In Silico Analysis of the Longevity and Timeline of Individual Germinal Center Reactions in a Primary Immune Response
Abstract
:1. Introduction
2. Materials and Methods
2.1. Model for Simulation of Multiple GCs
2.2. Experimental Data Used
2.3. Initiation of Representative GCs
2.4. Calculation of the Number of GCs and GC Lifetime
2.5. GC Simulations with Varying Antigen Availability
2.6. GC Simulations with Multiple Epitopes
2.7. Time Independent Random Variations
3. Results
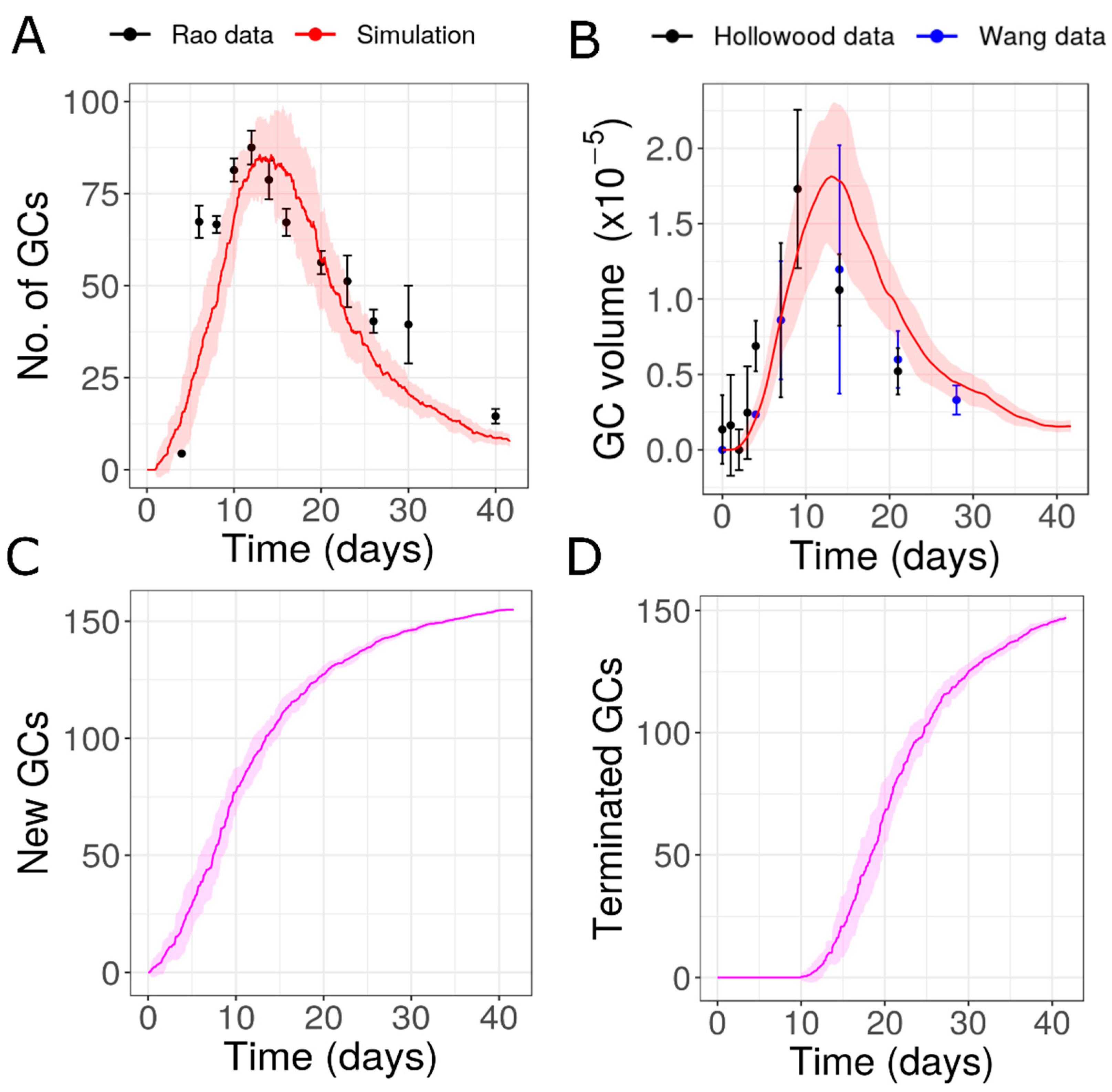
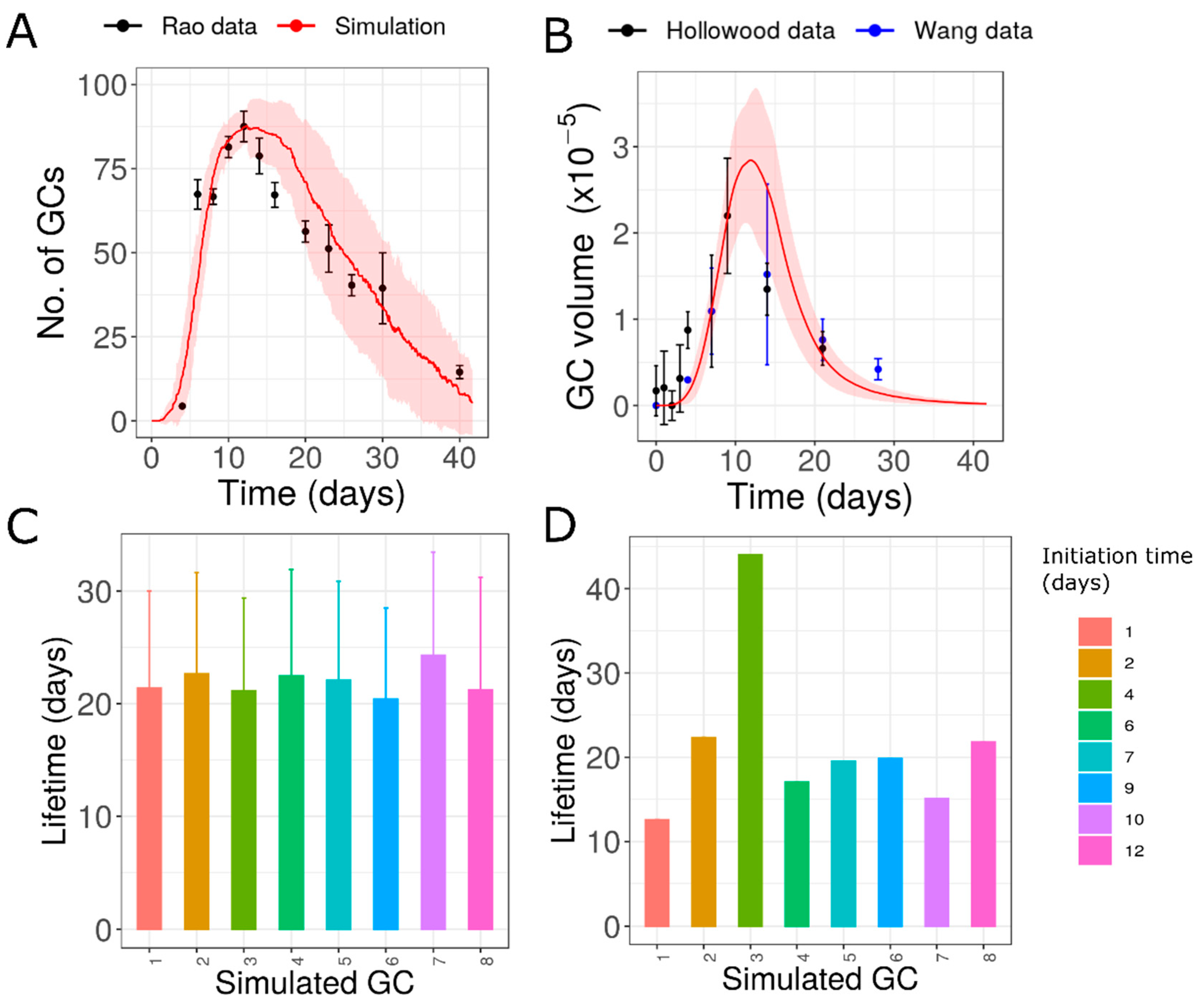
3.1. Extended Formation of New GCs with Similar Lifetimes (Hypothesis 2) Is Consistent with Data
3.2. Individual GCs Might Have Highly Variable Lifetimes
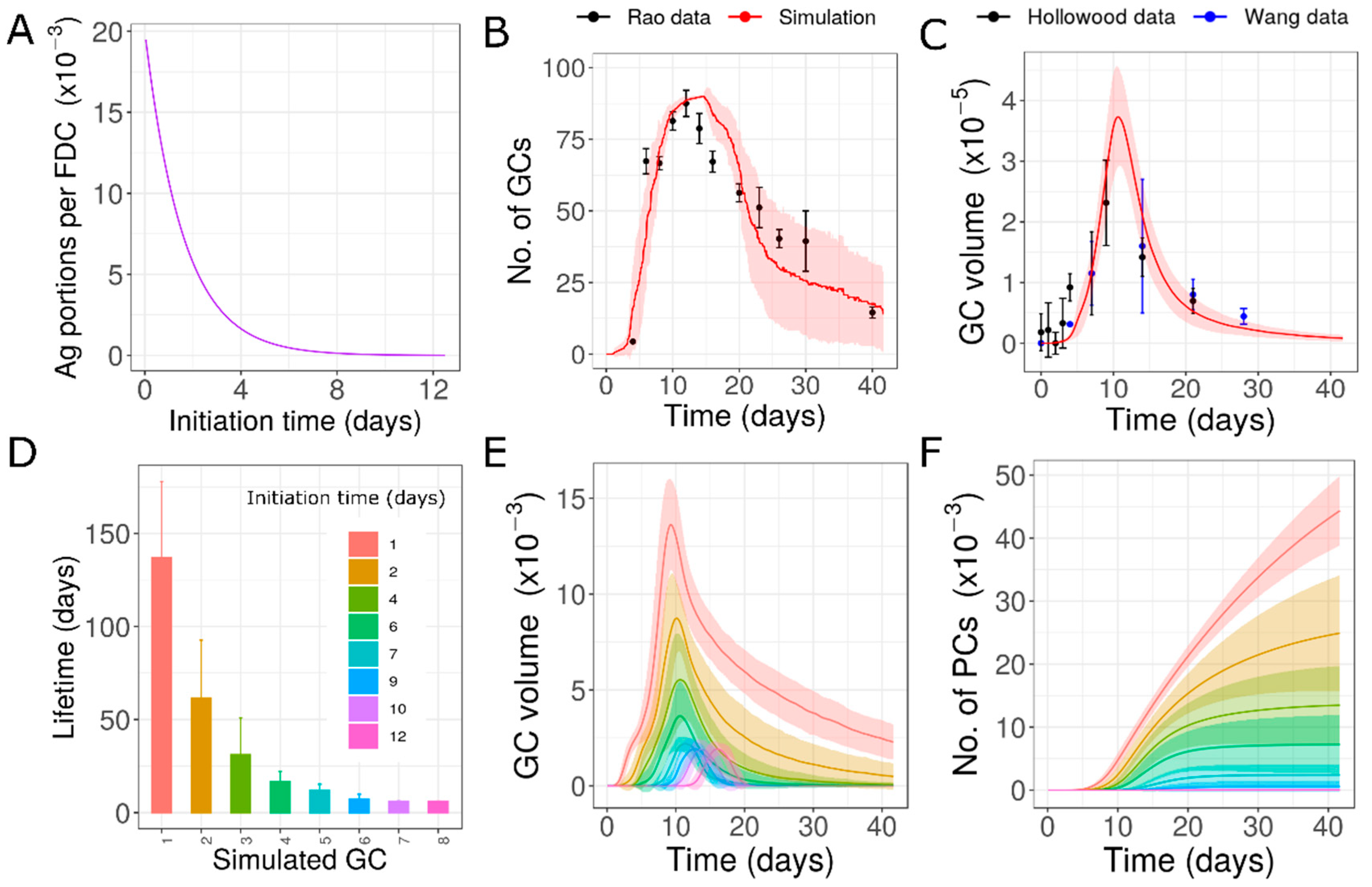
3.3. Variability Due to Different Antigen Availability (Hypothesis 3)
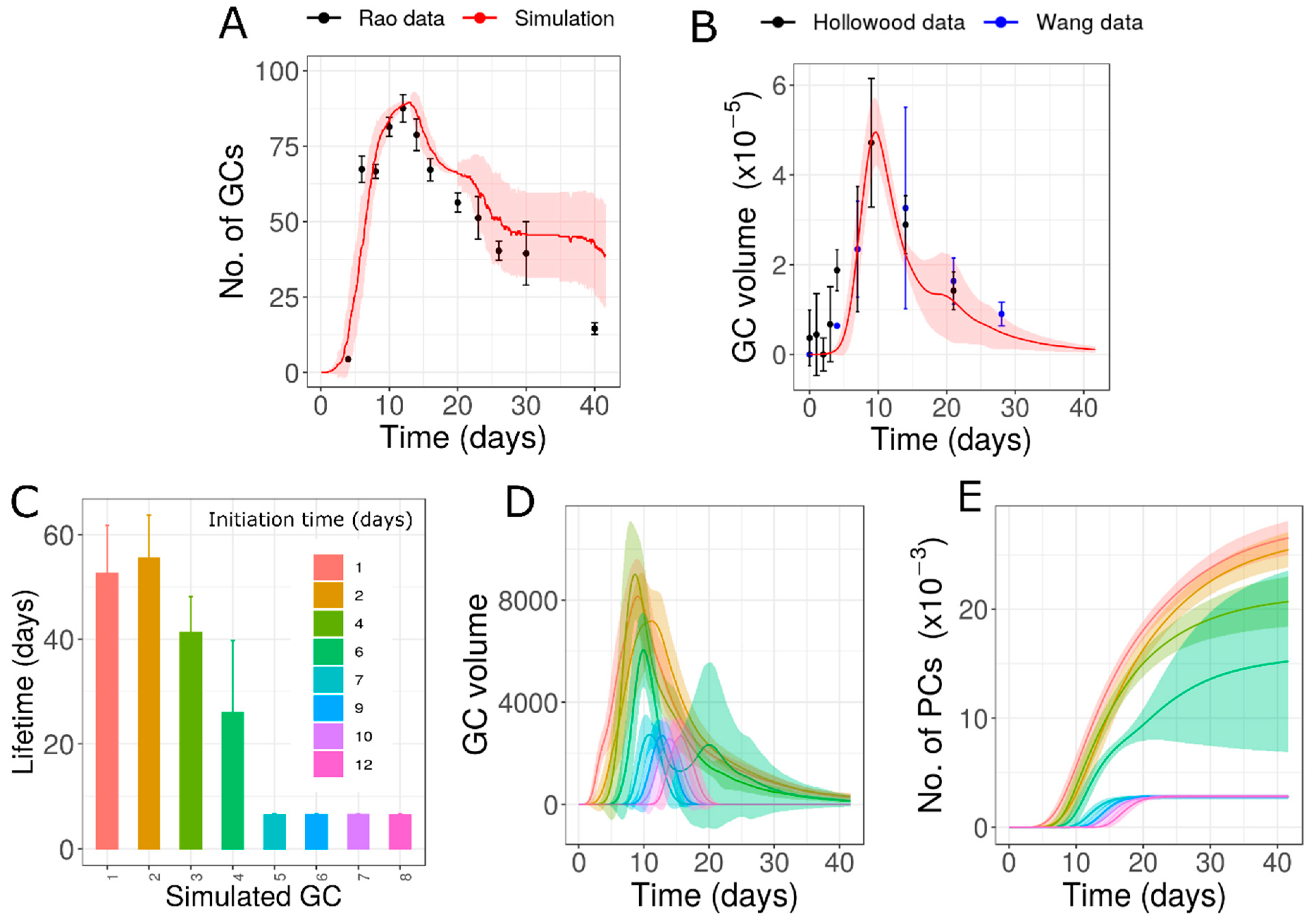
3.4. Lifetime Variability Due to Different Founder Cell Characteristics (Hypothesis 4)
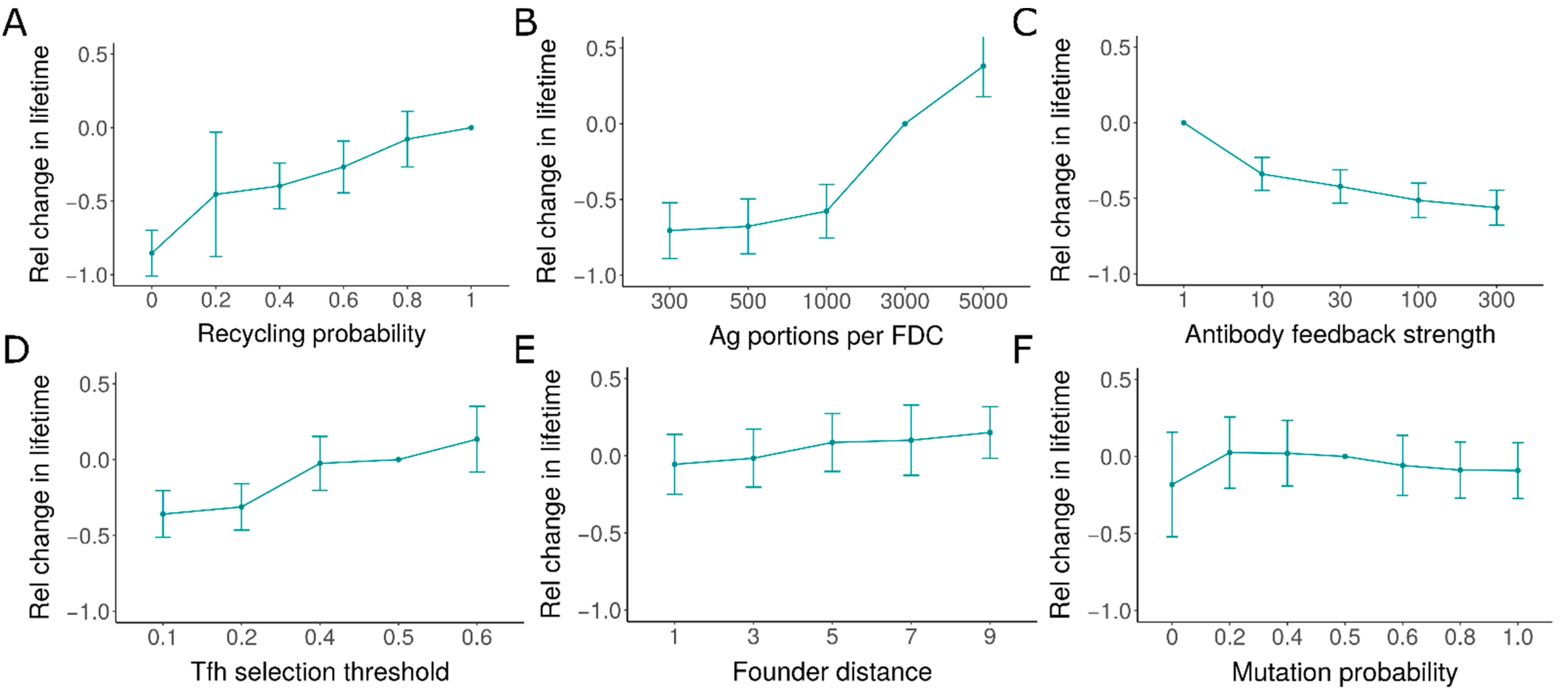
3.5. Initiation Time Independent Random Variation in GC Characteristics (Hypothesis 5)
3.6. Individual GCs Formed after NP-CGG Immunization
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- MacLennan, I.C.M. Germinal Centers. Annu. Rev. Immunol. 1994, 12, 117–139. [Google Scholar] [CrossRef]
- Victora, G.D.; Nussenzweig, M.C. Germinal Centers. Annu. Rev. Immunol. 2012, 30, 429–457. [Google Scholar] [CrossRef] [PubMed]
- Tas, J.M.J.; Mesin, L.; Pasqual, G.; Targ, S.; Jacobsen, J.T.; Mano, Y.M.; Chen, C.S.; Weill, J.-C.; Reynaud, C.-A.; Browne, E.P.; et al. Visualizing antibody affinity maturation in germinal centers. Science 2016, 351, 1048–1054. [Google Scholar] [CrossRef] [Green Version]
- Camacho, S.A.; Kosco-Vilbois, M.H.; Berek, C. The dynamic structure of the germinal center. Immunol. Today 1998, 19, 511–514. [Google Scholar] [CrossRef]
- Allen, C.D.C.; Ansel, K.M.; Low, C.; Lesley, R.; Tamamura, H.; Fujii, N.; Cyster, J.G. Germinal center dark and light zone organization is mediated by CXCR4 and CXCR5. Nat. Immunol. 2004, 5, 943–952. [Google Scholar] [CrossRef] [PubMed]
- Nossal, G.J.V. The molecular and cellular basis of affinity maturation in the antibody response. Cell 1992, 68, 1–2. [Google Scholar] [CrossRef]
- Victora, G.D.; Schwickert, T.A.; Fooksman, D.R.; Kamphorst, A.O.; Meyer-Hermann, M.; Dustin, M.L.; Nussenzweig, M.C. Germinal Center Dynamics Revealed by Multiphoton Microscopy with a Photoactivatable Fluorescent Reporter. Cell 2010, 143, 592–605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Finkin, S.; Hartweger, H.; Oliveira, T.Y.; Kara, E.E.; Nussenzweig, M.C. Protein Amounts of the MYC Transcription Factor Determine Germinal Center B Cell Division Capacity. Immunity 2019, 51, 324–336.e5. [Google Scholar] [CrossRef]
- Gitlin, A.D.; Shulman, Z.; Nussenzweig, M.C. Clonal selection in the germinal centre by regulated proliferation and hypermutation. Nature 2014, 509, 637–640. [Google Scholar] [CrossRef] [Green Version]
- Ersching, J.; Efeyan, A.; Mesin, L.; Jacobsen, J.T.; Pasqual, G.; Grabiner, B.C.; Dominguez-Sola, D.; Sabatini, D.M.; Victora, G.D. Germinal Center Selection and Affinity Maturation Require Dynamic Regulation of mTORC1 Kinase. Immunity 2017, 46, 1045–1058.e6. [Google Scholar] [CrossRef] [Green Version]
- Ise, W.; Fujii, K.; Shiroguchi, K.; Ito, A.; Kometani, K.; Takeda, K.; Kawakami, E.; Yamashita, K.; Suzuki, K.; Okada, T. T follicular helper cell-germinal center B cell interaction strength regulates entry into plasma cell or recycling germinal center cell fate. Immunity 2018, 48, 702–715. [Google Scholar] [CrossRef] [Green Version]
- Viant, C.; Weymar, G.H.J.; Escolano, A.; Chen, S.; Hartweger, H.; Cipolla, M.; Gazumyan, A.; Nussenzweig, M.C. Antibody Affinity Shapes the Choice between Memory and Germinal Center B Cell Fates. Cell 2020. [Google Scholar] [CrossRef]
- Hollowood, K.; Macartney, J. Cell kinetics of the germinal center reaction—A stathmokinetic study. Eur. J. Immunol. 1992, 22, 261–266. [Google Scholar] [CrossRef] [PubMed]
- Stoler-Barak, L.; Biram, A.; Davidzohn, N.; Addadi, Y.; Golani, O.; Shulman, Z. B cell dissemination patterns during the germinal center reaction revealed by whole-organ imaging. J. Exp. Med. 2019, 216, 2515–2530. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Zhang, J.; Lane, P.J.L.; Chan, E.Y.; Maclennan, I.C.M. Sites of specific B cell activation in primary and secondary responses to T cell-dependent and T cell-independent antigens. Eur. J. Immunol. 1991, 21, 2951–2962. [Google Scholar] [CrossRef]
- Mesin, L.; Ersching, J.; Victora, G.D. Germinal Center B Cell Dynamics. Immunity 2016, 45, 471–482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bachmann, M.F.; Odermatt, B.; Hengartner, H.; Zinkernagel, R.M. Induction of long-lived germinal centers associated with persisting antigen after viral infection. J. Exp. Med. 1996, 183, 2259–2269. [Google Scholar] [CrossRef] [Green Version]
- McHeyzer-Williams, M.G.; Ahmed, R. B cell memory and the long-lived plasma cell. Curr. Opin. Immunol. 1999, 11, 172–179. [Google Scholar] [CrossRef]
- Kelly, H.G.; Tan, H.-X.; Juno, J.A.; Esterbauer, R.; Ju, Y.; Jiang, W.; Wimmer, V.C.; Duckworth, B.C.; Groom, J.R.; Caruso, F. Self-assembling influenza nanoparticle vaccines drive extended germinal center activity and memory B cell maturation. JCI Insight 2020, 5, e136653. [Google Scholar] [CrossRef]
- Pedersen, G.K.; Wørzner, K.; Andersen, P.; Christensen, D. Vaccine Adjuvants Differentially Affect Kinetics of Antibody and Germinal Center Responses. Front. Immunol. 2020, 11, 2302. [Google Scholar] [CrossRef]
- Garg, A.K.; Desikan, R.; Dixit, N.M. Preferential Presentation of High-Affinity Immune Complexes in Germinal Centers Can Explain How Passive Immunization Improves the Humoral Response. Cell Rep. 2019, 29, 3946–3957.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cirelli, K.M.; Carnathan, D.G.; Nogal, B.; Martin, J.T.; Rodriguez, O.L.; Upadhyay, A.A.; Enemuo, C.A.; Gebru, E.H.; Choe, Y.; Viviano, F.; et al. Slow Delivery Immunization Enhances HIV Neutralizing Antibody and Germinal Center Responses via Modulation of Immunodominance. Cell 2019, 177, 1153–1171.e28. [Google Scholar] [CrossRef]
- Cirelli, K.M.; Crotty, S. Germinal center enhancement by extended antigen availability. Curr. Opin. Immunol. 2017, 47, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Tam, H.H.; Melo, M.B.; Kang, M.; Pelet, J.M.; Ruda, V.M.; Foley, M.H.; Hu, J.K.; Kumari, S.; Crampton, J.; Baldeon, A.D. Sustained antigen availability during germinal center initiation enhances antibody responses to vaccination. Proc. Natl. Acad. Sci. USA 2016, 113, E6639–E6648. [Google Scholar] [CrossRef] [Green Version]
- De Boer, R.J.; Perelson, A.S. How Germinal Centers Evolve Broadly Neutralizing Antibodies: The Breadth of the Follicular Helper T Cell Response. J. Virol. 2017, 91, e00983-17. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Mata-Fink, J.; Kriegsman, B.; Hanson, M.; Irvine, D.J.; Eisen, H.N.; Burton, D.R.; Wittrup, K.D.; Kardar, M.; Chakraborty, A.K. Manipulating the Selection Forces during Affinity Maturation to Generate Cross-Reactive HIV Antibodies. Cell 2015, 160, 785–797. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacob, J.; Kassir, R.; Kelsoe, G. In situ studies of the primary immune response to (4-hydroxy-3-nitrophenyl) acetyl. I. The architecture and dynamics of responding cell populations. J. Exp. Med. 1991, 173, 1165–1175. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hollowood, K.; Goodlad, J.R. Germinal centre cell kinetics. J. Pathol. A J. Pathol. Soc. Gt. Br. Irel. 1998, 185, 229–233. [Google Scholar] [CrossRef]
- Kulkarni, R.R.; Rasheed, M.A.U.; Bhaumik, S.K.; Ranjan, P.; Cao, W.; Davis, C.; Marisetti, K.; Thomas, S.; Gangappa, S.; Sambhara, S.; et al. Activation of the RIG-I pathway during influenza vaccination enhances the germinal center reaction, promotes T follicular helper cell induction, and provides a dose-sparing effect and protective immunity. J. Virol. 2014, 88, 13990–14001. [Google Scholar] [CrossRef] [Green Version]
- Quemeneur, L.; Angeli, V.; Chopin, M.; Jessberger, R. SWAP-70 deficiency causes high-affinity plasma cell generation despite impaired germinal center formation. Blood J. Am. Soc. Hematol. 2008, 111, 2714–2724. [Google Scholar] [CrossRef] [Green Version]
- Rao, S.P.; Vora, K.A.; Manser, T. Differential Expression of the Inhibitory IgG Fc Receptor FcγRIIB on Germinal Center Cells: Implications for Selection of High-Affinity B Cells. J. Immunol. 2002, 169, 1859–1868. [Google Scholar] [CrossRef] [Green Version]
- Szakal, A.K.; Taylor, J.K.; Smith, J.P.; Kosco, M.H.; Burton, G.F.; Tew, J.J. Kinetics of germinal center development in lymph nodes of young and aging immune mice. Anat. Rec. 1990, 227, 475–485. [Google Scholar] [CrossRef] [PubMed]
- Horie, K.; Chen, D.; Hoshi, H. Development of immune complex trapping: Experimental study of lymphoid follicles and germinal centers newly induced by exogenous stimulants in mouse popliteal lymph nodes. Histol. Histopathol. 1999, 14, 11–21. [Google Scholar] [PubMed]
- Firl, D.J.; Degn, S.E.; Padera, T.; Carroll, M.C. Capturing change in clonal composition amongst single mouse germinal centers. Elife 2018, 7, e33051. [Google Scholar] [CrossRef] [PubMed]
- Buchauer, L.; Wardemann, H. Calculating germinal centre reactions. Curr. Opin. Syst. Biol. 2019, 18, 1–8. [Google Scholar] [CrossRef]
- Meyer-Hermann, M.; Figge, M.T.; Toellner, K.-M. Germinal centres seen through the mathematical eye: B-cell models on the catwalk. Trends Immunol. 2009, 30, 157–164. [Google Scholar] [CrossRef]
- Meyer-Hermann, M.; Mohr, E.; Pelletier, N.; Zhang, Y.; Victora, G.D.; Toellner, K.-M. A theory of germinal center B cell selection, division, and exit. Cell Rep. 2012, 2, 162–174. [Google Scholar] [CrossRef] [Green Version]
- Meyer-Hermann, M. Overcoming the dichotomy of quantity and quality in antibody responses. J. Immunol. 2014, 193, 5414–5419. [Google Scholar] [CrossRef] [Green Version]
- Binder, S.C.; Meyer-Hermann, M. Implications of intravital imaging of murine germinal centers on the control of B cell selection and division. Front. Immunol. 2016, 7, 593. [Google Scholar] [CrossRef] [Green Version]
- Meyer-Hermann, M.; Binder, S.C.; Mesin, L.; Victora, G.D. Computer Simulation of Multi-Color Brainbow Staining and Clonal Evolution of B Cells in Germinal Centers. Front. Immunol. 2018, 9, 2020. [Google Scholar] [CrossRef]
- Perelson, A.S.; Oster, G.F. Theoretical studies of clonal selection: Minimal antibody repertoire size and reliability of self-non-self discrimination. J. Theor. Biol. 1979, 81, 645–670. [Google Scholar] [CrossRef]
- Berek, C.; Milstein, C. Mutation Drift and Repertoire Shift in the Maturation of the Immune Response. Immunol. Rev. 1987, 96, 23–41. [Google Scholar] [CrossRef]
- Thaunat, O.; Granja, A.G.; Barral, P.; Filby, A.; Montaner, B.; Collinson, L.; Martinez-Martin, N.; Harwood, N.E.; Bruckbauer, A.; Batista, F.D. Asymmetric Segregation of Polarized Antigen on B Cell Division Shapes Presentation Capacity. Science 2012, 335, 475–479. [Google Scholar] [CrossRef] [PubMed]
- Al-Qahtani, A.; Xu, Z.; Zan, H.; Walsh, C.M.; Casali, P. A role for DRAK2 in the germinal center reaction and the antibody response. Autoimmunity 2008, 41, 341–352. [Google Scholar] [CrossRef]
- Wang, Y.; Carter, R.H. CD19 regulates B cell maturation, proliferation, and positive selection in the FDC zone of murine splenic germinal centers. Immunity 2005, 22, 749–761. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meyer-Hermann, M. Injection of Antibodies against Immunodominant Epitopes Tunes Germinal Centers to Generate Broadly Neutralizing Antibodies. Cell Rep. 2019, 29, 1066–1073.e5. [Google Scholar] [CrossRef] [Green Version]
- ISO/IEC. ISO International Standard ISO/IEC 14882:2014(E)–Programming Language C++. Available online: https://isocpp.org/std/the-standard (accessed on 8 July 2021).
- R Core Team. A Language and Environment for Statistical Computing. Available online: https://www.R-project.org/ (accessed on 8 July 2021).
- Wickham, H. ggplot2: Elegant Graphics for Data Analysis. Available online: https://ggplot2.tidyverse.org (accessed on 8 July 2021).
- Hanna, M.G., Jr.; Francis, M.W.; Peters, L.C. Localization of 125I-labelled antigen in germinal centres of mouse spleen: Effects of competitive injection of specific or non-cross-reacting antigen. Immunology 1968, 15, 75–91. [Google Scholar]
- Tew, J.G.; Mandel, T.E.; Rice, P.L. Immune elimination and immune retention: The relationship between antigen retained in the foot and the elicitation of foodpad swelling. Immunology 1980, 40, 425–433. [Google Scholar]
- Zhang, Y.; Meyer-Hermann, M.; George, L.A.; Figge, M.T.; Khan, M.; Goodall, M.; Young, S.P.; Reynolds, A.; Falciani, F.; Waisman, A. Germinal center B cells govern their own fate via antibody feedback. J. Exp. Med. 2013, 210, 457–464. [Google Scholar] [CrossRef] [Green Version]
- Arulraj, T.; Binder, S.C.; Robert, P.A.; Meyer-Hermann, M. Synchronous Germinal Center Onset Impacts the Efficiency of Antibody Responses. Front. Immunol. 2019, 10, 2116. [Google Scholar] [CrossRef]
- Bergen, M.J.; Pan, C.-H.; Greer, C.E.; Legg, H.S.; Polo, J.M.; Griffin, D.E. Comparison of the immune responses induced by chimeric alphavirus-vectored and formalin-inactivated alum-precipitated measles vaccines in mice. PLoS ONE 2010, 5, e10297. [Google Scholar] [CrossRef] [Green Version]
- Shinall, S.M.; Gonzalez-Fernandez, M.; Noelle, R.J.; Waldschmidt, T.J. Identification of murine germinal center B cell subsets defined by the expression of surface isotypes and differentiation antigens. J. Immunol. 2000, 164, 5729–5738. [Google Scholar] [CrossRef] [Green Version]
- Wolniak, K.L.; Noelle, R.J.; Waldschmidt, T.J. Characterization of (4-Hydroxy-3-Nitrophenyl)Acetyl (NP)-Specific Germinal Center B Cells and Antigen-Binding B220−Cells after Primary NP Challenge in Mice. J. Immunol. 2006, 177, 2072–2079. [Google Scholar] [CrossRef] [Green Version]
- Dijkstra, C.D.; Van Tilburg, N.J.; Döpp, E.A. Ontogenetic aspects of immune-complex trapping in the spleen and popliteal lymph nodes of the rat. Cell Tissue Res. 1982, 223, 545–552. [Google Scholar] [CrossRef]
- Baumjohann, D.; Preite, S.; Reboldi, A.; Ronchi, F.; Ansel, K.M.; Lanzavecchia, A.; Sallusto, F. Persistent Antigen and Germinal Center B Cells Sustain T Follicular Helper Cell Responses and Phenotype. Immunity 2013, 38, 596–605. [Google Scholar] [CrossRef] [Green Version]
- Molari, M.; Eyer, K.; Baudry, J.; Cocco, S.; Monasson, R. Quantitative modeling of the effect of antigen dosage on B-cell affinity distributions in maturating germinal centers. Elife 2020, 9, e55678. [Google Scholar] [CrossRef]
- Mesin, L.; Schiepers, A.; Ersching, J.; Barbulescu, A.; Cavazzoni, C.B.; Angelini, A.; Okada, T.; Kurosaki, T.; Victora, G.D. Restricted Clonality and Limited Germinal Center Reentry Characterize Memory B Cell Reactivation by Boosting. Cell 2020, 180, 92–106.e11. [Google Scholar] [CrossRef] [Green Version]
- Dogan, I.; Bertocci, B.; Vilmont, V.; Delbos, F.; Mégret, J.; Storck, S.; Reynaud, C.-A.; Weill, J.-C. Multiple layers of B cell memory with different effector functions. Nat. Immunol. 2009, 10, 1292–1299. [Google Scholar] [CrossRef]
- Shulman, Z.; Gitlin, A.D.; Targ, S.; Jankovic, M.; Pasqual, G.; Nussenzweig, M.C.; Victora, G.D. T follicular helper cell dynamics in germinal centers. Science 2013, 341, 673–677. [Google Scholar] [CrossRef] [Green Version]
- Weisel, F.J.; Zuccarino-Catania, G.V.; Chikina, M.; Shlomchik, M.J. A temporal switch in the germinal center determines differential output of memory B and plasma cells. Immunity 2016, 44, 116–130. [Google Scholar] [CrossRef] [Green Version]
- Gatto, D.; Martin, S.W.; Bessa, J.; Pellicioli, E.; Saudan, P.; Hinton, H.J.; Bachmann, M.F. Regulation of memory antibody levels: The role of persisting antigen versus plasma cell life span. J. Immunol. 2007, 178, 67–76. [Google Scholar] [CrossRef] [Green Version]
- Schwickert, T.A.; Lindquist, R.L.; Shakhar, G.; Livshits, G.; Skokos, D.; Kosco-Vilbois, M.H.; Dustin, M.L.; Nussenzweig, M.C. In vivo imaging of germinal centres reveals a dynamic open structure. Nature 2007, 446, 83–87. [Google Scholar] [CrossRef]
- Havenar-Daughton, C.; Carnathan, D.G.; Boopathy, A.V.; Upadhyay, A.A.; Murrell, B.; Reiss, S.M.; Enemuo, C.A.; Gebru, E.H.; Choe, Y.; Dhadvai, P.; et al. Rapid Germinal Center and Antibody Responses in Non-human Primates after a Single Nanoparticle Vaccine Immunization. Cell Rep. 2019, 29, 1756–1766.e8. [Google Scholar] [CrossRef] [Green Version]
- Adachi, Y.; Onodera, T.; Yamada, Y.; Daio, R.; Tsuiji, M.; Inoue, T.; Kobayashi, K.; Kurosaki, T.; Ato, M.; Takahashi, Y. Distinct germinal center selection at local sites shapes memory B cell response to viral escape. J. Exp. Med. 2015, 212, 1709–1723. [Google Scholar] [CrossRef]


| Hypothesis No. | Hypothesis | Motivation | Fitting the Data | Outcome | Cost |
|---|---|---|---|---|---|
| 1 | GCs with similar characteristics (with new GCs until 12 days post immunization) | Basic assumption | No | Simulated GCs shutdown within a narrow period of time | - |
| 2 | GCs with similar characteristics (with new GCs for extended time periods) | Unknown | Yes | Prolonged contraction phase due to extended formation of new GCs | 10.777 |
| 3 | Decrease in initial antigen concentration as GC formation is delayed | Rapid clearance of free antigen over time after immunization [50,51] | Yes | Early formed GCs are long-lived and late ones are short-lived | 7.198 |
| 4 | Multiple epitopes with different founder cell specificities | Due to memory cells seeding late formed GCs or differences in the sequence/timing of activation of different clones | Yes | Different lifetimes in GCs due to unequal epitope proportions and different specificities | 11.645 or 7.262 |
| 5 | Initiation time independent variation (antigen concentration) | Unknown | Yes | In each simulation, individual GCs have large variability in lifetimes when compared to each other. GC lifetime is not correlated with the initiation time | 10.027 |
| 6 | Antibody feedback with single or multiple epitopes | Antibodies might be exchanged between GCs and they can influence kinetics [52,53] | No | Does not exactly reproduce data despite resulting in some variability | - |
| 7 | No. of founder cells | Unknown | No | Insufficient lifetime variability | - |
| 8 | Founder cell affinities | Due to memory cells entering late formed GCs or differences in the sequence/timing of activation | No | Insufficient lifetime variability | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Arulraj, T.; Binder, S.C.; Meyer-Hermann, M. In Silico Analysis of the Longevity and Timeline of Individual Germinal Center Reactions in a Primary Immune Response. Cells 2021, 10, 1736. https://doi.org/10.3390/cells10071736
Arulraj T, Binder SC, Meyer-Hermann M. In Silico Analysis of the Longevity and Timeline of Individual Germinal Center Reactions in a Primary Immune Response. Cells. 2021; 10(7):1736. https://doi.org/10.3390/cells10071736
Chicago/Turabian StyleArulraj, Theinmozhi, Sebastian C. Binder, and Michael Meyer-Hermann. 2021. "In Silico Analysis of the Longevity and Timeline of Individual Germinal Center Reactions in a Primary Immune Response" Cells 10, no. 7: 1736. https://doi.org/10.3390/cells10071736






