Impact of Tea Tree Cultivation on Soil Microbiota, Soil Organic Matter, and Nitrogen Cycling in Mountainous Plantations
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Site and Soil Sample Collection
2.2. Soil Physicochemical Properties and Enzyme Activity Analysis
2.3. Soil Total DNA Extraction
2.4. High-Throughput Sequencing Analysis of 16S/18S rDNA
2.5. Data Analysis
3. Results
3.1. Impact of Tea Cultivation on Terraced Soil’s Physicochemical Properties and Enzyme Activity
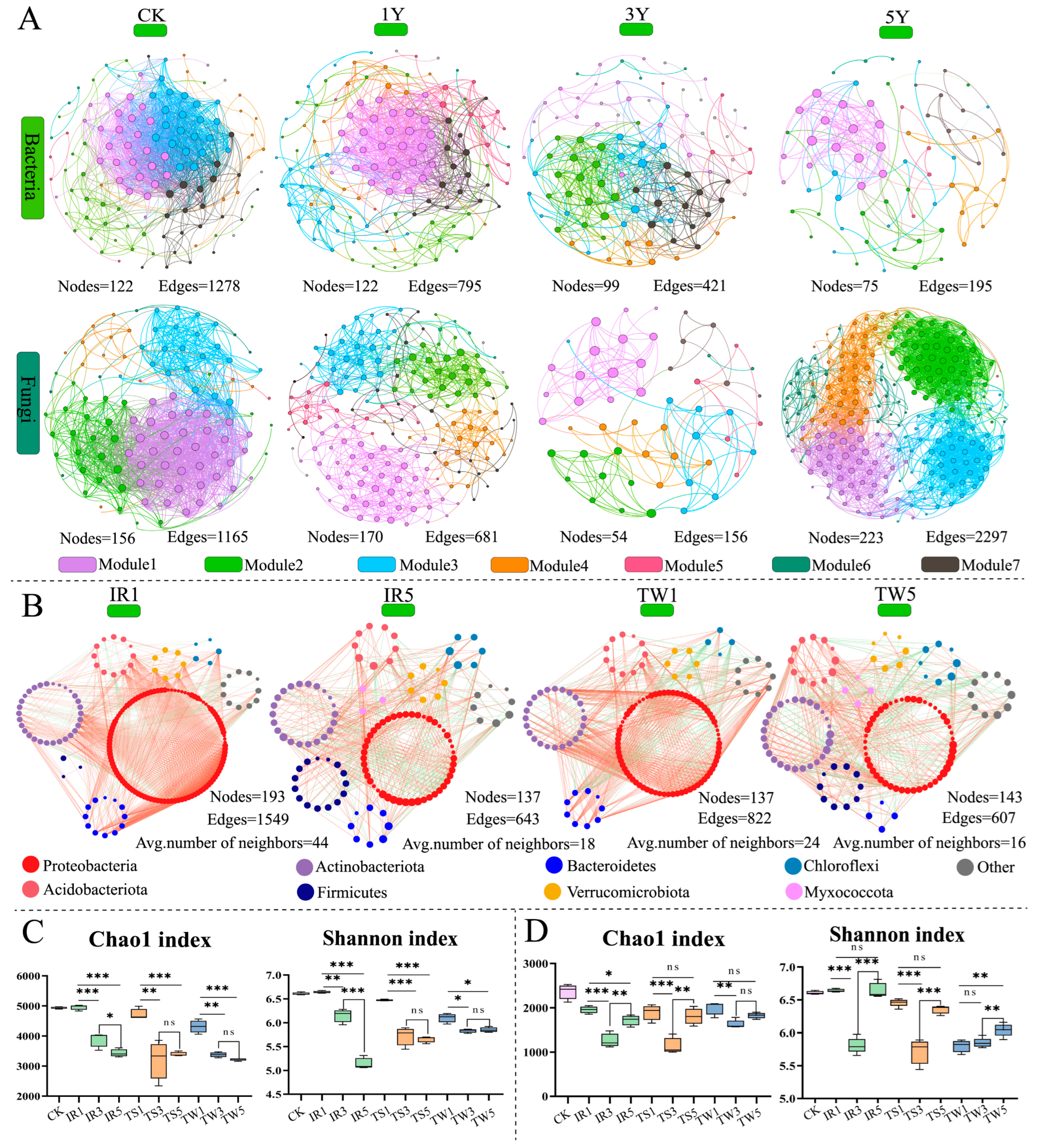
3.2. Impact of Tea Cultivation on Microbial Diversity in Mountainous Tea Plantations
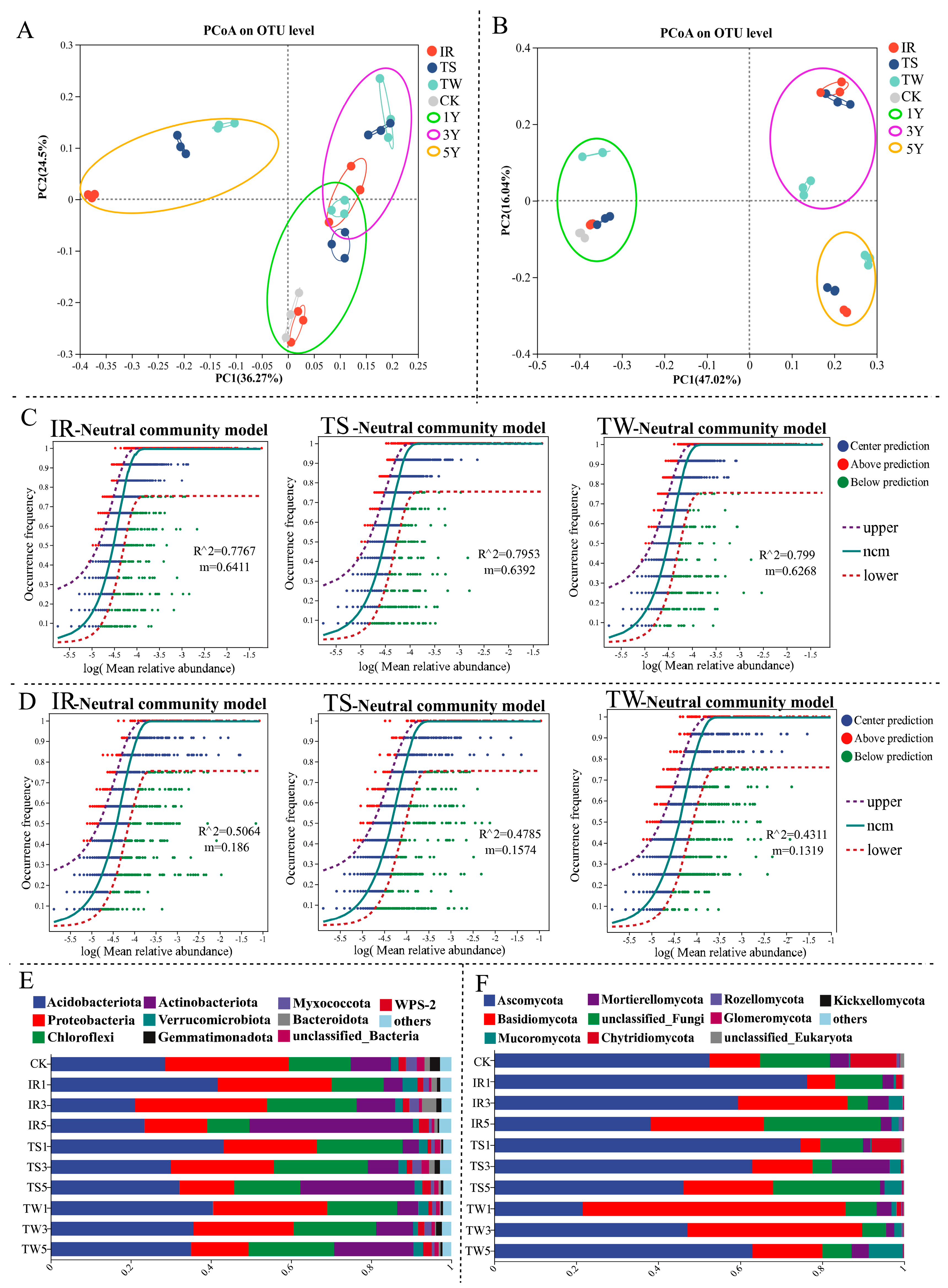
3.3. Impact of Tea Cultivation on Soil Microbial Community Construction and Dispersal in Mountainous Tea Plantations
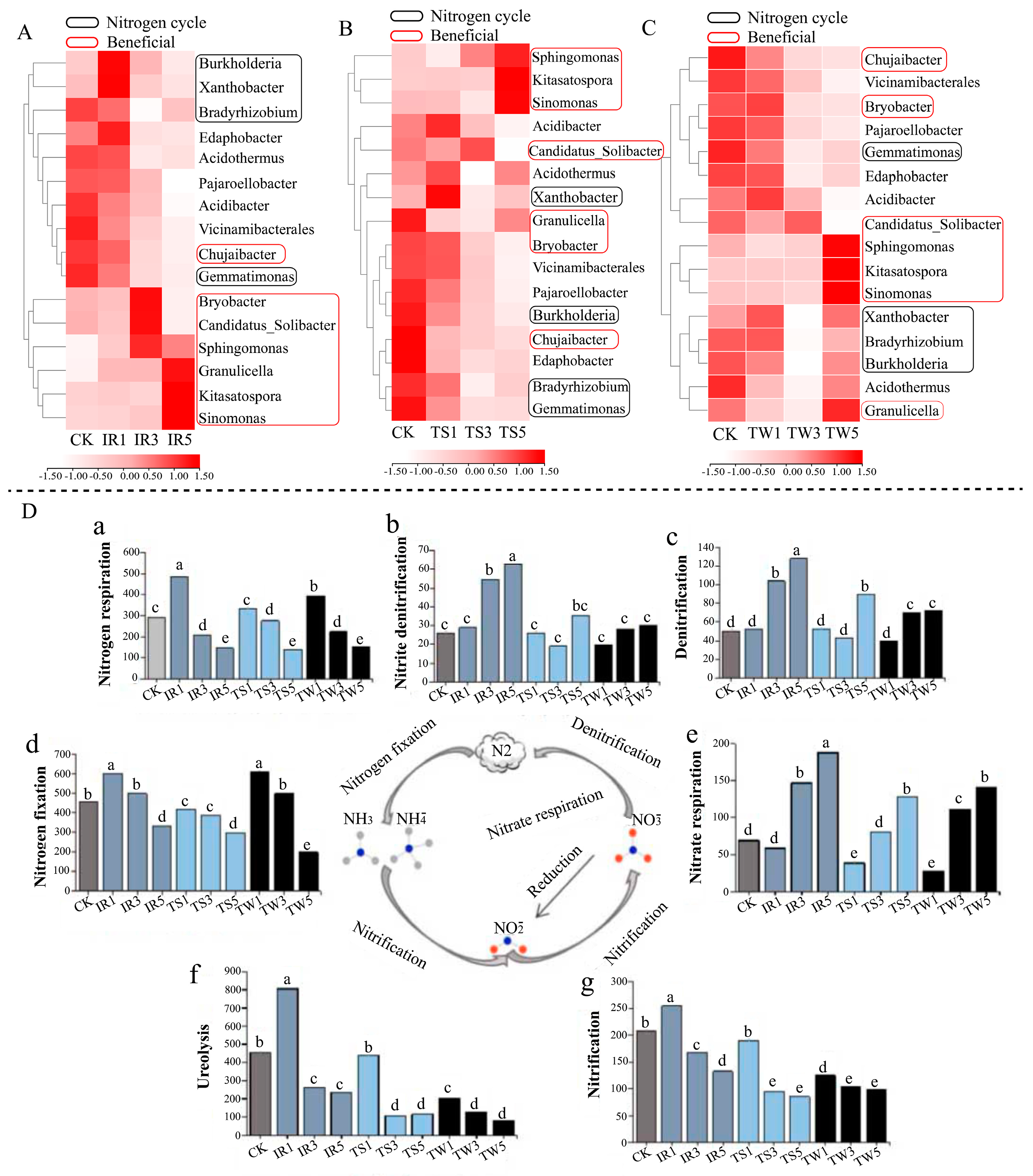
3.4. Effects of Tea Cultivation on Key Soil Bacterial Flora and Its Functions in Mountainous Tea Plantations
3.5. Effects of Tea Cultivation on Key Soil Fungal Flora and Its Functions in Mountainous Tea Plantations
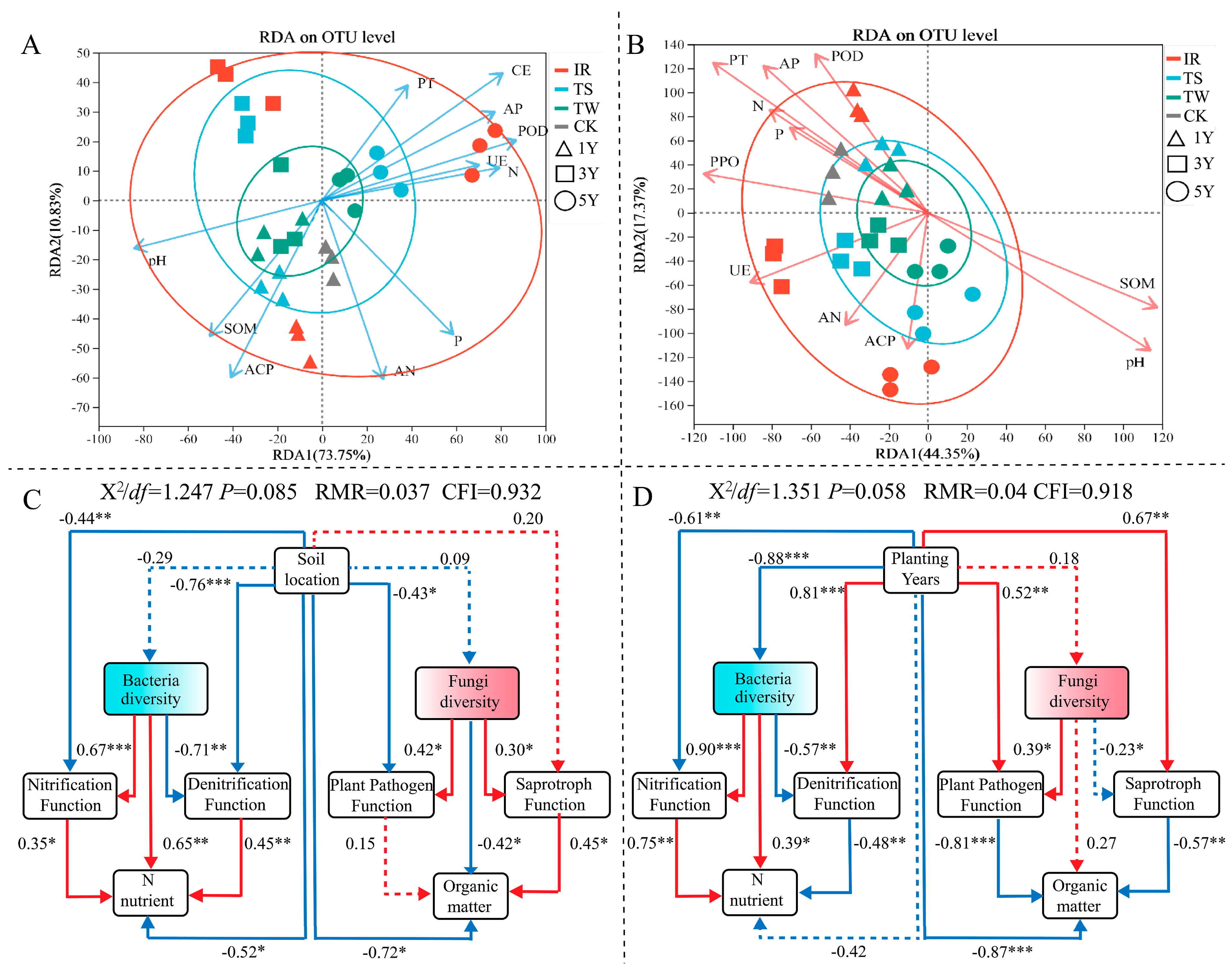
3.6. Correlation between Soil Microbial Community Structure and Soil Biochemical Properties
4. Discussion
4.1. Tea Cultivation Induces Specific Microbial Communities to Accelerate Soil Organic Matter Metabolism
4.2. Tea Tree Cultivation Regulates Soil Nitrogen Cycling Functional Bacteria and Reduces Soil Nitrogen Supply Capacity
4.3. Tea Cultivation Reshaped the Soil Microbial Community Diversity and Structure in Terraced Tea Plantations
4.4. Tea Tree Cultivation Increases the Abundance of Pathogenic Fungal Communities in Soil and Raises Plant Disease Risk
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Wu, H.; Long, X.; Geng, Y. Companion Plants of Tea: From Ancient to Terrace to Forest. Plants 2023, 12, 3061. [Google Scholar] [CrossRef]
- Deng, C.; Zhang, G.; Liu, Y.; Nie, X.; Li, Z.; Liu, J.; Zhu, D. Advantages and disadvantages of terracing: A comprehensive review. Int. Soil Water Conserv. Res. 2021, 9, 344–359. [Google Scholar] [CrossRef]
- Wen, B.; Ren, S.; Zhang, Y.; Duan, Y.; Shen, J.; Zhu, X.; Wang, Y.; Ma, Y.; Zou, Z.; Fang, W. Effects of geographic locations and topographical factors on secondary metabolites distribution in green tea at a regional scale. Food Control 2020, 110, 106979. [Google Scholar] [CrossRef]
- Zhao, M.; Zhao, J.; Yuan, J.; Hale, L.; Wen, T.; Huang, Q.; Vivanco, J.M.; Zhou, J.; Kowalchuk, G.A.; Shen, Q. Root exudates drive soil-microbe-nutrient feedbacks in response to plant growth. Plant Cell Environ. 2021, 44, 613–628. [Google Scholar] [CrossRef]
- Chen, H.; Li, D.; Mao, Q.; Xiao, K.; Wang, K. Resource limitation of soil microbes in karst ecosystems. Sci. Total Environ. 2019, 650, 241–248. [Google Scholar] [CrossRef]
- Zhang, T.A.; Chen, H.Y.H.; Ruan, H. Global negative effects of nitrogen deposition on soil microbes. ISME J. 2018, 12, 1817–1825. [Google Scholar] [CrossRef] [PubMed]
- Ke, P.-J.; Wan, J. Effects of soil microbes on plant competition: A perspective from modern coexistence theory. Ecol. Monogr. 2020, 90, e01391. [Google Scholar] [CrossRef]
- Ferreira, D.A.; da Silva, T.F.; Pylro, V.S.; Salles, J.F.; Andreote, F.D.; Dini-Andreote, F. Soil Microbial Diversity Affects the Plant-Root Colonization by Arbuscular Mycorrhizal Fungi. Microb. Ecol. 2021, 82, 100–103. [Google Scholar] [CrossRef] [PubMed]
- Kafle, A.; Cope, K.R.; Raths, R.; Krishna Yakha, J.; Subramanian, S.; Bücking, H.; Garcia, K. Harnessing Soil Microbes to Improve Plant Phosphate Efficiency in Cropping Systems. Agronomy 2019, 9, 127. [Google Scholar] [CrossRef]
- Hu, H.-Y.; Li, H.; Hao, M.-M.; Ren, Y.-N.; Zhang, M.-K.; Liu, R.-Y.; Zhang, Y.; Li, G.; Chen, J.-S.; Ning, T.-Y.; et al. Nitrogen fixation and crop productivity enhancements co-driven by intercrop root exudates and key rhizosphere bacteria. J. Appl. Ecol. 2021, 58, 2243–2255. [Google Scholar] [CrossRef]
- Li, J.; Sang, C.; Yang, J.; Qu, L.; Xia, Z.; Sun, H.; Jiang, P.; Wang, X.; He, H.; Wang, C. Stoichiometric imbalance and microbial community regulate microbial elements use efficiencies under nitrogen addition. Soil Biol. Biochem. 2021, 156, 108207. [Google Scholar] [CrossRef]
- Li, B.-B.; Roley, S.S.; Duncan, D.S.; Guo, J.; Quensen, J.F.; Yu, H.-Q.; Tiedje, J.M. Long-term excess nitrogen fertilizer increases sensitivity of soil microbial community to seasonal change revealed by ecological network and metagenome analyses. Soil Biol. Biochem. 2021, 160, 108349. [Google Scholar] [CrossRef]
- Yang, N.; Hua, J.; Zhang, J.; Liu, D.; Bhople, P.; Li, X.; Zhang, Y.; Ruan, H.; Xing, W.; Mao, L. Soil nutrients and plant diversity affect ectomycorrhizal fungal community structure and functional traits across three subalpine coniferous forests. Front. Microbiol. 2022, 13, 1016610. [Google Scholar] [CrossRef]
- Xue, D.; Yao, H.; Huang, C. Microbial Biomass, N Mineralization and Nitrification, Enzyme Activities, and Microbial Community Diversity in Tea Orchard Soils. Plant Soil 2006, 288, 319–331. [Google Scholar] [CrossRef]
- Zhao, J.; Wu, X.; Nie, C.; Wu, T.; Dai, W.; Liu, H.; Yang, R. Analysis of unculturable bacterial communities in tea orchard soils based on nested PCR-DGGE. World J. Microbiol. Biotechnol. 2012, 28, 1967–1979. [Google Scholar] [CrossRef] [PubMed]
- Guo, Z.-Y.; Kong, C.-H.; Wang, J.-G.; Wang, Y.-F. Rhizosphere isoflavones (daidzein and genistein) levels and their relation to the microbial community structure of mono-cropped soybean soil in field and controlled conditions. Soil Biol. Biochem. 2011, 43, 2257–2264. [Google Scholar] [CrossRef]
- Pramanik, P.; Phukan, M.; Ghosh, S.; Goswami, A.J. Pruned tea bushes secrete more root exudates to influence microbiological properties in soil. Arch. Agron. Soil Sci. 2018, 64, 1172–1180. [Google Scholar] [CrossRef]
- Zhang, X.; Long, H.; Huo, D.; Awan, M.I.; Shao, J.; Mahmood, A.; Liu, S.; Huang, J.; Parveen, A.; Aamer, M.; et al. Insights into the functional role of tea microbes on tea growth, quality and resistance against pests and diseases. Not. Bot. Horti Agrobot. Cluj-Napoca 2022, 50, 12915. [Google Scholar] [CrossRef]
- Lange, M.; Azizi-Rad, M.; Dittmann, G.; Lange, D.F.; Orme, A.M.; Schroeter, S.A.; Simon, C.; Gleixner, G. Stability and carbon uptake of the soil microbial community is determined by differences between rhizosphere and bulk soil. Soil Biol. Biochem. 2024, 189, 109280. [Google Scholar] [CrossRef]
- Huang, Y.; Chai, X.; Wang, X.; Gao, B.; Li, H.; Han, Z.; Xu, X.; Zhang, X.; Wu, T.; Wang, Y. Niche differentiation shapes the bacterial diversity and composition of apple. Hortic. Plant J. 2023, 9, 35–44. [Google Scholar] [CrossRef]
- Zhong, Y.; Yang, Y.; Liu, P.; Xu, R.; Rensing, C.; Fu, X.; Liao, H. Genotype and rhizobium inoculation modulate the assembly of soybean rhizobacterial communities. Plant Cell Environ. 2019, 42, 2028–2044. [Google Scholar] [CrossRef]
- Liu, J.; Shu, A.; Song, W.; Shi, W.; Li, M.; Zhang, W.; Li, Z.; Liu, G.; Yuan, F.; Zhang, S.; et al. Long-term organic fertilizer substitution increases rice yield by improving soil properties and regulating soil bacteria. Geoderma 2021, 404, 115287. [Google Scholar] [CrossRef]
- Ahirwal, J.; Maiti, S.K. Assessment of soil carbon pool, carbon sequestration and soil CO2 flux in unreclaimed and reclaimed coal mine spoils. Environ. Earth Sci. 2017, 77, 9. [Google Scholar] [CrossRef]
- Yao, H.; He, Z.; Wilson, M.J.; Campbell, C.D. Microbial Biomass and Community Structure in a Sequence of Soils with Increasing Fertility and Changing Land Use. Microb. Ecol. 2000, 40, 223–237. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Qu, L.; Yang, L.; Liu, D.; Morrissey, E.; Miao, R.; Liu, Z.; Wang, Q.; Fang, Y.; Bai, E. Large-scale importance of microbial carbon use efficiency and necromass to soil organic carbon. Glob. Chang. Biol. 2021, 27, 2039–2048. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Xu, Z.; Shi, S.; Lin, M. Lignocellulose degradation patterns, structural changes, and enzyme secretion by Inonotus obliquus on straw biomass under submerged fermentation. Bioresour. Technol. 2017, 241, 415–423. [Google Scholar] [CrossRef]
- Zhou, H.; Gao, Y.; Jia, X.; Wang, M.; Ding, J.; Cheng, L.; Bao, F.; Wu, B. Network analysis reveals the strengthening of microbial interaction in biological soil crust development in the Mu Us Sandy Land, northwestern China. Soil Biol. Biochem. 2020, 144, 107782. [Google Scholar] [CrossRef]
- Asaf, S.; Numan, M.; Khan, A.L.; Al-Harrasi, A. Sphingomonas: From diversity and genomics to functional role in environmental remediation and plant growth. Crit. Rev. Biotechnol. 2020, 40, 138–152. [Google Scholar] [CrossRef] [PubMed]
- Coloma, J.; Teeuwisse, L.; Afendi, M.; Hagedoorn, P.-L.; Hanefeld, U. Batch and Flow Nitroaldol Synthesis Catalysed by Granulicella tundricola Hydroxynitrile Lyase Immobilised on Celite R-633. Catalysts 2022, 12, 161. [Google Scholar] [CrossRef]
- Gillan, D.C.; Pan, H.; Roulez, A.; Wattiez, R. The metaphenome of a calaminiferous soil. Microbe 2023, 1, 100002. [Google Scholar] [CrossRef]
- Lee, L.H.; Azman, A.S.; Zainal, N.; Yin, W.F.; Mutalib, N.A.; Chan, K.G. Sinomonas humi sp. nov., an amylolytic actinobacterium isolated from mangrove forest soil. Int. J. Syst. Evol. Microbiol. 2015, 65, 996–1002. [Google Scholar] [CrossRef]
- Li, Y.; Wang, M.; Sun, Z.Z.; Xie, B.B. Comparative Genomic Insights into the Taxonomic Classification, Diversity, and Secondary Metabolic Potentials of Kitasatospora, a Genus Closely Related to Streptomyces. Front. Microbiol. 2021, 12, 683814. [Google Scholar] [CrossRef]
- Lin, Z.; Pang, S.; Zhou, Z.; Wu, X.; Li, J.; Huang, Y.; Zhang, W.; Lei, Q.; Bhatt, P.; Mishra, S.; et al. Novel pathway of acephate degradation by the microbial consortium ZQ01 and its potential for environmental bioremediation. J. Hazard. Mater. 2022, 426, 127841. [Google Scholar] [CrossRef]
- Costa, O.Y.A.; de Hollander, M.; Pijl, A.; Liu, B.; Kuramae, E.E. Cultivation-independent and cultivation-dependent metagenomes reveal genetic and enzymatic potential of microbial community involved in the degradation of a complex microbial polymer. Microbiome 2020, 8, 76. [Google Scholar] [CrossRef]
- Mielniczuk, E.; Skwaryło-Bednarz, B. Fusarium Head Blight, Mycotoxins and Strategies for Their Reduction. Agronomy 2020, 10, 509. [Google Scholar] [CrossRef]
- Schmidt, M.W.; Torn, M.S.; Abiven, S.; Dittmar, T.; Guggenberger, G.; Janssens, I.A.; Kleber, M.; Kögel-Knabner, I.; Lehmann, J.; Manning, D.A.; et al. Persistence of soil organic matter as an ecosystem property. Nature 2011, 478, 49–56. [Google Scholar] [CrossRef] [PubMed]
- Guzmán-Guzmán, P.; Porras-Troncoso, M.D.; Olmedo-Monfil, V.; Herrera-Estrella, A. Trichoderma Species: Versatile Plant Symbionts. Phytopathology 2019, 109, 6–16. [Google Scholar] [CrossRef] [PubMed]
- Summerell, B.A. Resolving Fusarium: Current Status of the Genus. Annu. Rev. Phytopathol. 2019, 57, 323–339. [Google Scholar] [CrossRef] [PubMed]
- Vaishnav, N.; Singh, A.; Adsul, M.; Dixit, P.; Sandhu, S.K.; Mathur, A.; Puri, S.K.; Singhania, R.R. Penicillium: The next emerging champion for cellulase production. Bioresour. Technol. Rep. 2018, 2, 131–140. [Google Scholar] [CrossRef]
- Vinale, F.; Sivasithamparam, K. Beneficial effects of Trichoderma secondary metabolites on crops. Phytother. Res. 2020, 34, 2835–2842. [Google Scholar] [CrossRef]
- Xj, Z.; Hu, Y.F.; Chen, X.; Wang, Y.H.; Fang, W.P.; Li, X.H. Endophytic fungi from Camellia sinensis show an antimicrobial activity against the rice blast pathogen Magnaporthe grisea. Phyton 2014, 83, 57–63. [Google Scholar] [CrossRef]
- Lin, S.; Liu, Z.; Wang, Y.; Li, J.; Wang, G.; Zhang, W.; Wang, H.; He, H. Soil acidification associated with changes in inorganic forms of N reduces the yield of tea (Camellia sinensis). Arch. Agron. Soil Sci. 2022, 69, 1660–1673. [Google Scholar] [CrossRef]
- Ruan, J.; Zhang, F.; Wong, M.H. Effect of nitrogen form and phosphorus source on the growth, nutrient uptake and rhizosphere soil property of Camellia sinensis L. Plant Soil 2000, 223, 65–73. [Google Scholar] [CrossRef]
- Cao, R.; Wu, F.Z.; Yang, W.Q.; Xu, Z.F.; Tani, B.; Wang, B.; Li, J.; Chang, C.H. Effects of altitudes on soil microbial biomass and enzyme activity in alpine-gorge regions. J. Appl. Ecol. 2016, 27, 1257–1264. [Google Scholar] [CrossRef]
- Cao, S.; Cao, G.; Feng, Q.; Han, G.; Lin, Y.; Yuan, J.; Wu, F.; Cheng, S. Alpine wetland ecosystem carbon sink and its controls at the Qinghai Lake. Environ. Earth Sci. 2017, 76, 210. [Google Scholar] [CrossRef]
- Zhang, H.; Wang, L.; Liu, H.; Zhao, J.; Li, G.; Wang, H.; Lai, X.; Li, J.; Xiu, W.; Yang, D. Nitrogen deposition combined with elevated precipitation is conducive to maintaining the stability of the soil fungal diversity on the Stipa baicalensis steppe. Soil Biol. Biochem. 2018, 117, 135–138. [Google Scholar] [CrossRef]
- Wang, X.; Feng, J.; Ao, G.; Qin, W.; Han, M.; Shen, Y.; Liu, M.; Chen, Y.; Zhu, B. Globally nitrogen addition alters soil microbial community structure, but has minor effects on soil microbial diversity and richness. Soil Biol. Biochem. 2023, 179, 108982. [Google Scholar] [CrossRef]
- Yang, Y.; Yang, Y.; Chen, X.; Liu, L.; Li, T.; Dou, Y.; Qiao, J.; Wang, Y.; An, S.; Chang, S.X. Nitrogen fertilization weakens the linkage between soil carbon and microbial diversity: A global meta-analysis. Glob. Chang. Biol. 2022, 28, 6446–6461. [Google Scholar] [CrossRef]
- Zhong, Y.; Liang, L.; Xu, R.; Xu, H.; Sun, L.; Liao, H. Intercropping tea plantations with soybean and rapeseed enhances nitrogen fixation through shifts in soil microbial communities. Front. Agr. Sci. Eng. 2022, 9, 344–355. [Google Scholar] [CrossRef]
- Juan Manuel, S.-Y. Xanthobacter autotrophicus an Endophytic Beneficial Bacterium for Wheat and Other Plants: A Short Review. In Current Trends in Wheat Research; Mahmood-ur-Rahman, A., Ed.; IntechOpen: London, UK, 2022; Chapter 4. [Google Scholar] [CrossRef]
- Ormeño-Orrillo, E.; Martínez-Romero, E. A Genomotaxonomy View of the Bradyrhizobium Genus. Front. Microbiol. 2019, 10, 450885. [Google Scholar] [CrossRef]
- Park, D.; Kim, H.; Yoon, S. Nitrous Oxide Reduction by an Obligate Aerobic Bacterium, Gemmatimonas aurantiaca Strain T-27. Appl. Environ. Microbiol. 2017, 83, e00502-17. [Google Scholar] [CrossRef] [PubMed]
- Sharma, V.; Bhattacharyya, S.; Kumar, R.; Kumar, A.; Ibañez, F.; Wang, J.; Guo, B.; Sudini, H.K.; Gopalakrishnan, S.; DasGupta, M.; et al. Molecular Basis of Root Nodule Symbiosis between Bradyrhizobium and ‘Crack-Entry’ Legume Groundnut (Arachis hypogaea L.). Plants 2020, 9, 276. [Google Scholar] [CrossRef] [PubMed]
- Zeffa, D.M.; Fantin, L.H.; Koltun, A.; de Oliveira, A.L.M.; Nunes, M.; Canteri, M.G.; Gonçalves, L.S.A. Effects of plant growth-promoting rhizobacteria on co-inoculation with Bradyrhizobium in soybean crop: A meta-analysis of studies from 1987 to 2018. PeerJ 2020, 8, e7905. [Google Scholar] [CrossRef] [PubMed]
- Nelson, M.B.; Martiny, A.C.; Martiny, J.B.H. Global biogeography of microbial nitrogen-cycling traits in soil. Proc. Natl. Acad. Sci. USA 2016, 113, 8033–8040. [Google Scholar] [CrossRef] [PubMed]
- Treseder, K.K.; Lennon, J.T. Fungal traits that drive ecosystem dynamics on land. Microbiol. Mol. Biol. Rev. 2015, 79, 243–262. [Google Scholar] [CrossRef]
- Jing, W.Y.; Jing, L.; Heng, G.J. Chemical transformation of soil nitrogen under the influence of iron: A review. J. China Agric. Univ. 2014, 2, 95–99. [Google Scholar] [CrossRef]
- Samarkin, V.A.; Madigan, M.T.; Bowles, M.W.; Casciotti, K.L.; Priscu, J.C.; McKay, C.P.; Joye, S.B. Abiotic nitrous oxide emission from the hypersaline Don Juan Pond in Antarctica. Nat. Geosci. 2010, 3, 341–344. [Google Scholar] [CrossRef]
- Chen, W.; Ren, K.; Isabwe, A.; Chen, H.; Liu, M.; Yang, J. Stochastic processes shape microeukaryotic community assembly in a subtropical river across wet and dry seasons. Microbiome 2019, 7, 138. [Google Scholar] [CrossRef]
- Gao, Z.; Han, M.; Hu, Y.; Li, Z.; Liu, C.; Wang, X.; Tian, Q.; Jiao, W.; Hu, J.; Liu, L.; et al. Effects of Continuous Cropping of Sweet Potato on the Fungal Community Structure in Rhizospheric Soil. Front. Microbiol. 2019, 10, 2269. [Google Scholar] [CrossRef]
- Li, A.; Wei, Y.; Sun, Z.; Fan, T.; Zhang, L. Analysis of bacterial and fungal community structure in replant strawberry rhizosphere soil with denaturing gradient gel electrophoresis. Afr. J. Biomed. Res. 2012, 11, 10962–10969. [Google Scholar] [CrossRef]
- Pervaiz, Z.H.; Iqbal, J.; Zhang, Q.; Chen, D.; Wei, H.; Saleem, M. Continuous Cropping Alters Multiple Biotic and Abiotic Indicators of Soil Health. Soil Syst. 2020, 4, 59. [Google Scholar] [CrossRef]
- Xu, Y.; Liu, J.; Liu, X.; Li, H.; Yang, Z.; Wang, H.; Huang, X.; Lan, L.; An, Y.; Li, L.; et al. Continuous cropping of alfalfa (Medicago sativa L.) reduces bacterial diversity and simplifies cooccurrence networks in aeolian sandy soil. Soil Ecol. Lett. 2022, 4, 131–143. [Google Scholar] [CrossRef]
- Ku, Y.; Han, X.; Lei, Y.; Zhang, M.; Zhao, Z. Different sensitivities and assembly mechanisms of the root-associated microbial communities of Robinia pseudoacacia to spatial variation at the regional scale. Plant Soil 2023, 486, 621–637. [Google Scholar] [CrossRef]
- Hu, Y.; Chen, M.; Yang, Z.; Cong, M.; Zhu, X.; Jia, H. Soil Microbial Community Response to Nitrogen Application on a Swamp Meadow in the Arid Region of Central Asia. Front. Microbiol. 2022, 12, 797306. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, Z.; Arafat, Y.; Lin, W. Studies on fungal communities and functional guilds shift in tea continuous cropping soils by high-throughput sequencing. Ann. Microbiol. 2020, 70, 7. [Google Scholar] [CrossRef]
- Zhang, H.; Yang, Y.; Mei, X.; Li, Y.; Wu, J.; Li, Y.; Wang, H.; Huang, H.; Yang, M.; He, X.; et al. Phenolic Acids Released in Maize Rhizosphere During Maize-Soybean Intercropping Inhibit Phytophthora Blight of Soybean. Front. Plant Sci. 2020, 11, 886. [Google Scholar] [CrossRef] [PubMed]
- Trivedi, P.; Leach, J.E.; Tringe, S.G.; Sa, T.; Singh, B.K. Plant-microbiome interactions: From community assembly to plant health. Nature reviews. Microbiology 2020, 18, 607–621. [Google Scholar] [CrossRef] [PubMed]
- Arafat, Y.; Ud Din, I.; Tayyab, M.; Jiang, Y.; Chen, T.; Cai, Z.; Zhao, H.; Lin, X.; Lin, W.; Lin, S. Soil Sickness in Aged Tea Plantation Is Associated with a Shift in Microbial Communities as a Result of Plant Polyphenol Accumulation in the Tea Gardens. Front. Plant Sci. 2020, 11, 601. [Google Scholar] [CrossRef]
- Zhu, B.; Li, Y.; Rensing, C.; Ye, J.; Qiu, J.; Li, Q.; Wu, L.; Lu, Q.; Lin, Y.; Jia, X. Improvement of phenolic acid autotoxicity in tea plantations by Pseudomonas fluorescens ZL22. J. Hazard. Mater. 2023, 458, 131957. [Google Scholar] [CrossRef]
- Dean, R.; Van Kan, J.A.; Pretorius, Z.A.; Hammond-Kosack, K.E.; Di Pietro, A.; Spanu, P.D.; Rudd, J.J.; Dickman, M.; Kahmann, R.; Ellis, J.; et al. The Top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 2012, 13, 414–430. [Google Scholar] [CrossRef]
- Gilardi, G.; Mocioni, M.; Gullino, M.L.; Guarnaccia, V. Curvularia americana and Curvularia tropicalis cause leaf and crown necrosis on Bermuda grass in Italy. Phytopathol. Mediterr. 2022, 61, 431–437. [Google Scholar] [CrossRef]
- Hou, Y.M.; Zhang, X.; Zhang, N.N.; Naklumpa, W.; Zhao, W.Y.; Liang, X.F.; Zhang, R.; Sun, G.Y.; Gleason, M.L. Genera Acremonium and Sarocladium Cause Brown Spot on Bagged Apple Fruit in China. Plant Dis. 2019, 103, 1889–1901. [Google Scholar] [CrossRef]
- Prasannath, K.; Shivas, R.G.; Galea, V.J.; Akinsanmi, O.A. Novel Botrytis and Cladosporium Species Associated with Flower Diseases of Macadamia in Australia. J. Fungi 2021, 7, 898. [Google Scholar] [CrossRef]
- Simão, R.C.; Gomes, S.L. Structure, expression, and functional analysis of the gene coding for calmodulin in the chytridiomycete Blastocladiella emersonii. J. Bacteriol. 2001, 183, 2280–2288. [Google Scholar] [CrossRef]
- Wu, H.-S.; Zhou, X.-D.; Shi, X.; Liu, Y.-D.; Wang, M.-Y.; Shang, X.-X.; Gu, D.-L.; Wang, W.-Z.; Wu, C.-W. In vitro responses of Fusarium oxysporum f. sp.niveum to phenolic acids in decaying watermelon tissues. Phytochem. Lett. 2014, 8, 171–178. [Google Scholar] [CrossRef]
- Yago, J.I.; Roh, J.H.; Bae, S.D.; Yoon, Y.N.; Kim, H.J.; Nam, M.H. The Effect of Seed-borne Mycoflora from Sorghum and Foxtail Millet Seeds on Germination and Disease Transmission. Mycobiology 2011, 39, 206–218. [Google Scholar] [CrossRef] [PubMed]
- Xie, H.; Chen, Z.; Feng, X.; Wang, M.; Luo, Y.; Wang, Y.; Xu, P. L-theanine exuded from Camellia sinensis roots regulates element cycling in soil by shaping the rhizosphere microbiome assembly. Sci. Total Environ. 2022, 837, 155801. [Google Scholar] [CrossRef]
- Arafat, Y.; Wei, X.; Jiang, Y.; Chen, T.; Saqib, H.S.A.; Lin, S.; Lin, W. Spatial Distribution Patterns of Root-Associated Bacterial Communities Mediated by Root Exudates in Different Aged Ratooning Tea Monoculture Systems. Int. J. Mol. Sci. 2017, 18, 1727. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Li, T.; Zheng, Z. Effect of tea plantation age on the distribution of soil organic carbon and nutrient within micro-aggregates in the hilly region of western Sichuan, China. Ecol. Eng. 2016, 90, 113–119. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shao, S.; Li, Y.; Li, Z.; Ma, X.; Zhu, Y.; Luo, Y.; Cai, P.; Jia, X.; Rensing, C.; Li, Q. Impact of Tea Tree Cultivation on Soil Microbiota, Soil Organic Matter, and Nitrogen Cycling in Mountainous Plantations. Agronomy 2024, 14, 638. https://doi.org/10.3390/agronomy14030638
Shao S, Li Y, Li Z, Ma X, Zhu Y, Luo Y, Cai P, Jia X, Rensing C, Li Q. Impact of Tea Tree Cultivation on Soil Microbiota, Soil Organic Matter, and Nitrogen Cycling in Mountainous Plantations. Agronomy. 2024; 14(3):638. https://doi.org/10.3390/agronomy14030638
Chicago/Turabian StyleShao, Shuaibo, Yuanping Li, Zhongwei Li, Xiaoxiao Ma, Yanqi Zhu, Yuqing Luo, Pumo Cai, Xiaoli Jia, Christopher Rensing, and Qisong Li. 2024. "Impact of Tea Tree Cultivation on Soil Microbiota, Soil Organic Matter, and Nitrogen Cycling in Mountainous Plantations" Agronomy 14, no. 3: 638. https://doi.org/10.3390/agronomy14030638




