Histone Epigenetic Signatures in Embryonic Limb Interdigital Cells Fated to Die
Abstract
:1. Introduction
2. Materials and Methods
2.1. Neutral Red Staining, TUNEL Assay, and Histochemical Detection of β-Gal Activity
2.2. Immunofluorescence and Confocal Microscopy
2.3. Experimental Manipulation of Interdigit Regression
2.4. In Situ Hybridization
2.5. Real Time Quantitative PCR (qPCR) for Gene Expression Analysis
3. Results
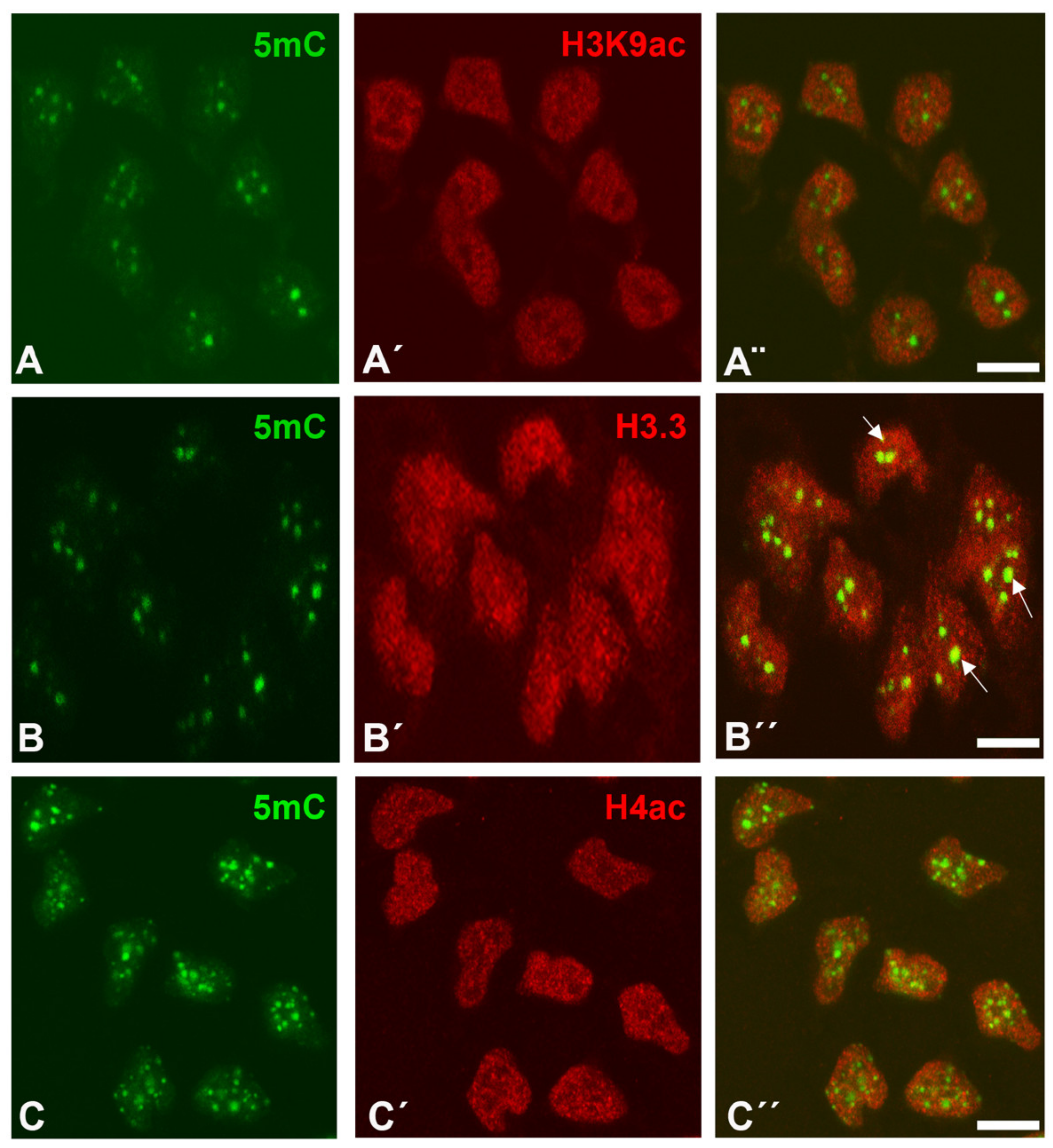
3.1. Post-Translational Histone Modifications (PTMs) of Interdigital Progenitors Fated to Die
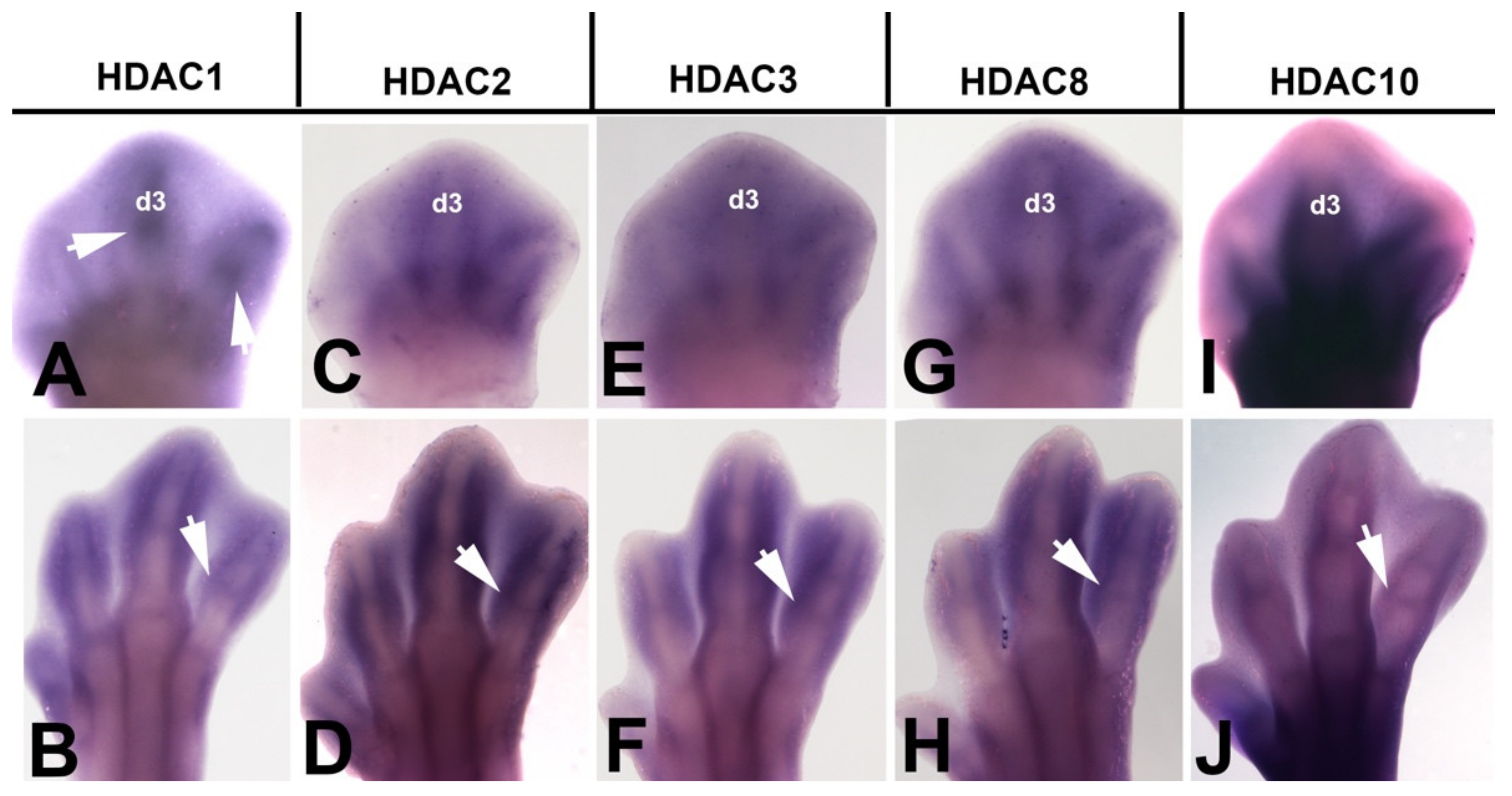
3.2. Expression Domains of HDACs in the Embryonic Limb
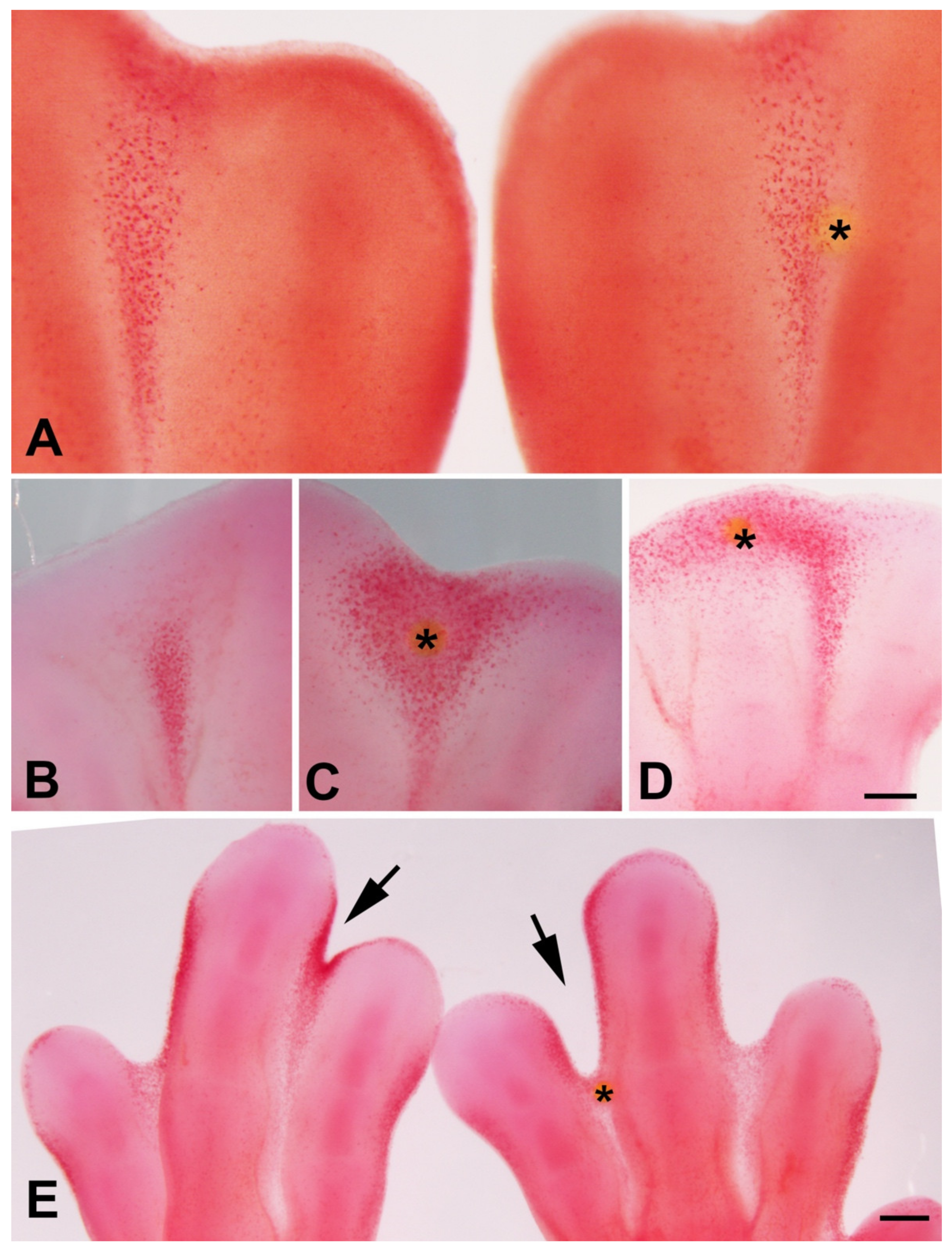
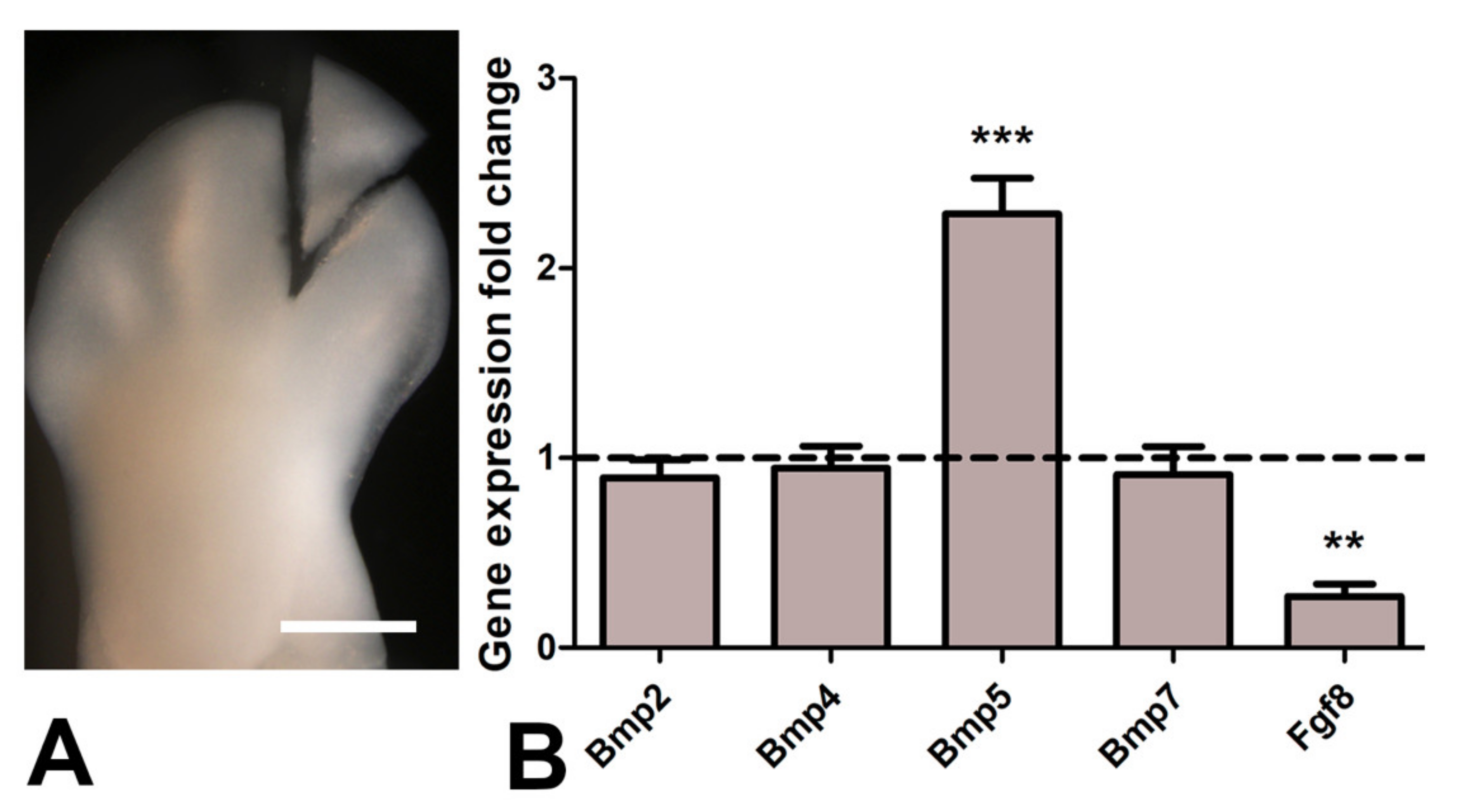
3.3. Inhibition of Histone Deacetylase and Cell Death
4. Discussion
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Spagnoli, A.; O’Rear, L.; Chandler, R.L.; Granero-Molto, F.; Mortlock, D.P.; Gorska, A.E.; Weis, J.A.; Longobardi, L.; Chytil, A.; Shimer, K.; et al. TGF-β signaling is essential for joint morphogenesis. J. Cell Biol. 2007, 117, 1105–1117. [Google Scholar] [CrossRef] [Green Version]
- Badugu, A.; Kraemer, C.; Germann, P.; Menshykau, D.; Iber, D. Digit patterning during limb development as a result of the BMP-receptor interaction. Sci. Rep. 2012, 2, 991. [Google Scholar] [CrossRef] [PubMed]
- Norrie, J.L.; Lewandowski, J.P.; Bouldin, C.M.; Amarnath, S.; Li, Q.; Vokes, M.S.; Ehrlich, L.I.R.; Harfe, B.D.; Vokes, S.A. Dynamics of BMP signaling in limb bud mesenchyme and polydactyly. Dev. Biol. 2014, 393, 270–281. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, M.; Chen, G.; Li, Y.-P. TGF-β and BMP signaling in osteoblast, skeletal development, and bone formation, homeostasis and disease. Bone Res. 2016, 4, 16009. [Google Scholar] [CrossRef] [PubMed]
- Montero, J.A.; Lorda-Diez, C.I.; Certal, A.C.; Moreno, N.; Rodriguez-Leon, J.; Torriglia, A.; Hurle, J.M. Coordinated and sequential activation of neutral and acidic DNases during interdigital cell death in the embryonic limb. Apoptosis 2010, 15, 1197–1210. [Google Scholar] [CrossRef]
- Storer, M.; Mas, A.; Robert-Moreno, A.; Pecoraro, M.; Ortells, M.C.; Di Giacomo, V.; Yosef, R.; Pilpel, N.; Krizhanovsky, V.; Sharpe, J.; et al. Senescence Is a Developmental Mechanism that Contributes to Embryonic Growth and Patterning. Cell 2013, 155, 1119–1130. [Google Scholar] [CrossRef] [Green Version]
- Zuzarte-Luis, V.; Montero, J.A.; Kawakami, Y.; Izpisua-Belmonte, J.C.; Hurle, J.M. Lysosomal cathepsins in embryonic programmed cell death. Dev. Biol. 2007, 301, 205–217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montero, J.A.; Lorda-Diez, C.I.; Hurle, J.M. Confluence of Cellular Degradation Pathways During Interdigital Tissue Remodeling in Embryonic Tetrapods. Front. Cell Dev. Biol. 2020, 8, 593761. [Google Scholar] [CrossRef] [PubMed]
- Salas-Vidal, E.; Lomelí, H.; Castro-Obregón, S.; Cuervo, R.; Escalante-Alcalde, D.; Covarrubias, L. Reactive Oxygen Species Participate in the Control of Mouse Embryonic Cell Death. Exp. Cell Res. 1998, 238, 136–147. [Google Scholar] [CrossRef] [PubMed]
- Schnabel, D.; Salas-Vidal, E.; Narváez, V.; Sánchez-Carbente, M.D.R.; Hernández-García, D.; Cuervo, R.; Covarrubias, L. Expression and regulation of antioxidant enzymes in the developing limb support a function of ROS in interdigital cell death. Dev. Biol. 2006, 291, 291–299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cordeiro, I.R.; Kabashima, K.; Ochi, H.; Munakata, K.; Nishimori, C.; Laslo, M.; Hanken, J.; Tanaka, M. Environmental Oxygen Exposure Allows for the Evolution of Interdigital Cell Death in Limb Patterning. Dev. Cell 2019, 50, 155–166.e4. [Google Scholar] [CrossRef]
- Montero, J.A.; Sanchez-Fernandez, C.; Diez, C.I.L.; Garcia-Porrero, J.A.; Hurle, J.M. DNA damage precedes apoptosis during the regression of the interdigital tissue in vertebrate embryos. Sci. Rep. 2016, 6, 35478. [Google Scholar] [CrossRef] [PubMed]
- Montero, J.A.; Lorda-Diez, C.; Sanchez-Fernandez, C.; Hurle, J. Cell death in the developing vertebrate limb: A Locally regulated mechanism contributing to musculoskeletal tissue morphogenesis and differentiation. Dev. Dyn. 2020. [Google Scholar] [CrossRef] [PubMed]
- Sidler, C.; Kovalchuk, O.; Kovalchuk, I. Epigenetic Regulation of Cellular Senescence and Aging. Front. Genet. 2017, 8, 138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanchez-Fernandez, C.; Lorda-Diez, C.I.; García-Porrero, J.A.; Montero, J.A.; Hurlé, J.M. UHRF genes regulate programmed interdigital tissue regression and chondrogenesis in the embryonic limb. Cell Death Dis. 2019, 10, 1–14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanchez-Fernandez, C.; Lorda-Diez, C.I.; Hurlé, J.M.; Montero, J.A. The methylation status of the embryonic limb skeletal progenitors determines their cell fate in chicken. Commun. Biol. 2020, 3, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Cavalli, G.; Heard, E. Advances in epigenetics link genetics to the environment and disease. Nat. Cell Biol. 2019, 571, 489–499. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Z.; Li, S.; Subramaniam, S.; Shyy, J.Y.-J.; Chien, S. Epigenetic Regulation: A New Frontier for Biomedical Engineers. Ann. Rev. Biomed. Eng. 2017, 19, 195–219. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Harris, C.J.; Zhong, Z.; Chen, W.; Liu, R.; Jia, B.; Wang, Z.; Li, S.; Jacobsen, S.E.; Du, J. Mechanistic insights into plant SUVH family H3K9 methyltransferases and their binding to context-biased non-CG DNA methylation. Proc. Natl. Acad. Sci. USA 2018, 115, E8793–E8802. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Nikolai, B.C.; Gates, L.A.; Jung, S.Y.; Siwak, E.B.; Zheng, Z.; Rice, A.P.; O’Malley, B.W.; Feng, Q. Crosstalk between histone modifications indicates that inhibition of arginine methyltransferase CARM1 activity reverses HIV latency. Nucleic Acids Res. 2017, 45, 9348–9360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramakrishnan, S.; Pokhrel, S.; Palani, S.; Pflueger, C.; Parnell, T.J.; Cairns, B.R.; Bhaskara, S.; Chandrasekharan, M.B. Counteracting H3K4 methylation modulators Set1 and Jhd2 co-regulate chromatin dynamics and gene transcription. Nat. Commun. 2016, 7, 11949. [Google Scholar] [CrossRef] [Green Version]
- Ortega, E.; Rengachari, S.; Ibrahim, Z.; Hoghoughi, N.; Gaucher, J.; Holehouse, A.S.; Khochbin, S.; Panne, D. Transcription factor dimerization activates the p300 acetyltransferase. Nat. Cell Biol. 2018, 562, 538–544. [Google Scholar] [CrossRef] [PubMed]
- Heintzman, N.D.; Stuart, R.K.; Hon, G.; Fu, Y.; Ching, C.W.; Hawkins, R.D.; Barrera, L.O.; Van Calcar, S.; Qu, C.; A Ching, K.; et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 2007, 39, 311–318. [Google Scholar] [CrossRef]
- Chrun, E.S.; Modolo, F.; Daniel, F.I. Histone modifications: A review about the presence of this epigenetic phenomenon in carcinogenesis. Pathol. Res. Pract. 2017, 213, 1329–1339. [Google Scholar] [CrossRef]
- Di Bernardo, G.; Squillaro, T.; Dell’Aversana, C.; Miceli, M.; Cipollaro, M.; Cascino, A.; Altucci, L.; Galderisi, U. Histone Deacetylase Inhibitors Promote Apoptosis and Senescence in Human Mesenchymal Stem Cells. Stem Cells Dev. 2009, 18, 573–582. [Google Scholar] [CrossRef] [PubMed]
- Milstone, Z.J.; Lawson, G.; Trivedi, C.M. Histone deacetylase 1 and 2 are essential for murine neural crest proliferation, pharyngeal arch development, and craniofacial morphogenesis. Dev. Dyn. 2017, 246, 1015–1026. [Google Scholar] [CrossRef] [Green Version]
- Samardzija, M.; Corna, A.; Gomez-Sintes, R.; Jarboui, M.A.; Armento, A.; Roger, J.E.; Petridou, E.; Haq, W.; Paquet-Durand, F.; Zrenner, E.; et al. HDAC inhibition ameliorates cone survival in retinitis pigmentosa mice. bioRxiv 2019. [Google Scholar] [CrossRef]
- Sancho-Pelluz, J.; Alavi, M.; Sahaboglu, A.; Kustermann, S.; Farinelli, P.; Azadi, S.; Van Veen, T.; Romero, F.J.; Paquet-Durand, F.; Ekström, P. Excessive HDAC activation is critical for neurodegeneration in the rd1 mouse. Cell Death Dis. 2010, 1, e24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daly, C.; Yin, J.; Kennedy, B.N. Histone Deacetylase: Therapeutic Targets in Retinal Degeneration. Adv. Exp. Med. Biol. 2015, 854, 455–461. [Google Scholar] [CrossRef]
- Zhao, W.; Dai, F.; Bonafede, A.; Schäfer, S.; Jung, M.; Yusuf, F.; Gamel, A.J.; Wang, J.; Brand-Saberi, B. Histone Deacetylase Inhibitor, Trichostatin A, Affects Gene Expression Patterns during Morphogenesis of Chicken Limb Buds in vivo. Cells Tissues Organs 2009, 190, 121–134. [Google Scholar] [CrossRef] [PubMed]
- Cotney, J.; Leng, J.; Oh, S.; Demare, L.E.; Reilly, S.K.; Gerstein, M.B.; Noonan, J.P. Chromatin state signatures associated with tissue-specific gene expression and enhancer activity in the embryonic limb. Genome Res. 2012, 22, 1069–1080. [Google Scholar] [CrossRef] [Green Version]
- Rosello-Diez, A.; Arques, C.G.; Delgado, I.; Giovinazzo, G.; Torres, M. Diffusible signals and epigenetic timing cooperate in late proximo-distal limb patterning. Development 2014, 141, 1534–1543. [Google Scholar] [CrossRef] [Green Version]
- Peluso, S.; Douglas, A.; Hill, A.; De Angelis, C.; Moore, B.L.; Grimes, G.; Petrovich, G.; Essafi, A.; Hill, R.E. Fibroblast growth factors (FGFs) prime the limb specific Shh enhancer for chromatin changes that balance histone acetylation mediated by E26 transformation-specific (ETS) factors. eLife 2017, 6, e28590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Allas, L.; Boumédiene, K.; Baugé, C. Epigenetic dynamic during endochondral ossification and articular cartilage development. Bone 2019, 120, 523–532. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Zhang, J.H.; Xu, M. DNA fragmentation in apoptosis. Cell Res. 2000, 10, 205–211. [Google Scholar] [CrossRef] [PubMed]
- Dobrucki, J.; Darzynkiewicz, Z. Chromatin condensation and sensitivity of DNA in situ to denaturation during cell cycle and apoptosis--a confocal microscopy study. Micron 2001, 32, 645–652. [Google Scholar] [CrossRef]
- Lu, X.; Wang, L.; Yu, C.; Yu, D.; Yu, G. Histone Acetylation Modifiers in the Pathogenesis of Alzheimer´s Disease. Front. Cell. Neurosci. 2015, 9, 226. [Google Scholar] [CrossRef] [Green Version]
- Allis, C.D.; Jenuwein, T. The molecular hallmarks of epigenetic control. Nat. Rev. Genet. 2016, 17, 487–500. [Google Scholar] [CrossRef]
- Gräff, J.; Tsai, L.-H. Histone acetylation: Molecular mnemonics on the chromatin. Nat. Rev. Neurosci. 2013, 14, 97–111. [Google Scholar] [CrossRef]
- Eslaminejad, M.B.; Fani, N.; Shahhoseini, M. Epigenetic regulation of osteogenic and chondrogenic differentiation of mesenchymal stem cells in culture. Cell J. 2013, 15, 1–10. [Google Scholar]
- Yang, X.-J.; Grégoire, S. Class II Histone Deacetylases: From Sequence to Function, Regulation, and Clinical Implication. Mol. Cell. Biol. 2005, 25, 2873–2884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haberland, M.; Carrer, M.; Mokalled, M.H.; Montgomery, R.L.; Olson, E.N. Redundant Control of Adipogenesis by Histone Deacetylases 1 and 2. J. Biol. Chem. 2010, 285, 14663–14670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoshida, M.; Horinouchi, S.; Beppu, T. Trichostatin A and trapoxin: Novel chemical probes for the role of histone acetylation in chromatin structure and function. BioEssays 1995, 17, 423–430. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanian, S.; Verner, E.; Buggy, J.J. Isoform-specific histone deacetylase inhibitors: The next step? Cancer Lett. 2009, 280, 211–221. [Google Scholar] [CrossRef]
- Chiba, T.; Yokosuka, O.; Arai, M.; Tada, M.; Fukai, K.; Imazeki, F.; Kato, M.; Seki, N.; Saisho, H. Identification of genes up-regulated by histone deacetylase inhibition with cDNA microarray and exploration of epigenetic alterations on hepatoma cells. J. Hepatol. 2004, 41, 436–445. [Google Scholar] [CrossRef] [PubMed]
- Montero, J.A.; Gañan, Y.; Macias, D.; Rodriguez-Leon, J.; Sanz-Ezquerro, J.J.; Merino, R.; Chimal-Monroy, J.; Nieto, M.A.; Hurle, J.M. Role of FGFs in the control of programmed cell death during limb development. Development 2001, 128, 2075–2084. [Google Scholar] [PubMed]
- Zhang, M.; Zhao, J.; Lv, Y.; Wang, W.; Feng, C.; Zou, W.; Su, L.; Jiao, J. Histone Variants and Histone Modifications in Neurogenesis. Trends Cell Biol. 2020, 30, 869–880. [Google Scholar] [CrossRef]
- Mikkelsen, T.S.; Ku, M.; Jaffe, D.B.; Issac, B.; Lieberman, E.; Giannoukos, G.; Alvarez, P.; Brockman, W.; Kim, T.-K.; Koche, R.P.; et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 2007, 448, 553–560. [Google Scholar] [CrossRef] [PubMed]
- Henikoff, S. Labile H3.3+H2A.Z nucleosomes mark ’nucleosome-free regions’. Nat. Genet. 2009, 41, 865–866. [Google Scholar] [CrossRef] [PubMed]
- Szenker, E.; Ray-Gallet, D.; Almouzni, G. The double face of the histone variant H3.3. Cell Res. 2011, 21, 421–434. [Google Scholar] [CrossRef] [Green Version]
- Barrero, M.J.; Izpisua Belmonte, J.C. Epigenetic Mechanisms Controlling Mesodermal Specification. In StemBook; Harvard Stem Cell Institute: Cambridge, MA, USA, 2012. [Google Scholar] [PubMed]
- Harikumar, A.; Meshorer, E. Chromatin remodeling and bivalent histone modifications in embryonic stem cells. Embo Rep. 2015, 16, 1609–1619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mas, G.; Blanco, E.; Ballaré, C.; Sansó, M.; Spill, Y.G.; Hu, D.; Aoi, Y.; Le Dily, F.; Shilatifard, A.; Marti-Renom, M.A.; et al. Promoter bivalency favors an open chromatin architecture in embryonic stem cells. Nat. Genet. 2018, 50, 1452–1462. [Google Scholar] [CrossRef] [PubMed]
- Goldberg, A.D.; Banaszynski, L.A.; Noh, K.-M.; Lewis, P.W.; Elsaesser, S.J.; Stadler, S.; Dewell, S.; Law, M.; Guo, X.; Li, X.; et al. Distinct Factors Control Histone Variant H3.3 Localization at Specific Genomic Regions. Cell 2010, 140, 678–691. [Google Scholar] [CrossRef] [Green Version]
- Hurle, J.M.; Ganan, Y. Formation of extra-digits induced by surgical removal of the apical ectodermal ridge of the chick embryo leg bud in the stages previous to the onset of interdigital cell death. Anat. Embryol. 1987, 176, 393–399. [Google Scholar] [CrossRef] [PubMed]
- Macias, D.; Gañan, Y.; Sampath, T.K.; Piedra, M.E.; Ros, M.A.; Hurle, J.M. Role of BMP-2 and OP-1 (BMP-7) in programmed cell death and skeletogenesis during chick limb development. Development 1997, 124, 1109–1117. [Google Scholar] [PubMed]
- Montero, J.A.; Lorda-Diez, C.I.; Gañan, Y.; Macias, D.; Hurle, J.M. Activin/TGFβ and BMP crosstalk determines digit chondrogenesis. Dev. Biol. 2008, 321, 343–356. [Google Scholar] [CrossRef] [Green Version]
- Zaret, K.S. Cell fate conversion: A chromatin remodeling checkpoint revealed. Cell Res. 2017, 27, 598–599. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, S.; Zhu, L.; Gao, Y.; Zhang, X.; Yan, Y.; Cen, J.; Li, R.; Zeng, R.; Liao, L.; Hou, C.; et al. Baf60b-mediated ATM-p53 activation blocks cell identity conversion by sensing chromatin opening. Cell Res. 2017, 27, 642–656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takata, H.; Hanafusa, T.; Mori, T.; Shimura, M.; Iida, Y.; Yoshikawa, K.; Yoshikawa, Y.; Maeshima, K. Chromatin Compaction Protects Genomic DNA from Radiation Damage. PLoS ONE 2013, 8, e75622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adam, S.; Polo, S.E.; Almouzni, G. Transcription Recovery after DNA Damage Requires Chromatin Priming by the H3.3 Histone Chaperone HIRA. Cell 2013, 155, 94–106. [Google Scholar] [CrossRef] [Green Version]
- Meyer, B.; Fabbrizi, M.R.; Raj, S.; Zobel, C.L.; Hallahan, D.E.; Sharma, G.G. Histone H3 lysine 9 acetylation obstructs ATM activation and promotes ionizing radiation sensitivity in normal stem cells. Stem Cell Rep. 2016, 7, 1013–1022. [Google Scholar] [CrossRef] [PubMed]
- Sharma, G.G.; So, S.; Gupta, A.; Kumar, R.; Cayrou, C.; Avvakumov, N.; Bhadra, U.; Pandita, R.K.; Porteus, M.H.; Chen, D.J.; et al. MOF and Histone H4 Acetylation at Lysine 16 Are Critical for DNA Damage Response and Double-Strand Break Repair. Mol. Cell. Biol. 2010, 30, 3582–3595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ge, Z.; Nair, D.; Guan, X.; Rastogi, N.; Freitas, M.A.; Parthun, M.R. Sites of Acetylation on Newly Synthesized Histone H4 Are Required for Chromatin Assembly and DNA Damage Response Signaling. Mol. Cell. Biol. 2013, 33, 3286–3298. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sanchez-Fernandez, C.; Lorda-Diez, C.I.; Duarte-Olivenza, C.; Hurle, J.M.; Montero, J.A. Histone Epigenetic Signatures in Embryonic Limb Interdigital Cells Fated to Die. Cells 2021, 10, 911. https://doi.org/10.3390/cells10040911
Sanchez-Fernandez C, Lorda-Diez CI, Duarte-Olivenza C, Hurle JM, Montero JA. Histone Epigenetic Signatures in Embryonic Limb Interdigital Cells Fated to Die. Cells. 2021; 10(4):911. https://doi.org/10.3390/cells10040911
Chicago/Turabian StyleSanchez-Fernandez, Cristina, Carlos I. Lorda-Diez, Cristina Duarte-Olivenza, Juan M. Hurle, and Juan A. Montero. 2021. "Histone Epigenetic Signatures in Embryonic Limb Interdigital Cells Fated to Die" Cells 10, no. 4: 911. https://doi.org/10.3390/cells10040911





