGGGGTAATCCTAATTTT | 111 |
| HRM4A_U-13 | - | D | ATTGTTCGTTCCGTTTTTGG | ACTCCCTCCGTCCCATAATA | 114 |
| HRM4A_U-14 | - | D | CCCTCTGTAAAGAAATATAAGACCGT | TGGATGCAGCTAACTCGAAA | 107 |
| Marker ID | Chr | Dist (bp) | Gene ID | Description | Sum of TPMs |
|---|---|---|---|---|---|
| HRM4A_2791416 | 4A | −814 | TraesCS4A01G003600 | Alpha/beta-Hydrolases superfamily protein | 8.79 |
| −5141 | TraesCS4A01G003500 | Thionin-like protein | 124.0 | ||
| HRM4A_36371442 | 4A | −2736 | TraesCS4A01G043500 | STAS domain-containing protein | 47.32 |
| HRM4A_38654555 | 4A | −9961 | TraesCS4A01G047100 | Activating signal cointegrator 1 complex subunit 2 | 12.70 |
| −14,885 | TraesCS4A01G047000 | Formin-like protein | 4.87 | ||
| HRM4A_56502024 | 4A | −417 | TraesCS4A01G060200 | BHLH domain-containing protein | 10.54 |
| −5039 | TraesCS4A01G060100 | Uncharacterized protein | 50.64 | ||
| HRM4A_67413676 | 4A | 975 | TraesCS4A01G069200 | Armadillo repeat only | 21.26 |
| HRM4A_99034796 | 4A | −5609 | TraesCS4A01G092400 | SH3 domain-containing protein | 43.46 |
| HRM4A_109848074 | 4A | 97 | TraesCS4A01G098300 | Xylosyltransferase 1 | 21.59 |
| HRM4A_141111880 | 4A | −3308 | TraesCS4A01G116200 | MYND-type domain-containing protein | 50.62 |
| HRM4A_162423177 | 4A | 9 | TraesCS4A01G126300 | Transcription factor TGA2.2 * | 83.14 |
| HRM4A_176868209 | 4A | −406 | TraesCS4A01G132000 | Uncharacterized protein | 17.27 |
| HRM4A_183985221 | 4A | −5474 | TraesCS4A01G134900 | Lung seven transmembrane receptor family protein, expressed * | 44.02 |
| HRM4A_317085557 | 4A | −14,884 | TraesCS4A01G157000 | SAP domain-containing protein | 127.49 |
| HRM4A_460238681 | 4A | −9041 | TraesCS4A01G182900 | SPRING type domain-containing protein * | 11.42 |
| HRM4A_548541053 | 4A | −11,850 | TraesCS4A01G239500 | ATP-dependent RNA helicase dhx8 * | 19.65 |
| HRM4A_583704598 | 4A | 3305 | TraesCS4A01G271900 | Histone-lysine N-methyltransferase | 11.28 |
| HRM4A_617938526 | 4A | −5243 | TraesCS4A01G335200 | Protein kinase domain-containing protein * | 10.09 |
| HRM4A_681664894 | 4A | −3025 | TraesCS4A01G408900 | Protein PIR * | 33.28 |
| HRM4A_683608822 | 4A | −2685 | TraesCS4A01G410700 | Ras-related protein RABC2a * | 82.16 |
| HRM4A_716986193 | 4A | −3576 | TraesCS4A01G671200LC | Peptidase M20/M25/M40 family protein | 104.94 |
| HRM4A_U-2 | 4A | −9642 | TraesCS4A01G287200 | C2 calcium/lipid-binding and GRAM domain containing protein | 2.72 |
| Marker ID | Chr | Dist (bp) | Gene ID | Description | Sum of TPMs |
|---|---|---|---|---|---|
| HRM3B_124761338 | 3B | 6356 | TraesCS3B01G138700 | Ribonucleoside-diphosphate reductase | 7.48 |
| 8268 | TraesCS3B01G185800LC | Serine/threonine-protein phosphatase 6 regulatory subunit 1 | 7.25 | ||
| 14,040 | TraesCS3B01G185900LC | LEM3 (Ligand-effect modulator 3)-like | 180.07 | ||
| HRM3B_203288704 | 3B | 227,619 | TraesCS3B01G262900LC | Retrovirus-related Pol polyprotein LINE-1 | 7.05 |
| 260,457 | TraesCS3B01G190500 | Polynucleotidyl transferase ribonuclease H-like superfamily protein | 28.66 | ||
| 266,554 | TraesCS3B01G190600 | Beta-amylase | 118.13 | ||
| HRM3B_273339424 | 3B | −89,674 | TraesCS3B01G221100 | Protein kinase superfamily protein | 43.92 |
| −90,463 | TraesCS3B01G308500LC | translation initiation factor 3 subunit H1 | 22.30 | ||
| −101,322 | TraesCS3B01G221000 | Kelch repeat protein, putative | 4.08 | ||
| −286,319 | TraesCS3B01G308400LC | Ribonuclease H-like superfamily protein | 29.01 | ||
| HRM3B_331497483 | 3B | 55,223 | TraesCS3B01G228700 | carboxyl-terminal peptidase, putative (DUF239) | 6.57 |
| HRM3B_465802537 | 3B | 83,072 | TraesCS3B01G290200 | Glycosyltransferase | 9.98 |
| 125,805 | TraesCS3B01G290300 | ABC transporter B family protein | 19.57 | ||
| 296,991 | TraesCS3B01G290400 | zein-binding protein (Protein of unknown function, DUF593) | 34.58 | ||
| HRM3B_609364064 | 3B | −24,580 | TraesCS3B01G387400 | SAC3/GANP/Nin1/mts3/eIF-3 p25 family protein | 15.46 |
| −31,936 | TraesCS3B01G575000LC | Myosin-1 | 48.25 |
| Marker ID | Chr | MQTL | Dist (bp) | Gene ID | Description |
|---|---|---|---|---|---|
| HRM4A_716986193 | 4A | 32 | −3576 | TraesCS4A01G671200LC | Peptidase M20/M25/M40 family protein |
| HRM4A_714743756 | 4A | close to QTL in MQTL32 | −5397 | TraesCS4A01G665300LC | LINE-1 reverse transcriptase-like protein |
| −6886 | TraesCS4A01G665200LC | C4-dicarboxylate transport protein | |||
| −12,058 | TraesCS4A01G665100LC | NBS-LRR disease resistance protein | |||
| −14,064 | TraesCS4A01G665000LC | Disease resistance protein (NBS-LRR class) family | |||
| 14,217 | TraesCS4A01G665400LC | NBS-LRR disease resistance protein | |||
| 17,933 | TraesCS4A01G665500LC | Transposase | |||
| HRM4A_583704598 | 4A | between MQTL30 and 31 | −2715 | TraesCS4A01G271800 | Kinase family protein |
| 3305 | TraesCS4A01G271900 | Histone-lysine N-methyltransferase | |||
| −6092 | TraesCS4A01G431700LC | Serine/threonine-protein kinase mph1 | |||
| HRM_4A660524139 | 4A | close to QTL in MQTL31 | |||
| HRM4A_702156718 | 4A | close to QTL in MQTL32 | −1999 | TraesCS4A01G638600LC | Retrotransposon protein, putative, unclassified |
| 14,448 | TraesCS4A01G638700LC | Kinase, putative | |||
| HRM4A_618105078 | 4A | 31 | −2442 | TraesCS4A01G497800LC | Receptor-like protein kinase |
| −11,813 | TraesCS4A01G497700LC | Protein FAR1-RELATED SEQUENCE 5 | |||
| 16,466 | TraesCS4A01G335600 | NBS-LRR-like resistance protein | |||
| −19,471 | TraesCS4A01G335500 | NBS-LRR disease resistance protein | |||
| HRM4A_617938526 | 4A | 31 | −5243 | TraesCS4A01G335200 | Protein kinase domain-containing protein * |
| HRM4A_460238681 | 4A | 30 | 132 | TraesCS4A01G183000 | Protein kinase domain-containing protein |
| −9041 | TraesCS4A01G182900 | SP-RING-type domain-containing protein * | |||
| HRM4A_317085557 | 4A | 30 | −14,884 | TraesCS4A01G157000 | SAP domain-containing protein |
| HRM4A_683608822 | 4A | 32 | −2685 | TraesCS4A01G410700 * | Ras-related protein RABC2a * |
| HRM3B_609364064 | 3B | 26 | 52,100 | TraesCS3B01G575100LC | Transposon protein, putative, CACTA, En/Spm sub-class |
| −24,580 | TraesCS3B01G387400 | SAC3/GANP/Nin1/mts3/eIF-3 p25 family protein | |||
| −31,936 | TraesCS3B01G575000LC | Myosin-1 | |||
| −53,463 | TraesCS3B01G574900LC | Endonuclease/exonuclease/phosphatase family protein | |||
| −59,628 | TraesCS3B01G387300 | PP2A regulatory subunit TAP46 | |||
| HRM3B_465802537 | 3B | 26 | 83,072 | TraesCS3B01G290200 * | Glycosyltransferase |
| −88,899 | TraesCS3B01G451200LC | phospholipase-like protein (PEARLI 4) family protein | |||
| −95,341 | TraesCS3B01G451100LC | Retrotransposon protein, putative, unclassified | |||
| −98,315 | TraesCS3B01G451000LC | Ubiquilin-1 | |||
| −106,901 | TraesCS3B01G290100 | Ribose-5-phosphate isomerase A | |||
| 125,805 | TraesCS3B01G290300 * | ABC transporter B family protein | |||
| 138,676 | TraesCS3B01G451300LC | Polynucleotidyl transferase, ribonuclease H-like superfamily protein | |||
| −144,473 | TraesCS3B01G450900LC | Protein regulator of cytokinesis 1 | |||
| −147,938 | TraesCS3B01G450800LC | Beta-hexosaminidase | |||
| 161,237 | TraesCS3B01G451400LC | Endonuclease/exonuclease/phosphatase family protein | |||
| 296,991 | TraesCS3B01G290400 | zein-binding protein (Protein of unknown function, DUF593) | |||
| 297,913 | TraesCS3B01G290500 | Potassium channel |
| Marker ID | HRM Type | No. of Alleles | PIC |
|---|---|---|---|
| HRM4A_2791416 | A | 2 | 0.336 |
| HRM4A_38654555 | A | 2 | 0.566 |
| HRM4A_67413676 | A | 3 | 0.584 |
| HRM4A_141912346 | A | 5 * | 0.595 |
| HRM4A_109848074 | A | 2 | 0.544 |
| HRM4A_618105078 | B | 4 * | 0.677 |
| HRM4A_716986193 | C | 4 | 0.602 |
| HRM4A_U-3 | C | 2 * | 0.242 |
| HRM4A_714743756 | A | 2 * | 0.368 |
| HRM4A_U-4 | A | 3 * | 0.474 |
| HRM4A_583704598 | A | 7 * | 0.571 |
| HRM4A_660524139 | A | 4 * | 0.57 |
| HRM4A_702156718 | A | 4 * | 0.563 |
| Mean | 3.38 | 0.515 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mérida-García, R.; Gálvez, S.; Paux, E.; Dorado, G.; Pascual, L.; Giraldo, P.; Hernandez, P. High Resolution Melting and Insertion Site-Based Polymorphism Markers for Wheat Variability Analysis and Candidate Genes Selection at Drought and Heat MQTL Loci. Agronomy 2020, 10, 1294. https://doi.org/10.3390/agronomy10091294
Mérida-García R, Gálvez S, Paux E, Dorado G, Pascual L, Giraldo P, Hernandez P. High Resolution Melting and Insertion Site-Based Polymorphism Markers for Wheat Variability Analysis and Candidate Genes Selection at Drought and Heat MQTL Loci. Agronomy. 2020; 10(9):1294. https://doi.org/10.3390/agronomy10091294
Chicago/Turabian StyleMérida-García, Rosa, Sergio Gálvez, Etienne Paux, Gabriel Dorado, Laura Pascual, Patricia Giraldo, and Pilar Hernandez. 2020. "High Resolution Melting and Insertion Site-Based Polymorphism Markers for Wheat Variability Analysis and Candidate Genes Selection at Drought and Heat MQTL Loci" Agronomy 10, no. 9: 1294. https://doi.org/10.3390/agronomy10091294