Metabolic Alterations in a Drosophila Model of Parkinson’s Disease Based on DJ-1 Deficiency
Abstract
:1. Introduction
2. Materials and Methods
2.1. Drosophila Strains
2.2. Metabolite Extraction
2.3. NMR Analysis
2.4. Metabolite Assignment and Quantification
2.5. Measurement of ATP Levels
2.6. Enzymatic Activity Assays
2.7. RT-qPCR Analyses
2.8. Statistical Analysis and Data Representation
3. Results
3.1. Impact of DJ-1β Loss on the General Metabolic Profile
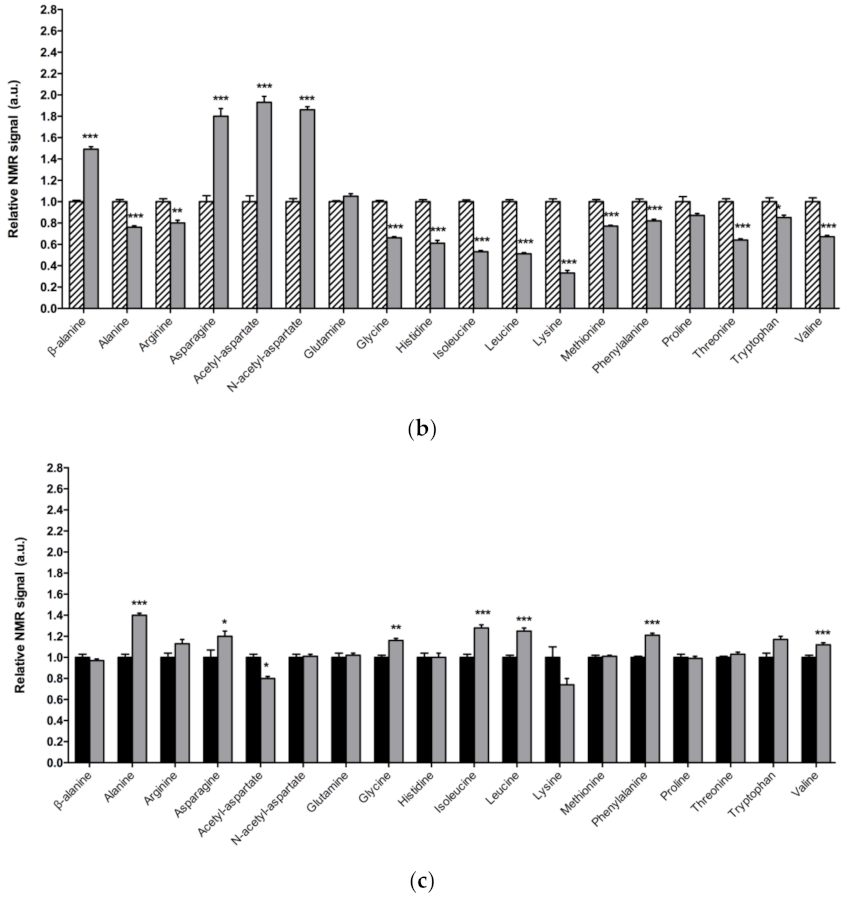
3.2. Alterations in Amino Acid Content in DJ-1β Mutant Flies
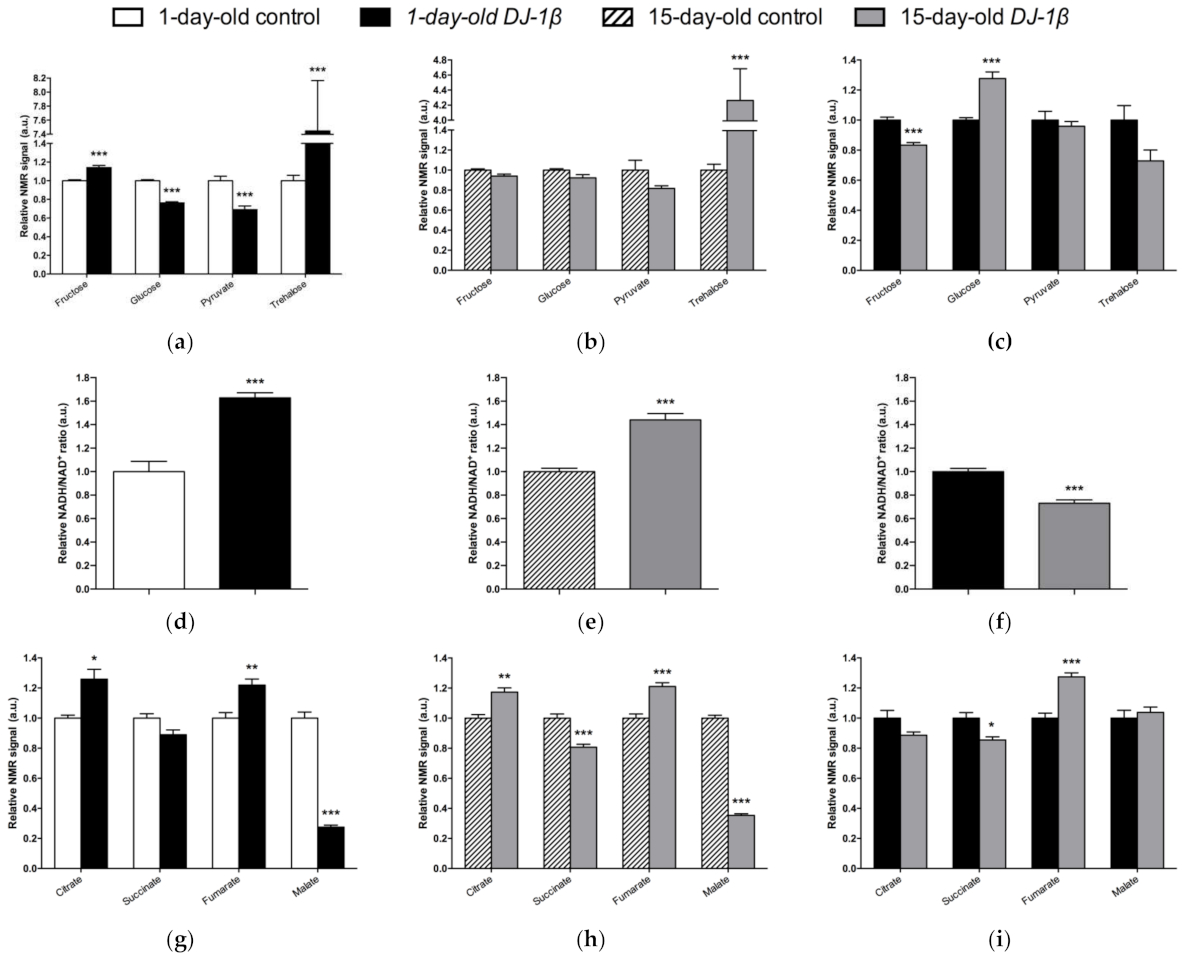
3.3. DJ-1β Deficiency Leads to Changes in Carbohydrate Metabolism
3.4. Switch from TCA Cycle to Glycolysis in DJ-1β Mutant Flies
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Castillo, X.; Castro-Obregón, S.; Gutiérrez-Becker, B.; Gutiérrez-Ospina, G.; Karalis, N.; Khalil, A.A.; Lopez-Noguerola, J.S.; Rodríguez, L.L.; Martínez-Martínez, E.; Perez-Cruz, C.; et al. Re-thinking the etiological framework of neurodegeneration. Front. Neurosci. 2019, 13, 728. [Google Scholar] [CrossRef] [Green Version]
- Bloem, B.R.; Okun, M.S.; Klein, C. Parkinson’s disease. Lancet 2021, 397, 2284–2303. [Google Scholar] [CrossRef]
- Tolosa, E.; Garrido, A.; Scholz, S.W.; Poewe, W. Challenges in the diagnosis of Parkinson’s disease. Lancet Neurol. 2021, 20, 385–397. [Google Scholar] [CrossRef]
- Poewe, W.; Seppi, K.; Tanner, C.M.; Halliday, G.M.; Brundin, P.; Volkmann, J.; Schrag, A.-E.; Lang, A.E. Parkinson disease. Nat. Rev. Dis. Prim. 2017, 3, 17013. [Google Scholar] [CrossRef] [PubMed]
- Panicker, N.; Ge, P.; Dawson, V.L.; Dawson, T.M. The cell biology of Parkinson’s disease. J. Cell Biol. 2021, 220, e202012095. [Google Scholar] [CrossRef]
- Maiti, P.; Manna, J.; Dunbar, G.L. Current understanding of the molecular mechanisms in Parkinson’s disease: Targets for potential treatments. Transl. Neurodegener. 2017, 6, 28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Solana-Manrique, C.; Sanz, F.J.; Ripollés, E.; Bañó, M.C.; Torres, J.; Muñoz-Soriano, V.; Paricio, N. Enhanced activity of glycolytic enzymes in Drosophila and human cell models of Parkinson’s disease based on DJ-1 deficiency. Free Radic. Biol. Med. 2020, 158, 137–148. [Google Scholar] [CrossRef] [PubMed]
- Anandhan, A.; Jacome, M.S.; Lei, S.; Hernandez-Franco, P.; Pappa, A.; Panayiotidis, M.I.; Powers, R.; Franco, R. Metabolic dysfunction in Parkinson’s disease: Bioenergetics, redox homeostasis and central carbon metabolism. Brain Res. Bull. 2017, 133, 12–30. [Google Scholar] [CrossRef]
- Weissbach, A.; Wittke, C.; Kasten, M.; Klein, C. “Atypical” Parkinson’s disease-genetic. Int. Rev. Neurobiol. 2019, 149, 207–235. [Google Scholar] [CrossRef]
- Niemann, N.; Jankovic, J. Juvenile parkinsonism: Differential diagnosis, genetics, and treatment. Parkinsonism Relat. Disord. 2019, 67, 74–89. [Google Scholar] [CrossRef] [PubMed]
- Bonifati, V.; Rizzu, P.; van Baren, M.J.; Schaap, O.; Breedveld, G.J.; Krieger, E.; Dekker, M.C.J.; Squitieri, F.; Ibanez, P.; Joosse, M.; et al. Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science 2003, 299, 256–259. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biosa, A.; Sanchez-Martinez, A.; Filograna, R.; Terriente-Felix, A.; Alam, S.M.; Beltramini, M.; Bubacco, L.; Bisaglia, M.; Whitworth, A.J. Superoxide dismutating molecules rescue the toxic effects of PINK1 and parkin loss. Hum. Mol. Genet. 2018, 27, 1618–1629. [Google Scholar] [CrossRef]
- Van der Vlag, M.; Havekes, R.; Heckman, P.R.A. The contribution of Parkin, PINK1 and DJ-1 genes to selective neuronal degeneration in Parkinson’s disease. Eur. J. Neurosci. 2020, 52, 3256–3268. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ariga, H.; Takahashi-Niki, K.; Kato, I.; Maita, H.; Niki, T.; Iguchi-Ariga, S.M.M. Neuroprotective function of DJ-1 in Parkinson’s disease. Oxid. Med. Cell. Longev. 2013, 2013, 683920. [Google Scholar] [CrossRef] [Green Version]
- Repici, M.; Giorgini, F. DJ-1 in Parkinson’s disease: Clinical insights and therapeutic perspectives. J. Clin. Med. 2019, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalia, L.V.; Lang, A.E. Parkinson’s disease. Lancet 2015, 386, 896–912. [Google Scholar] [CrossRef]
- Lotankar, S.; Prabhavalkar, K.S.; Bhatt, L.K. Biomarkers for Parkinson’s disease: Recent advancement. Neurosci. Bull. 2017, 33, 585–597. [Google Scholar] [CrossRef]
- Havelund, J.F.; Heegaard, N.H.H.; Færgeman, N.J.K.; Gramsbergen, J.B. Biomarker research in Parkinson’s disease using metabolite profiling. Metabolites 2017, 7, 42. [Google Scholar] [CrossRef] [Green Version]
- Binder, T.; Hobert, M.A.; Pfrommer, T.; Leks, E.; Granert, O.; Weigl, B.; Ethofer, T.; Erb, M.; Wilke, M.; Maetzler, W.; et al. Increased functional connectivity in a population at risk of developing Parkinson’s disease. Parkinsonism Relat. Disord. 2021, 92, 1–6. [Google Scholar] [CrossRef]
- Li, X.; Fan, X.; Yang, H.; Liu, Y. Review of metabolomics-based biomarker research for Parkinson’s disease. Mol. Neurobiol. 2021, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Shukla, A.K.; Ratnasekhar, C.; Pragya, P.; Chaouhan, H.S.; Patel, D.K.; Chowdhuri, D.K.; Mudiam, M.K.R. Metabolomic analysis provides insights on paraquat-induced Parkinson-like symptoms in Drosophila melanogaster. Mol. Neurobiol. 2016, 53, 254–269. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.H.; Ivanisevic, J.; Siuzdak, G. Metabolomics: Beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 2016, 17, 451–459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhinderwala, F.; Lei, S.; Woods, J.; Rose, J.; Marshall, D.D.; Riekeberg, E.; Leite, A.D.L.; Morton, M.; Dodds, E.D.; Franco, R.; et al. Metabolomics Analyses from Tissues in Parkinson’s Disease. Methods Mol. Biol. 2019, 1996, 217–257. [Google Scholar] [CrossRef]
- Solana-Manrique, C.; Moltó, M.D.; Calap-Quintana, P.; Sanz, F.J.; Llorens, J.V.; Paricio, N. Drosophila as a model system for the identification of pharmacological therapies in neurodegenerative diseases. In Insights into Human Neurodegeneration: Lessons Learnt from Drosophila; Mutsuddi, M., Mukrherjee, A., Eds.; Springer Nature Pte Ltd.: Singapore, 2019; pp. 433–467. [Google Scholar]
- Aryal, B.; Lee, Y. Disease model organism for Parkinson disease: Drosophila melanogaster. BMB Rep. 2019, 52, 250–258. [Google Scholar] [CrossRef] [Green Version]
- Lavara-Culebras, E.; Muñoz-Soriano, V.; Gómez-Pastor, R.; Matallana, E.; Paricio, N. Effects of pharmacological agents on the lifespan phenotype of Drosophila DJ-1beta mutants. Gene 2010, 462, 26–33. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lavara-Culebras, E.; Paricio, N. Drosophila DJ-1 mutants are sensitive to oxidative stress and show reduced lifespan and motor deficits. Gene 2007, 400, 158–165. [Google Scholar] [CrossRef] [PubMed]
- Casani, S.; Gómez-Pastor, R.; Matallana, E.; Paricio, N. Antioxidant compound supplementation prevents oxidative damage in a Drosophila model of Parkinson’s disease. Free Radic. Biol. Med. 2013, 61, 151–160. [Google Scholar] [CrossRef]
- Zhou, S.; Morgante, F.; Geisz, M.S.; Ma, J.; Anholt, R.R.H.; Mackay, T.F.C. Systems genetics of the Drosophila metabolome. Genome Res. 2020, 30, 392–405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yon, M.; Decoville, M.; Sarou-Kanian, V.; Fayon, F.; Birman, S. Spatially-resolved metabolic profiling of living Drosophila in neurodegenerative conditions using 1H magic angle spinning NMR. Sci. Rep. 2020, 10, 9516. [Google Scholar] [CrossRef] [PubMed]
- Yakhine-Diop, S.M.S.; Morales-García, J.A.; Niso-Santano, M.; González-Polo, R.A.; Uribe-Carretero, E.; Martinez-Chacon, G.; Durand, S.; Maiuri, M.C.; Aiastui, A.; Zulaica, M.; et al. Metabolic alterations in plasma from patients with familial and idiopathic Parkinson’s disease. Aging 2020, 12, 16690–16708. [Google Scholar] [CrossRef]
- López Del Amo, V.; Palomino-Schätzlein, M.; Seco-Cervera, M.; García-Giménez, J.L.; Pallardó, F.V.; Pineda-Lucena, A.; Galindo, M.I. A Drosophila model of GDAP1 function reveals the involvement of insulin signalling in the mitochondria-dependent neuromuscular degeneration. Biochim. Biophys. Acta Mol. Basis Dis. 2017, 1863, 801–809. [Google Scholar] [CrossRef]
- Bertrand, M.; Decoville, M.; Meudal, H.; Birman, S.; Landon, C. Metabolomic Nuclear Magnetic Resonance studies at presymptomatic and symptomatic stages of Huntington’s disease on a Drosophila model. J. Proteome Res. 2020, 19, 4034–4045. [Google Scholar] [CrossRef] [PubMed]
- Park, J.; Kim, S.Y.; Cha, G.-H.; Lee, S.B.; Kim, S.; Chung, J. Drosophila DJ-1 mutants show oxidative stress-sensitive locomotive dysfunction. Gene 2005, 361, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Sanz, F.J.; Solana-Manrique, C.; Muñoz-soriano, V.; Calap-quintana, P.; Moltó, M.D.; Paricio, N. Identification of potential therapeutic compounds for Parkinson’s disease using Drosophila and human cell models. Free Radic. Biol. Med. 2017, 108, 683–691. [Google Scholar] [CrossRef]
- Sanz, F.J.; Solana-Manrique, C.; Torres, J.; Masiá, E.; Vicent, M.J.; Paricio, N. A high-throughput chemical screen in DJ-1β mutant flies identifies Zaprinast as a potential Parkinson’s disease treatment. Neurotherapeutics 2021, 25, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Solana-Manrique, C.; Muñoz-Soriano, V.; Sanz, F.J.; Paricio, N. Oxidative modification impairs SERCA activity in Drosophila and human cell models of Parkinson’s disease. Biochim. Biophys. Acta Mol. Basis Dis. 2021, 1867, 166152. [Google Scholar] [CrossRef] [PubMed]
- Jacob, D.; Deborde, C.; Lefebvre, M.; Maucourt, M.; Moing, A. NMRProcFlow: A graphical and interactive tool dedicated to 1D spectra processing for NMR-based metabolomics. Metabolomics 2017, 13, 36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hollywood, K.A.; Shadi, I.T.; Goodacre, R. Monitoring the succinate dehydrogenase activity isolated from mitochondria by surface enhanced Raman scattering. J. Phys. Chem. C 2010, 114, 7308–7313. [Google Scholar] [CrossRef] [Green Version]
- Villa-Cuesta, E.; Rand, D.M. Preparation of Mitochondrial Enriched Fractions for Metabolic Analysis in Drosophila. J. Vis. Exp. 2015, 103. [Google Scholar] [CrossRef] [Green Version]
- Messaoudi, N.; Gautier, V.; Dairou, J.; Mihoub, M.; Lelandais, G.; Bouloc, P.; Landoulsi, A.; Richarme, G. Fermentation and alternative respiration compensate for NADH dehydrogenase deficiency in a prokaryotic model of DJ-1-associated Parkinsonism. Microbiology 2015, 161, 2220–2231. [Google Scholar] [CrossRef] [PubMed]
- Meiser, J.; Delcambre, S.; Wegner, A.; Jäger, C.; Ghelfi, J.; D’Herouel, A.F.; Dong, X.; Weindl, D.; Stautner, C.; Nonnenmacher, Y.; et al. Loss of DJ-1 impairs antioxidant response by altered glutamine and serine metabolism. Neurobiol. Dis. 2016, 89, 112–125. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Requejo-Aguilar, R.; Bolaños, J.P. Mitochondrial control of cell bioenergetics in Parkinson’s disease. Free Radic. Biol. Med. 2016, 100, 123–137. [Google Scholar] [CrossRef] [Green Version]
- Neinast, M.; Murashige, D.; Arany, Z. Branched chain amino acids. Annu. Rev. Physiol. 2019, 81, 139–164. [Google Scholar] [CrossRef]
- Tosukhowong, P.; Boonla, C.; Dissayabutra, T.; Kaewwilai, L.; Muensri, S.; Chotipanich, C.; Joutsa, J.; Rinne, J.; Bhidayasiri, R. Biochemical and clinical effects of Whey protein supplementation in Parkinson’s disease: A pilot study. J. Neurol. Sci. 2016, 367, 162–170. [Google Scholar] [CrossRef]
- Szabó, N.; Kincses, Z.T.; Toldi, J.; Vécsei, L. Altered tryptophan metabolism in Parkinson’s disease: A possible novel therapeutic approach. J. Neurol. Sci. 2011, 310, 256–260. [Google Scholar] [CrossRef]
- Figura, M.; Kuśmierska, K.; Bucior, E.; Szlufik, S.; Koziorowski, D.; Jamrozik, Z.; Janik, P. Serum amino acid profile in patients with Parkinson’s disease. PLoS ONE 2018, 13, e0191670. [Google Scholar] [CrossRef]
- Dunn, L.; Allen, G.F.; Mamais, A.; Ling, H.; Li, A.; Duberley, K.E.; Hargreaves, I.P.; Pope, S.; Holton, J.L.; Lees, A.; et al. Dysregulation of glucose metabolism is an early event in sporadic Parkinson’s disease. Neurobiol. Aging 2014, 35, 1111–1115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borghammer, P.; Hansen, S.B.; Eggers, C.; Chakravarty, M.; Vang, K.; Aanerud, J.; Hilker, R.; Heiss, W.-D.; Rodell, A.; Munk, O.L.; et al. Glucose metabolism in small subcortical structures in Parkinson’s disease. Acta Neurol. Scand. 2012, 125, 303–310. [Google Scholar] [CrossRef] [PubMed]
- Murali Mahadevan, H.; Hashemiaghdam, A.; Ashrafi, G.; Harbauer, A.B. Mitochondria in neuronal health: From energy metabolism to Parkinson’s disease. Adv. Biol. 2021, 5, e2100663. [Google Scholar] [CrossRef]
- Wu, J.; Jin, Z.; Zheng, H.; Yan, L.-J. Sources and implications of NADH/NAD(+) redox imbalance in diabetes and its complications. Diabetes Metab. Syndr. Obes. 2016, 9, 145–153. [Google Scholar] [CrossRef] [Green Version]
- Cheng, Z.; Tsuda, M.; Kishita, Y.; Sato, Y.; Aigaki, T. Impaired energy metabolism in a Drosophila model of mitochondrial aconitase deficiency. Biochem. Biophys. Res. Commun. 2013, 433, 145–150. [Google Scholar] [CrossRef]
- Cecchini, G. Function and structure of complex II of the respiratory chain. Annu. Rev. Biochem. 2003, 72, 77–109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quinlan, C.L.; Orr, A.L.; Perevoshchikova, I.V.; Treberg, J.R.; Ackrell, B.A.; Brand, M.D. Mitochondrial complex II can generate reactive oxygen species at high rates in both the forward and reverse reactions. J. Biol. Chem. 2012, 287, 27255–27264. [Google Scholar] [CrossRef] [Green Version]
- Pesi, R.; Balestri, F.; Ipata, P.L. Metabolic interaction between urea cycle and citric acid cycle shunt: A guided approach. Biochem. Mol. Biol. Educ. 2018, 46, 182–185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garabadu, D.; Agrawal, N.; Sharma, A.; Sharma, S. Mitochondrial metabolism: A common link between neuroinflammation and neurodegeneration. Behav. Pharmacol. 2019, 30, 642–652. [Google Scholar] [CrossRef] [PubMed]
- Bhaumik, P.; Koski, M.K.; Bergmann, U.; Wierenga, R.K. Structure determination and refinement at 2.44 A resolution of argininosuccinate lyase from Escherichia coli. Acta Crystallogr. D Biol. Crystallogr. 2004, 60, 1964–1970. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andersen, A.D.; Binzer, M.; Stenager, E.; Gramsbergen, J.B. Cerebrospinal fluid biomarkers for Parkinson’s disease—A systematic review. Acta Neurol. Scand. 2017, 135, 34–56. [Google Scholar] [CrossRef] [PubMed]
- Kumari, S.; Kumaran, S.S.; Goyal, V.; Sharma, R.K.; Sinha, N.; Dwivedi, S.N.; Srivastava, A.K.; Jagannathan, N.R. Identification of potential urine biomarkers in idiopathic parkinson’s disease using NMR. Clin. Chim. Acta 2020, 510, 442–449. [Google Scholar] [CrossRef] [PubMed]
- Kumari, S.; Kumaran, S.S.; Goyal, V.; Bose, S.; Jain, S.; Dwivedi, S.N.; Srivastava, A.K.; Jagannathan, N.R. Metabolomic analysis of serum using proton NMR in 6-OHDA experimental PD model and patients with PD. Neurochem. Int. 2020, 134, 104670. [Google Scholar] [CrossRef] [PubMed]
- LeWitt, P. Recent advances in CSF biomarkers for Parkinson’s disease. Parkinsonism Relat. Disord. 2012, 18, S49–S51. [Google Scholar] [CrossRef]
- LeWitt, P.A.; Li, J.; Lu, M.; Guo, L.; Auinger, P. Parkinson Study Group—DATATOP Investigators Metabolomic biomarkers as strong correlates of Parkinson disease progression. Neurology 2017, 88, 862–869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trezzi, J.-P.; Galozzi, S.; Jaeger, C.; Barkovits, K.; Brockmann, K.; Maetzler, W.; Berg, D.; Marcus, K.; Betsou, F.; Hiller, K.; et al. Distinct metabolomic signature in cerebrospinal fluid in early parkinson’s disease. Mov. Disord. 2017, 32, 1401–1408. [Google Scholar] [CrossRef]
- Tufi, R.; Gandhi, S.; de Castro, I.P.; Lehmann, S.; Angelova, P.R.; Dinsdale, D.; Deas, E.; Plun-Favreau, H.; Nicotera, P.; Abramov, A.Y.; et al. Enhancing nucleotide metabolism protects against mitochondrial dysfunction and neurodegeneration in a PINK1 model of Parkinson’s disease. Nat. Cell Biol. 2014, 16, 157–166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shao, Y.; Le, W. Recent advances and perspectives of metabolomics-based investigations in Parkinson’s disease. Mol. Neurodegener. 2019, 14, 3. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sinclair, E.; Trivedi, D.K.; Sarkar, D.; Walton-Doyle, C.; Milne, J.; Kunath, T.; Rijs, A.M.; de Bie, R.M.A.; Goodacre, R.; Silverdale, M.; et al. Metabolomics of sebum reveals lipid dysregulation in Parkinson’s disease. Nat. Commun. 2021, 12, 1592. [Google Scholar] [CrossRef] [PubMed]
- Barichella, M.; Cereda, E.; Pinelli, G.; Iorio, L.; Caroli, D.; Masiero, I.; Ferri, V.; Cassani, E.; Bolliri, C.; Caronni, S.; et al. Muscle-targeted nutritional support for rehabilitation in patients with parkinsonian syndrome. Neurology 2019, 93, e485–e496. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Riano, C.; Saiz, J.; Barbas, C.; Bergareche, A.; Huerta, J.M.; Ardanaz, E.; Konjevod, M.; Mondragon, E.; Erro, M.E.; Chirlaque, M.D.; et al. Prognostic biomarkers of Parkinson’s disease in the Spanish EPIC cohort: A multiplatform metabolomics approach. NPJ Park. Dis. 2021, 7, 73. [Google Scholar] [CrossRef]
- Ullah, R.; Jo, M.H.; Riaz, M.; Alam, S.I.; Saeed, K.; Ali, W.; Rehman, I.U.; Ikram, M.; Kim, M.O. Glycine, the smallest amino acid, confers neuroprotection against D-galactose-induced neurodegeneration and memory impairment by regulating c-Jun N-terminal kinase in the mouse brain. J. Neuroinflamm. 2020, 17, 303. [Google Scholar] [CrossRef]
- García-Ayuso, D.; Di Pierdomenico, J.; Valiente-Soriano, F.J.; Martínez-Vacas, A.; Agudo-Barriuso, M.; Vidal-Sanz, M.; Picaud, S.; Villegas-Pérez, M.P. β-alanine supplementation induces taurine depletion and causes alterations of the retinal nerve fiber layer and axonal transport by retinal ganglion cells. Exp. Eye Res. 2019, 188, 107781. [Google Scholar] [CrossRef]
- Kirov, I.I.; Sollberger, M.; Davitz, M.S.; Glodzik, L.; Soher, B.J.; Babb, J.S.; Monsch, A.U.; Gass, A.; Gonen, O. Global brain volume and N-acetyl-aspartate decline over seven decades of normal aging. Neurobiol. Aging 2021, 98, 42–51. [Google Scholar] [CrossRef]
- Okarmus, J.; Havelund, J.F.; Ryding, M.; Schmidt, S.I.; Bogetofte, H.; Heon-Roberts, R.; Wade-Martins, R.; Cowley, S.A.; Ryan, B.J.; Færgeman, N.J.; et al. Identification of bioactive metabolites in human iPSC-derived dopaminergic neurons with PARK2 mutation: Altered mitochondrial and energy metabolism. Stem Cell Rep. 2021, 16, 1510–1526. [Google Scholar] [CrossRef]
- Zilberter, Y.; Zilberter, M. The vicious circle of hypometabolism in neurodegenerative diseases: Ways and mechanisms of metabolic correction. J. Neurosci. Res. 2017, 95, 2217–2235. [Google Scholar] [CrossRef] [Green Version]
- Hong, C.T.; Chau, K.-Y.; Schapira, A.H.V. Meclizine-induced enhanced glycolysis is neuroprotective in Parkinson disease cell models. Sci. Rep. 2016, 6, 25344. [Google Scholar] [CrossRef]
- Díaz-García, C.M.; Yellen, G. Neurons rely on glucose rather than astrocytic lactate during stimulation. J. Neurosci. Res. 2019, 97, 883–889. [Google Scholar] [CrossRef] [Green Version]
- Chakraborty, S.; Nian, F.-S.; Tsai, J.-W.; Karmenyan, A.; Chiou, A. Quantification of the metabolic state in cell-model of Parkinson’s disease by fluorescence lifetime imaging microscopy. Sci. Rep. 2016, 6, 19145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tapias, V.; McCoy, J.L.; Greenamyre, J.T. Phenothiazine normalizes the NADH/NAD+ ratio, maintains mitochondrial integrity and protects the nigrostriatal dopamine system in a chronic rotenone model of Parkinson’s disease. Redox Biol. 2019, 24, 101164. [Google Scholar] [CrossRef]
- Cantu, D.; Schaack, J.; Patel, M. Oxidative inactivation of mitochondrial aconitase results in iron and H2O2-mediated neurotoxicity in rat primary mesencephalic cultures. PLoS ONE 2009, 4, e7095. [Google Scholar] [CrossRef] [Green Version]
- Jodeiri Farshbaf, M.; Kiani-Esfahani, A. Succinate dehydrogenase: Prospect for neurodegenerative diseases. Mitochondrion 2018, 42, 77–83. [Google Scholar] [CrossRef] [PubMed]
- Ikenaka, K.; Suzuki, M.; Mochizuki, H.; Nagai, Y. Lipids as trans-acting effectors for α-synuclein in the pathogenesis of Parkinson’s disease. Front. Neurosci. 2019, 13, 693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Simunovic, F.; Yi, M.; Wang, Y.; Macey, L.; Brown, L.T.; Krichevsky, A.M.; Andersen, S.L.; Stephens, R.M.; Benes, F.M.; Sonntag, K.C. Gene expression profiling of substantia nigra dopamine neurons: Further insights into Parkinson’s disease pathology. Brain J. Neurol. 2009, 132, 1795–1809. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Edson, A.J.; Hushagen, H.A.; Frøyset, A.K.; Elda, I.; Khan, E.A.; Di Stefano, A.; Fladmark, K.E. Dysregulation in the brain protein profile of zebrafish lacking the Parkinson’s disease-related protein DJ-1. Mol. Neurobiol. 2019, 56, 8306–8322. [Google Scholar] [CrossRef] [PubMed]
- Griffin, J.W.D.; Bradshaw, P.C. Amino acid catabolism in Alzheimer’s disease brain: Friend or foe? Oxid. Med. Cell. Longev. 2017, 2017, 5472792. [Google Scholar] [CrossRef] [PubMed] [Green Version]

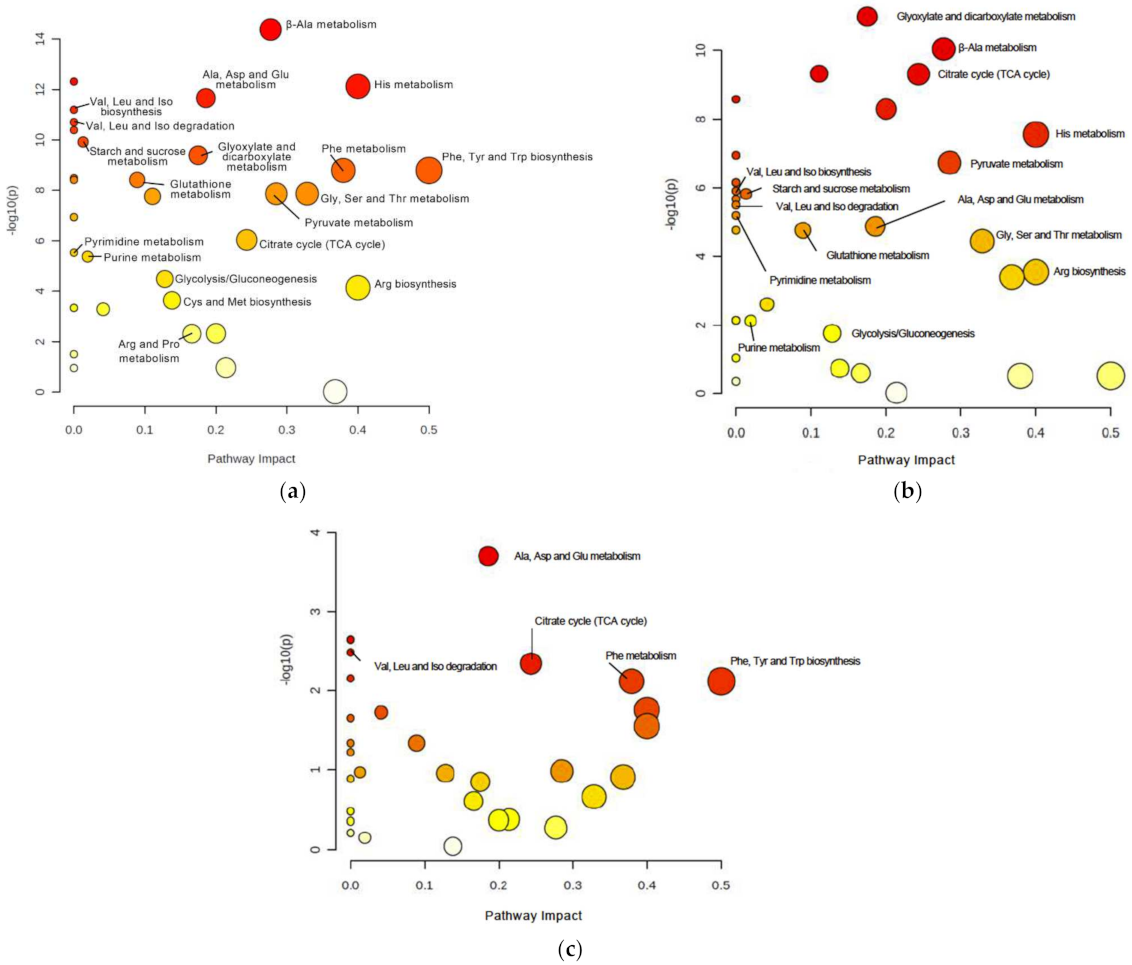



| Pathway | Number of Differential Metabolites/Totals | Raw p-Value | p-Value FDR Corrected | Impact |
|---|---|---|---|---|
| beta-Alanine metabolism | 2/14 | 4.21 × 10−15 | 1.43 × 10−13 | 0.28 |
| Histidine metabolism | 2/9 | 7.51 × 10−13 | 8.51 × 10−12 | 0.40 |
| Alanine, aspartate and glutamate metabolism | 6/23 | 2.20 × 10−12 | 1.87 × 10−11 | 0.19 |
| Glyoxylate and dicarboxylate metabolism | 6/24 | 4.07 × 10−10 | 1.54 × 10−19 | 0.17 |
| Phenylalanine, tyrosine and tryptophan biosynthesis | 1/4 | 1.61 × 10−9 | 4.98 × 10−9 | 0.50 |
| Phenylalanine metabolism | 1/7 | 1.61 × 10−9 | 4.98 × 10−9 | 0.38 |
| Glutathione metabolism | 1/26 | 3.84 × 10−9 | 9.33 × 10−9 | 0.09 |
| Glycine, serine and threonine metabolism | 3/30 | 1.35 × 10−8 | 2.90 × 10−8 | 0.33 |
| Pyruvate metabolism | 4/22 | 1.36 × 10−8 | 2.90 × 10−8 | 0.28 |
| Glycerophospholipid metabolism | 2/32 | 1.73 × 10−8 | 3.36 × 10−8 | 0.11 |
| Citrate cycle (TCA cycle) | 5/20 | 9.,20 × 10−7 | 1.56 × 10−6 | 0.24 |
| Glycolysis/Gluconeogenesis | 4/26 | 3.25 × 10−5 | 4.81 × 10−5 | 0.13 |
| Arginine biosynthesis | 3/12 | 7.25 × 10−5 | 1.03 × 10−4 | 0.40 |
| Cysteine and methionine metabolism | 2/32 | 2.29 × 10−4 | 3.11 × 10−4 | 0.14 |
| Taurine and hypotaurine metabolism | 1/7 | 4.72 × 10−3 | 5.48 × 10−3 | 0.20 |
| Arginine and proline metabolism | 2/31 | 4.84 × 10−3 | 5.48 × 10−3 | 0.17 |
| Pathway | Number of Differential Metabolites/Totals | Raw p-Value | p-Value FDR Corrected | Impact |
|---|---|---|---|---|
| Glyoxylate and dicarboxylate metabolism | 6/24 | 1.03 × 10−11 | 3.50 × 10−10 | 0.17 |
| beta-Alanine metabolism | 2/14 | 8.98 × 10−11 | 1.53 × 10−9 | 0.28 |
| Glycerophospholipid metabolism | 2/32 | 4.74 × 10−10 | 4.15 × 10−9 | 0.11 |
| Citrate cycle (TCA cycle) | 5/20 | 4.89 × 10−10 | 4.15 × 10−9 | 0.24 |
| Taurine and hypotaurine metabolism | 1/7 | 5.08 × 10−9 | 2.88 × 10−8 | 0.20 |
| Histidine metabolism | 2/9 | 2.79 × 10−8 | 1.36 × 10−7 | 0.40 |
| Pyruvate metabolism | 4/22 | 1.88 × 10−7 | 7.09 × 10−7 | 0.28 |
| Alanine, aspartate and glutamate metabolism | 6/23 | 1.33 × 10−5 | 2.66 × 10−5 | 0.19 |
| Glycine, serine and threonine metabolism | 3/30 | 3.65 × 10−5 | 6.20 × 10−5 | 0.33 |
| Arginine biosynthesis | 3/12 | 2.89 × 10−4 | 4.67 × 10−4 | 0.40 |
| Nicotinate and nicotinemide metabolism | 1/9 | 4.03 × 10−4 | 6.23 × 10−4 | 0.37 |
| Tyrosine metabolism | 2/33 | 2.52 × 10−3 | 3.72 × 10−3 | 0.04 |
| Purine metabolism | 3/63 | 7.64 × 10−3 | 9.98 × 10−3 | 0.02 |
| Glycolysis/Gluconeogenesis | 4/26 | 1.76 × 10−2 | 2.21 × 10−2 | 0.13 |
| Pathway | Number of Differential Metabolites/Totals | Raw p-Value | p-Value FDR Corrected | Impact |
|---|---|---|---|---|
| Glycerophospholipid metabolism | 2/32 | 1.85 × 10−5 | 6.29 × 10−4 | 0.11 |
| Alanine, aspartate and glutamate | 6/23 | 1.98 × 10−4 | 3.36 × 10−3 | 0.19 |
| Citrate cycle (TCA cycle) | 5/20 | 4.54 × 10−3 | 2.57 × 10−2 | 0.24 |
| Phenylalanine, tyrosine and tryptophan biosynthesis | 1/4 | 7.60 × 10−3 | 2.87 × 10−2 | 0.50 |
| Phenylalanine metabolism | 1/7 | 7.60 × 10−3 | 2.87 × 10−2 | 0.38 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Solana-Manrique, C.; Sanz, F.J.; Torregrosa, I.; Palomino-Schätzlein, M.; Hernández-Oliver, C.; Pineda-Lucena, A.; Paricio, N. Metabolic Alterations in a Drosophila Model of Parkinson’s Disease Based on DJ-1 Deficiency. Cells 2022, 11, 331. https://doi.org/10.3390/cells11030331
Solana-Manrique C, Sanz FJ, Torregrosa I, Palomino-Schätzlein M, Hernández-Oliver C, Pineda-Lucena A, Paricio N. Metabolic Alterations in a Drosophila Model of Parkinson’s Disease Based on DJ-1 Deficiency. Cells. 2022; 11(3):331. https://doi.org/10.3390/cells11030331
Chicago/Turabian StyleSolana-Manrique, Cristina, Francisco José Sanz, Isabel Torregrosa, Martina Palomino-Schätzlein, Carolina Hernández-Oliver, Antonio Pineda-Lucena, and Nuria Paricio. 2022. "Metabolic Alterations in a Drosophila Model of Parkinson’s Disease Based on DJ-1 Deficiency" Cells 11, no. 3: 331. https://doi.org/10.3390/cells11030331






