The Small GTPase Rab7 Regulates Antigen Processing in B Cells in a Possible Interplay with Autophagy Machinery
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells and Transfections
| Plasmids |
 |
| Antibodies |
 |
2.2. Sample Preparation for Confocal Microscopy
2.3. Image Acquisition and Processing
2.4. Transmission Electron Microscopy (TEM) Sample Preparation and Acquisition
2.5. Structured Illumination Microscopy (SIM) Acquisition
2.6. DQ-Ova Assay
2.7. Antigen Presentation Measured by ELISA
2.8. Antigen Internalisation by Flow Cytometry
2.9. Microscopy Data Analysis, Statistical Analysis, and Illustrations
3. Results
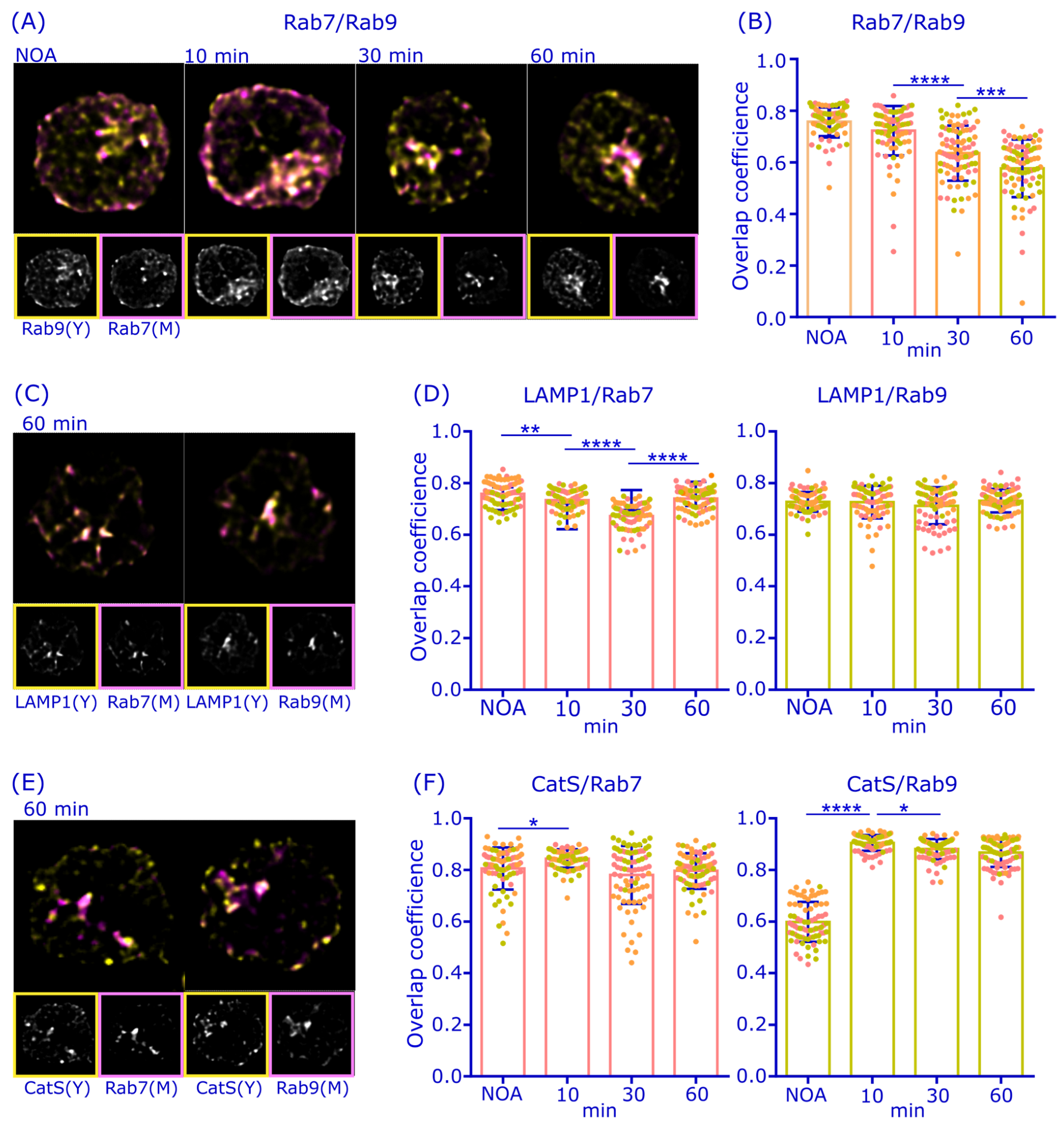
3.1. Rab7 and Rab9 Largely Colocalize but Show Distinct Responses to Antigen Activation in B Cells
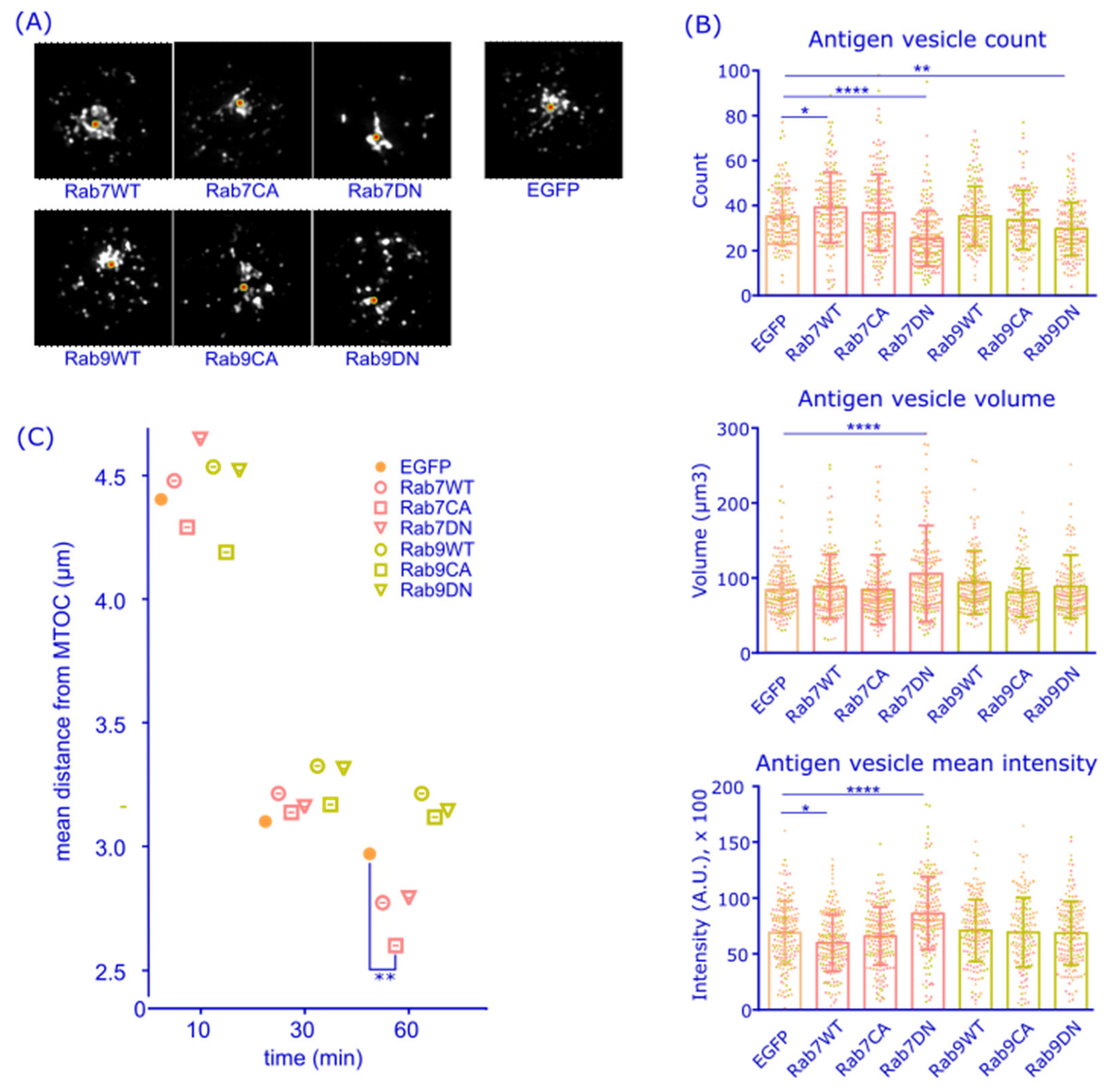
3.2. Rab7 May Coordinate Perinuclear Antigen Gathering
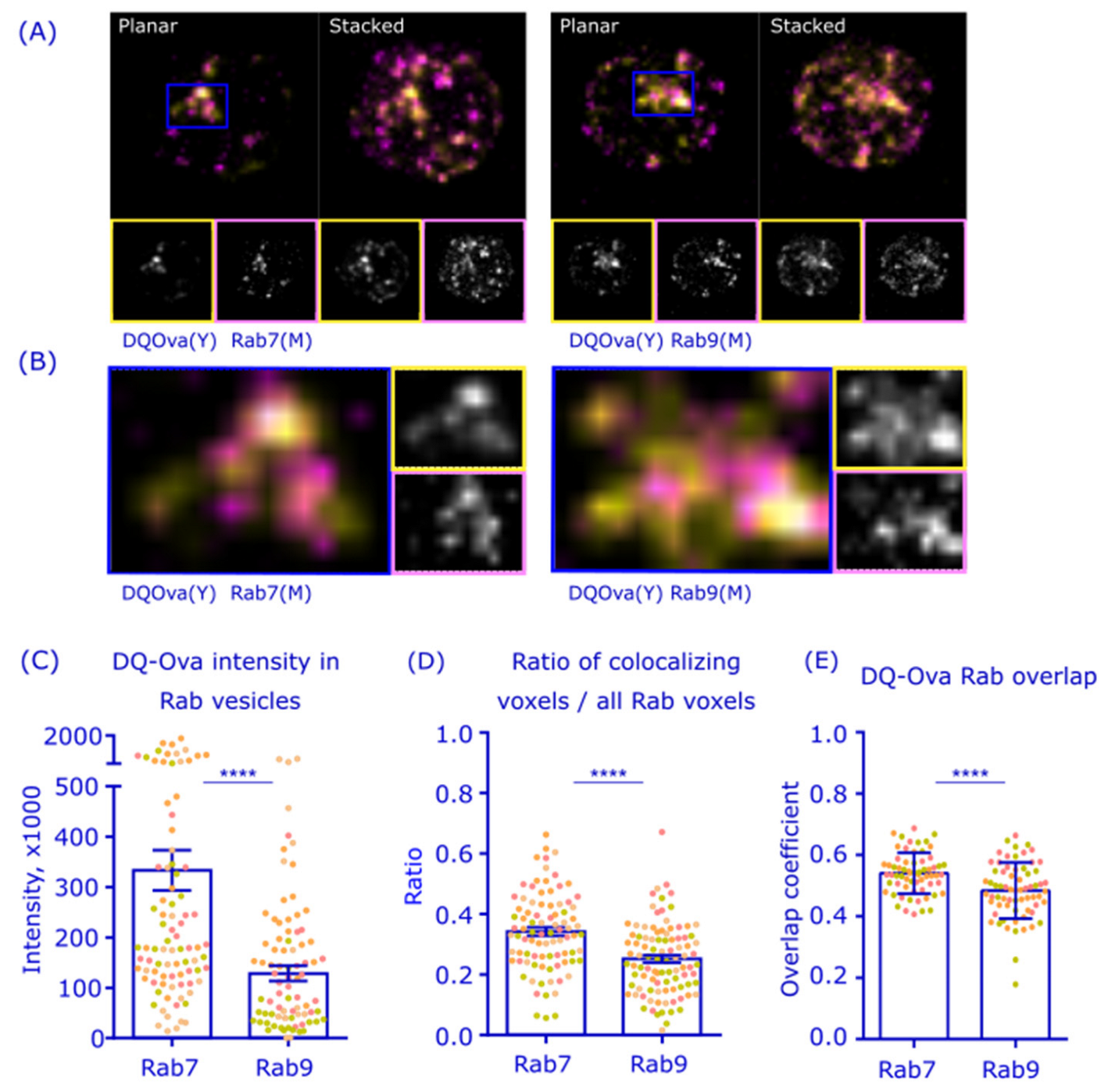
3.3. Antigen Processing Activity Prefers Rab7+ over Rab9+ Vesicles
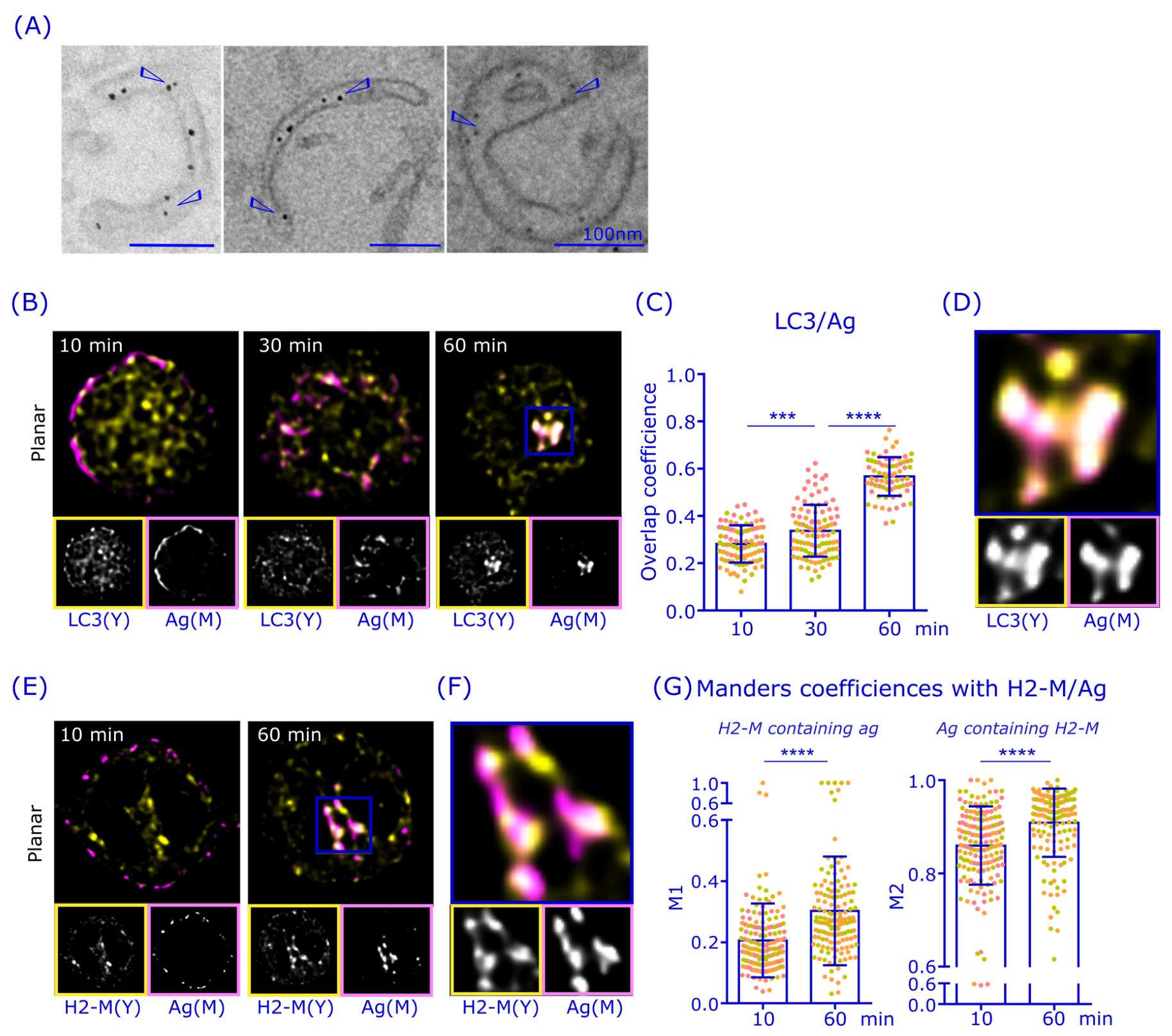
3.4. Antigen Is Gathered in LC3+ and H2-M+ Perinuclear Compartments
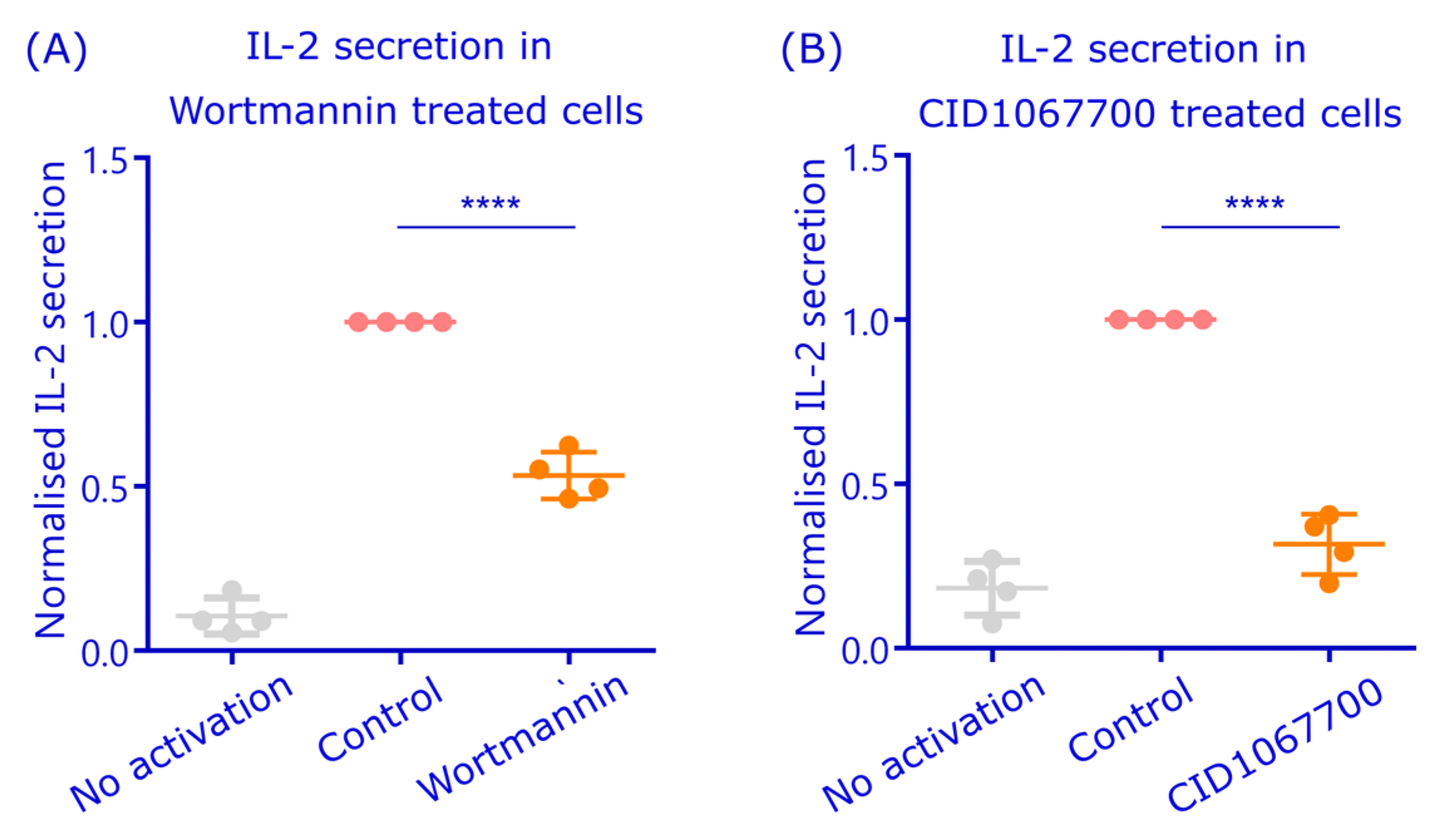
3.5. Disturbing Rab7 and Autophagosome Functions Inhibit Peptide Antigen Presentation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Adler, L.N.; Jiang, W.; Bhamidipati, K.; Millican, M.; Macaubas, C.; Hung, S.C.; Mellins, E.D. The other function: Class II-restricted antigen presentation by B cells. Front. Immunol. 2017, 8, 319. [Google Scholar] [CrossRef] [PubMed]
- McHeyzer-Williams, M.; Okitsu, S.; Wang, N.; McHeyzer-Williams, L. Molecular programming of B cell memory. Nat. Rev. Immunol. 2012, 12, 24–34. [Google Scholar] [CrossRef] [PubMed]
- Lankar, D.; Vincent-Schneider, H.; Briken, V.; Yokozeki, T.; Raposo, G.; Bonnerot, C. Dynamics of major histocompatibility complex class II compartments during B cell receptor—Mediated cell activation. J. Exp. Med. 2002, 195, 461–472. [Google Scholar] [CrossRef]
- Unanue, E.R.; Turk, V.; Neefjes, J. Variations in MHC Class II Antigen Processing and Presentation in Health and Disease. Annu. Rev. Immunol. 2016, 34, 265–297. [Google Scholar] [CrossRef] [PubMed]
- Avalos, A.M.; Ploegh, H.L. Early BCR events and antigen capture, processing, and loading on MHC class II on B cells. Front. Immunol. 2014, 5, 92. [Google Scholar] [CrossRef] [PubMed]
- Watts, C. The exogenous pathway for antigen presentation on major histocompatibility complex class II and CD1 molecules. Nat. Immunol. 2004, 5, 685–692. [Google Scholar] [CrossRef] [PubMed]
- West, M.A.; Lucocq, J.M.; Watts, C. Antigen processing and class II MHC peptide-loading compartments in human B-lymphoblastoid cells. Nature 1994, 369, 147–151. [Google Scholar] [CrossRef]
- Whitmire, J.K.; Asano, M.S.; Kaech, S.M.; Sarkar, S.; Hannum, L.G.; Shlomchik, M.J.; Ahmed, R. Requirement of B Cells for Generating CD4 + T Cell Memory. J. Immunol. 2009, 182, 1868–1876. [Google Scholar] [CrossRef]
- Aluvihare, V.R.; Khamlichi, A.A.; Williams, G.T.; Adorini, L.; Neuberger, M.S. Acceleration of intracellular targeting of antigen by the B-cell antigen receptor: Importance depends on the nature of the antigen-antibody interaction. EMBO J. 1997, 16, 3553–3562. [Google Scholar] [CrossRef]
- Vascotto, F.; Le Roux, D.; Lankar, D.; Faure-André, G.; Vargas, P.; Guermonprez, P.; Lennon-Duménil, A.M. Antigen presentation by B lymphocytes: How receptor signaling directs membrane trafficking. Curr. Opin. Immunol. 2007, 19, 93–98. [Google Scholar] [CrossRef]
- Driessen, C.; Bryant, R.A.R.; Lennon-Duménil, A.M.; Villadangos, J.A.; Bryant, P.W.; Shi, G.P.; Chapman, H.A.; Ploegh, H.L. Cathepsin S controls the trafficking and maturation of MHC class II molecules in dendritic cells. J. Cell Biol. 1999, 147, 775–790. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, L. DM and DO shape the repertoire of peptide-MHC-class-II complexes. Curr. Opin. Immunol. 2005, 17, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Van Lith, M.; van Ham, M.; Griekspoor, A.; Tjin, E.; Verwoerd, D.; Calafat, J.; Janssen, H.; Reits, E.; Pastoors, L.; Neefjes, J. Regulation of MHC Class II Antigen Presentation by Sorting of Recycling HLA-DM/DO and Class II within the Multivesicular Body. J. Immunol. 2001, 167, 884–892. [Google Scholar] [CrossRef] [PubMed]
- Pierre, P.; Denzin, L.K.; Hammond, C.; Drake, J.R.; Amigorena, S.; Cresswell, P.; Mellman, I. HLA-DM is localized to conventional and unconventional MHC class II-Containing endocytic compartments. Immunity 1996, 4, 229–239. [Google Scholar] [CrossRef] [PubMed]
- Wandinger-Ness, A.; Zerial, M. Rab proteins and the compartmentalization of the endosomal system. Cold Spring Harb. Perspect. Biol. 2014, 6, a022616. [Google Scholar] [CrossRef] [PubMed]
- Mellman, I. Endocytosis and Molecular Sorting. Annu. Rev. Cell Dev. Biol. 1996, 12, 575–625. [Google Scholar] [CrossRef] [PubMed]
- Stenmark, H. Rab GTPases as coordinators of vesicle traffic. Nat. Rev. Mol. Cell Biol. 2009, 10, 513–525. [Google Scholar] [CrossRef]
- Scott, C.C.; Vacca, F.; Gruenberg, J. Endosome maturation, transport and functions. Semin. Cell Dev. Biol. 2014, 31, 2–10. [Google Scholar] [CrossRef]
- Hutagalung, A.H.; Novick, P.J. Role of Rab GTPases in Membrane Traffic and Cell Physiology. Physiol. Rev. 2013, 91, 119–149. [Google Scholar] [CrossRef]
- McNally, K.E.; Faulkner, R.; Steinberg, F.; Gallon, M.; Ghai, R.; Pim, D.; Langton, P.; Pearson, N.; Danson, C.M.; Nägele, H.; et al. Retriever is a multiprotein complex for retromer-independent endosomal cargo recycling. Nat. Cell Biol. 2017, 19, 1214–1225. [Google Scholar] [CrossRef]
- Weeratunga, S.; Paul, B.; Collins, B.M. Recognising the signals for endosomal trafficking. Curr. Opin. Cell Biol. 2020, 65, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Solinger, J.A.; Rashid, H.O.; Prescianotto-Baschong, C.; Spang, A. FERARI is required for Rab11-dependent endocytic recycling. Nat. Cell Biol. 2020, 22, 213–224. [Google Scholar] [CrossRef]
- Delevoye, C.; Marks, M.S.; Raposo, G. Lysosome-related organelles as functional adaptations of the endolysosomal system. Curr. Opin. Cell Biol. 2019, 59, 147–158. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Pérez, S.; Vainio, M.; Kuokkanen, E.; Šuštar, V.; Petrov, P.; Forstén, S.; Paavola, V.; Rajala, J.; Awoniyi, L.O.; Sarapulov, A.V.; et al. B cells rapidly target antigen and surface-derived MHCII into peripheral degradative compartments. J. Cell Sci. 2020, 133, jcs235192. [Google Scholar] [CrossRef] [PubMed]
- Satpathy, S.; Wagner, S.A.; Beli, P.; Gupta, R.; Kristiansen, T.A.; Malinova, D.; Francavilla, C.; Tolar, P.; Bishop, G.A.; Hostager, B.S.; et al. Systems-wide analysis of BCR signalosomes and downstream phosphorylation and ubiquitylation. Mol. Syst. Biol. 2015, 11, 810. [Google Scholar] [CrossRef] [PubMed]
- Lam, T.; Kulp, D.V.; Wang, R.; Lou, Z.; Taylor, J.; Rivera, C.E.; Yan, H.; Zhang, Q.; Wang, Z.; Zan, H.; et al. Small Molecule Inhibition of Rab7 Impairs B Cell Class Switching and Plasma Cell Survival to Dampen the Autoantibody Response in Murine Lupus. J. Immunol. 2016, 197, 3792–3805. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Fernandez, M.; Wang, J.; Wu, S.; Wang, R.; Lou, Z.; Moroney, J.B.; Rivera, C.E.; Taylor, J.R.; Gan, H.; et al. B Cell Endosomal RAB7 Promotes TRAF6 K63 Polyubiquitination and NF-κB Activation for Antibody Class-Switching. J. Immunol. 2020, 204, 1146–1157. [Google Scholar] [CrossRef] [PubMed]
- Pone, E.J.; Lam, T.; Lou, Z.; Wang, R.; Chen, Y.; Liu, D.; Edinger, A.L.; Xu, Z.; Casali, P. B Cell Rab7 Mediates Induction of Activation-Induced Cytidine Deaminase Expression and Class-Switching in T-Dependent and T-Independent Antibody Responses. J. Immunol. 2015, 194, 3065–3078. [Google Scholar] [CrossRef]
- Williams, G.T.; Peaker, C.J.G.; Patel, K.J.; Neuberger, M.S. The a/b sheath and its cytoplasmic tyrosines are required for signaling by the B-cell antigen receptor but not for capping or for serine/threonine-kinase recruitment. Proc. Natl. Acad. Sci. USA 1994, 91, 474–478. [Google Scholar] [CrossRef]
- Adorini, L.; Guery, J.; Fuchs, S.; Ortiz-Navarrete, V.; Hämmerling, G.J.; Momburf, F. Processing of endogenously synthesized hen egg-white lysozyme retained in the endoplasmic reticulum or in secretory form gives rise to a similar but not identical set of epitopes recognized by class II-restricted T cells. J. Immunol. 1993, 151, 3567–3586. [Google Scholar] [CrossRef]
- Šuštar, V.; Vainio, M.; Mattila, P.K. Visualization and quantitative analysis of the actin cytoskeleton upon B cell activation. In B Cell Receptor Signaling: Methods and Protocols; Humana Press: New York, NY, USA, 2018; pp. 243–257. Available online: http://link.springer.com/10.1007/978-1-4939-7474-0_18 (accessed on 1 February 2018).
- Kucera, A.; Bakke, O.; Progida, C. The multiple roles of Rab9 in the endolysosomal system. Commun. Integr. Biol. 2016, 9, e1204498. [Google Scholar] [CrossRef] [PubMed]
- Kucera, A.; Borg Distefano, M.; Berg-Larsen, A.; Skjeldal, F.; Repnik, U.; Bakke, O.; Progida, C. Spatiotemporal Resolution of Rab9 and CI-MPR Dynamics in the Endocytic Pathway. Traffic 2016, 17, 211–229. [Google Scholar] [CrossRef] [PubMed]
- Lombardi, D.; Soldatil, T.; Riederer, M.A.; Goda, Y.; Zerial, M.; Pfeffer, S.R. Rab9 functions in transport between late endosomes and the trans Golgi network. EMBO J. 1993, 12, 677–682. [Google Scholar] [CrossRef] [PubMed]
- Dong, B.; Kakihara, K.; Otani, T.; Wada, H.; Hayashi, S. Rab9 and retromer regulate retrograde trafficking of luminal protein required for epithelial tube length control. Nat. Commun. 2013, 4, 1312–1358. [Google Scholar] [CrossRef] [PubMed]
- Huotari, J.; Helenius, A. Endosome maturation. EMBO J. 2011, 30, 3481–3500. [Google Scholar] [CrossRef] [PubMed]
- Bucci, C.; Thomsen, P.; Nicoziani, P.; McCarthy, J.; Van Deurs, B. Rab7: A key to lysosome biogenesis. Mol. Biol. Cell. 2000, 11, 467–480. [Google Scholar] [CrossRef] [PubMed]
- Cantalupo, G.; Alifano, P.; Roberti, V.; Bruni, C.B.; Bucci, C. Rab-interacting lysosomal protein (RILP): The Rab7 effector required for transport to lysosomes. EMBO J. 2001, 20, 683–693. [Google Scholar] [CrossRef] [PubMed]
- Riederer, M.A.; Soldati, T.; Shapiro, A.D.; Lin, J.; Pfeffer, S.R. Lysosome biogenesis requires rab9 function and receptor recycling from endosomes to the trans-Golgi network. J. Cell Biol. 1994, 125, 573–582. [Google Scholar] [CrossRef]
- Janicka, M.; Chindemi, P.A.; Hu, W.; Regoeczi, E. Effect of transferrin on the degradation of glycoproteins bearing a hybrid or high-mannose glycan by alveolar macrophages. Exp. Cell Res. 1994, 215, 17–22. [Google Scholar] [CrossRef]
- Kawabata, T.; Yoshimori, T. Autophagosome biogenesis and human health. Cell Discov. 2020, 6, 33. [Google Scholar] [CrossRef]
- Barral, D.C.; Staiano, L.; Almeida, C.G.; Cutler, D.F.; Eden, E.R.; Futter, C.E.; Galione, A.; Marques, A.R.; Medina, D.L.; Napolitano, G.; et al. Current methods to analyse lysosome morphology, positioning, motility and function. Traffic 2022, 23, 238–269. [Google Scholar] [CrossRef]
- Cai, C.Z.; Yang, C.; Zhuang, X.X.; Yuan, N.N.; Wu, M.Y.; Tan, J.Q.; Song, J.X.; Cheung, K.H.; Su, H.; Wang, Y.T.; et al. NRBF2 is a RAB7 effector required for autophagosome maturation and mediates the association of APP-CTFs with active form of RAB7 for degradation. Autophagy 2021, 17, 1112–1130. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.G.; Zhang, H. Autophagosome maturation: An epic journey from the ER to lysosomes. J. Cell Biol. 2019, 218, 757–770. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.G.; Codogno, P.; Zhang, H. Machinery, regulation and pathophysiological implications of autophagosome maturation. Nat. Rev. Mol. Cell Biol. 2021, 22, 733–750. [Google Scholar] [CrossRef] [PubMed]
- Kuchitsu, Y.; Homma, Y.; Fujita, N.; Fukuda, M. Rab7 knockout unveiled regulated autolysosome maturation induced by glutamine starvation. J. Cell Sci. 2018, 131, jcs215442. [Google Scholar] [CrossRef] [PubMed]
- Guerra, F.; Bucci, C. Multiple Roles of the Small GTPase Rab7. Cells 2016, 5, 34. [Google Scholar] [CrossRef]
- Arcaro, A.; Wymann, M.P. Wortmannin is a potent phosphatidylinositol 3-kinase inhibitor: The role of phosphatidylinositol 3,4,5-trisphosphate in neutrophil responses. Biochem. J. 1993, 296, 297–301. [Google Scholar] [CrossRef]
- Obara, K.; Noda, T.; Niimi, K.; Ohsumi, Y. Transport of phosphatidylinositol 3-phosphate into the vacuole via autophagic membranes in Saccharomyces cerevisiae. Genes Cells 2008, 13, 537–547. [Google Scholar] [CrossRef]
- Axe, E.L.; Walker, S.A.; Manifava, M.; Chandra, P.; Roderick, H.L.; Habermann, A.; Griffiths, G.; Ktistakis, N.T. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell Biol. 2008, 182, 685–701. [Google Scholar] [CrossRef]
- Agola, J.O.; Hong, L.; Surviladze, Z.; Ursu, O.; Waller, A.; Strouse, J.J.; Simpson, D.S.; Schroeder, C.E.; Oprea, T.I.; Golden, J.E.; et al. A competitive nucleotide binding inhibitor: In vitro characterization of Rab7 GTPase inhibition. ACS Chem. Biol. 2012, 7, 1095–1108. [Google Scholar] [CrossRef]
- Yuseff, M.I.; Pierobon, P.; Reversat, A.; Lennon-Duménil, A.M. How B cells capture, process and present antigens: A crucial role for cell polarityTitle. Nat. Rev. Immunol. 2013, 13, 475–486. [Google Scholar] [CrossRef] [PubMed]
- Barbero, P.; Bittova, L.; Pfeffer, S.R. Visualization of Rab9-mediated vesicle transport from endosomes to the trans-Golgi in living cells. J. Cell Biol. 2002, 156, 511–518. [Google Scholar] [CrossRef] [PubMed]
- Hernández-Pérez, S.; Mattila, P.K. A specific hybridisation internalisation probe (SHIP) enables precise live-cell and super-resolution imaging of internalized cargo. Sci. Rep. 2022, 12, 620. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.G.; Stevens, M.W.; So, L.; Edinger, A.L. Reciprocal effects of rab7 deletion in activated and neglected T cells. Autophagy 2013, 9, 1009–1023. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, M.G.; Munafó, D.B.; Berón, W.; Colombo, M.I. Rab7 is required for the normal progression of the autophagic pathway in mammalian cells. J. Cell Sci. 2004, 117, 2687–2697. [Google Scholar] [CrossRef] [PubMed]
- Jäger, S.; Bucci, C.; Tanida, I.; Ueno, T.; Kominami, E.; Saftig, P.; Eskelinen, E.L. Role for Rab7 in maturation of late autophagic vacuoles. J. Cell Sci. 2004, 117, 4837–4848. [Google Scholar] [CrossRef] [PubMed]
- Feng, Y.; He, D.; Yao, Z.; Klionsky, D.J. The machinery of macroautophagy. Cell Res. 2014, 24, 24–41. [Google Scholar] [CrossRef]
- Martinez-Martin, N.; Maldonado, P.; Gasparrini, F.; Frederico, B.; Aggarwal, S.; Gaya, M.; Tsui, C.; Burbage, M.; Keppler, S.J.; Montaner, B.; et al. A switch from canonical to noncanonical autophagy shapes B cell responses. Science 2017, 355, 641–647. [Google Scholar] [CrossRef]
- Chen, M.; Hong, M.J.; Sun, H.; Wang, L.; Shi, X.; Gilbert, B.E.; Corry, D.B.; Kheradmand, F.; Wang, J. Essential role for autophagy in the maintenance of immunological memory against influenza infection. Nat. Med. 2014, 20, 503–510. [Google Scholar] [CrossRef]
- Renard, H.-F.; Simunovic, M.; Lemière, J.; Boucrot, E.; Garcia-Castillo, M.D.; Arumugam, S.; Chambon, V.; Lamaze, C.; Wunder, C.; Kenworthy, A.K.; et al. Endophilin-A2 functions in membrane scission in clathrin-independent endocytosis. Nature 2014, 517, 493–496. [Google Scholar] [CrossRef]
- Schmid, D.; Pypaert, M.; Münz, C. MHC class II antigen loading compartments continuously receive input from autophagosomes. Immunity 2007, 26, 79–92. [Google Scholar] [CrossRef] [PubMed]
- Münz, C. Canonical and Non-Canonical Functions of the Autophagy Machinery in MHC Restricted Antigen Presentation. Front. Immunol. 2022, 13, 868888. [Google Scholar] [CrossRef]
- Watanabe, K.; Tsubata, T. Autophagy connects antigen receptor signaling to costimulatory signaling in B lymphocytes. Autophagy 2009, 5, 108–110. [Google Scholar] [CrossRef] [PubMed]
- Galluzzi, L.; Green, D.R. Autophagy-Independent Functions of the Autophagy Machinery. Cell 2019, 177, 1682–1699. [Google Scholar] [CrossRef] [PubMed]
- Arbogast, F.; Arnold, J.; Hammann, P.; Kuhn, L.; Chicher, J.; Murera, D.; Weishaar, J.; Muller, S.; Fauny, J.D.; Gros, F. ATG5 is required for B cell polarization and presentation of particulate antigens. Autophagy 2019, 15, 280–294. [Google Scholar] [CrossRef] [PubMed]
- Eskelinen, E.L. Maturation of autophagic vacuoles in Mammalian cells. Autophagy 2005, 1, 1–10. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Runsala, M.; Kuokkanen, E.; Uski, E.; Šuštar, V.; Balci, M.Ö.; Rajala, J.; Paavola, V.; Mattila, P.K. The Small GTPase Rab7 Regulates Antigen Processing in B Cells in a Possible Interplay with Autophagy Machinery. Cells 2023, 12, 2566. https://doi.org/10.3390/cells12212566
Runsala M, Kuokkanen E, Uski E, Šuštar V, Balci MÖ, Rajala J, Paavola V, Mattila PK. The Small GTPase Rab7 Regulates Antigen Processing in B Cells in a Possible Interplay with Autophagy Machinery. Cells. 2023; 12(21):2566. https://doi.org/10.3390/cells12212566
Chicago/Turabian StyleRunsala, Marika, Elina Kuokkanen, Eveliina Uski, Vid Šuštar, Meryem Özge Balci, Johanna Rajala, Vilma Paavola, and Pieta K. Mattila. 2023. "The Small GTPase Rab7 Regulates Antigen Processing in B Cells in a Possible Interplay with Autophagy Machinery" Cells 12, no. 21: 2566. https://doi.org/10.3390/cells12212566




