WAVE2 Regulates Actin-Dependent Processes Induced by the B Cell Antigen Receptor and Integrins
Abstract
:1. Introduction
2. Materials and Methods
2.1. B Cells and siRNA Transfection
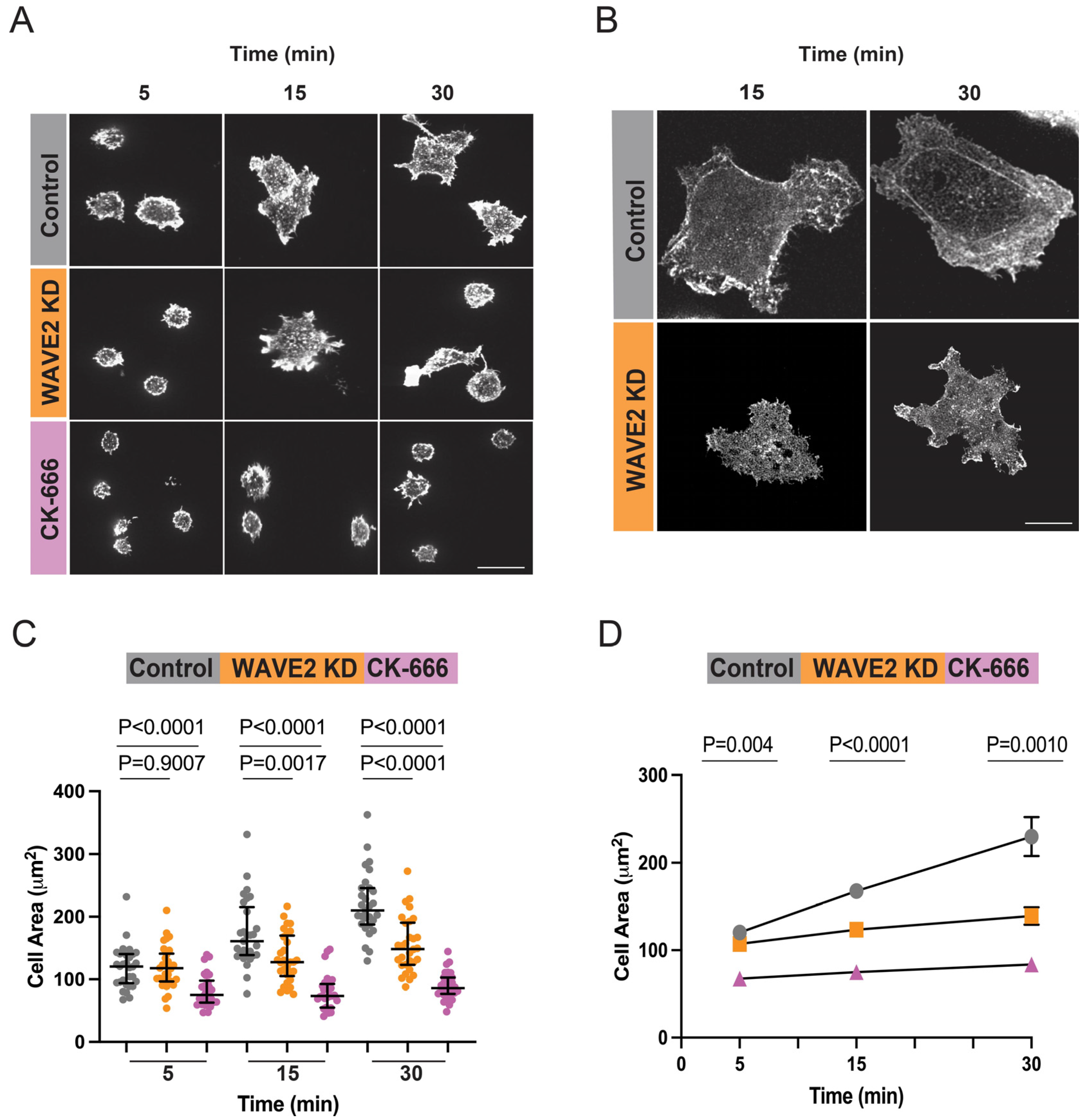
2.2. Cell Spreading Analysis
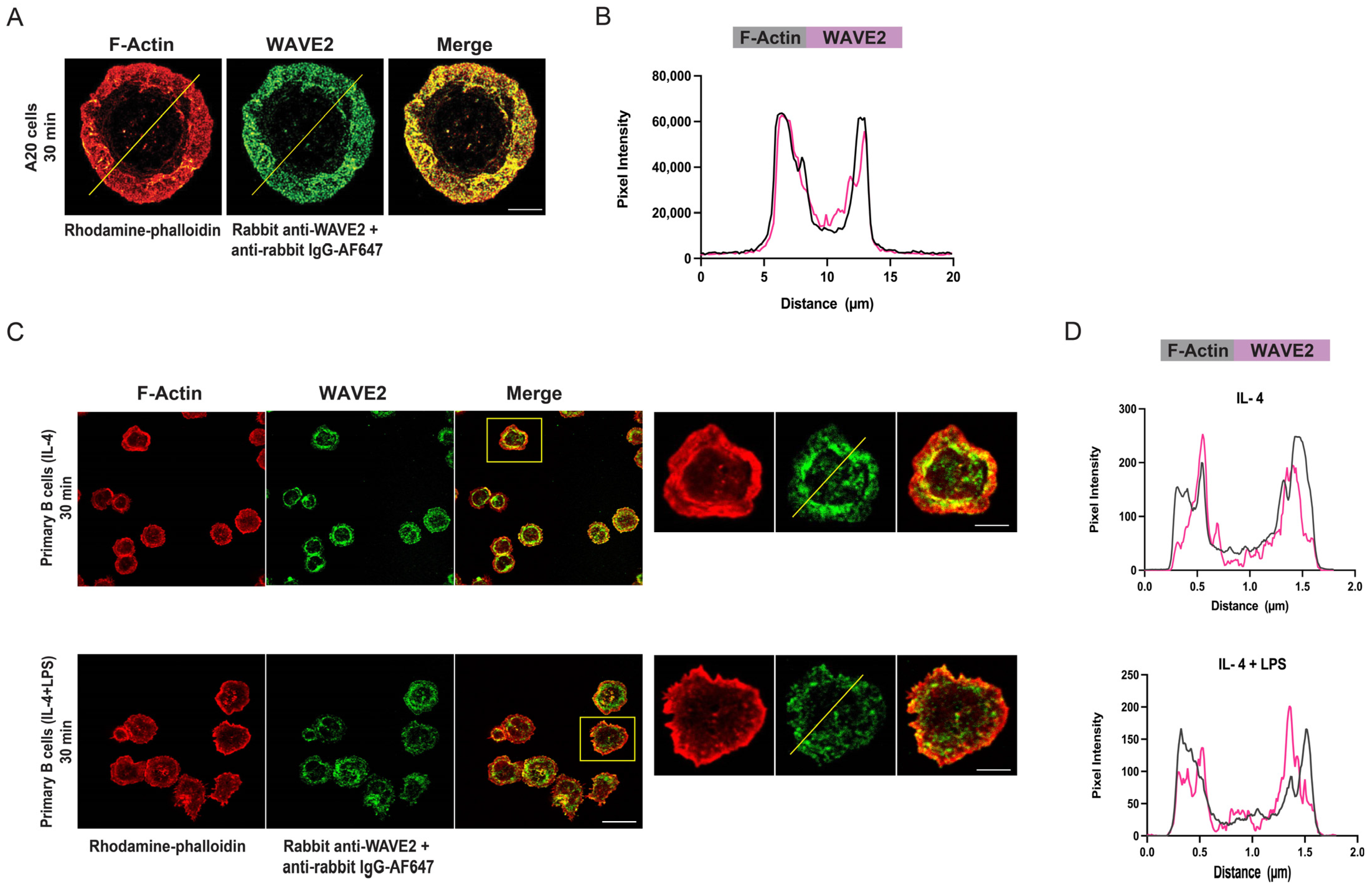
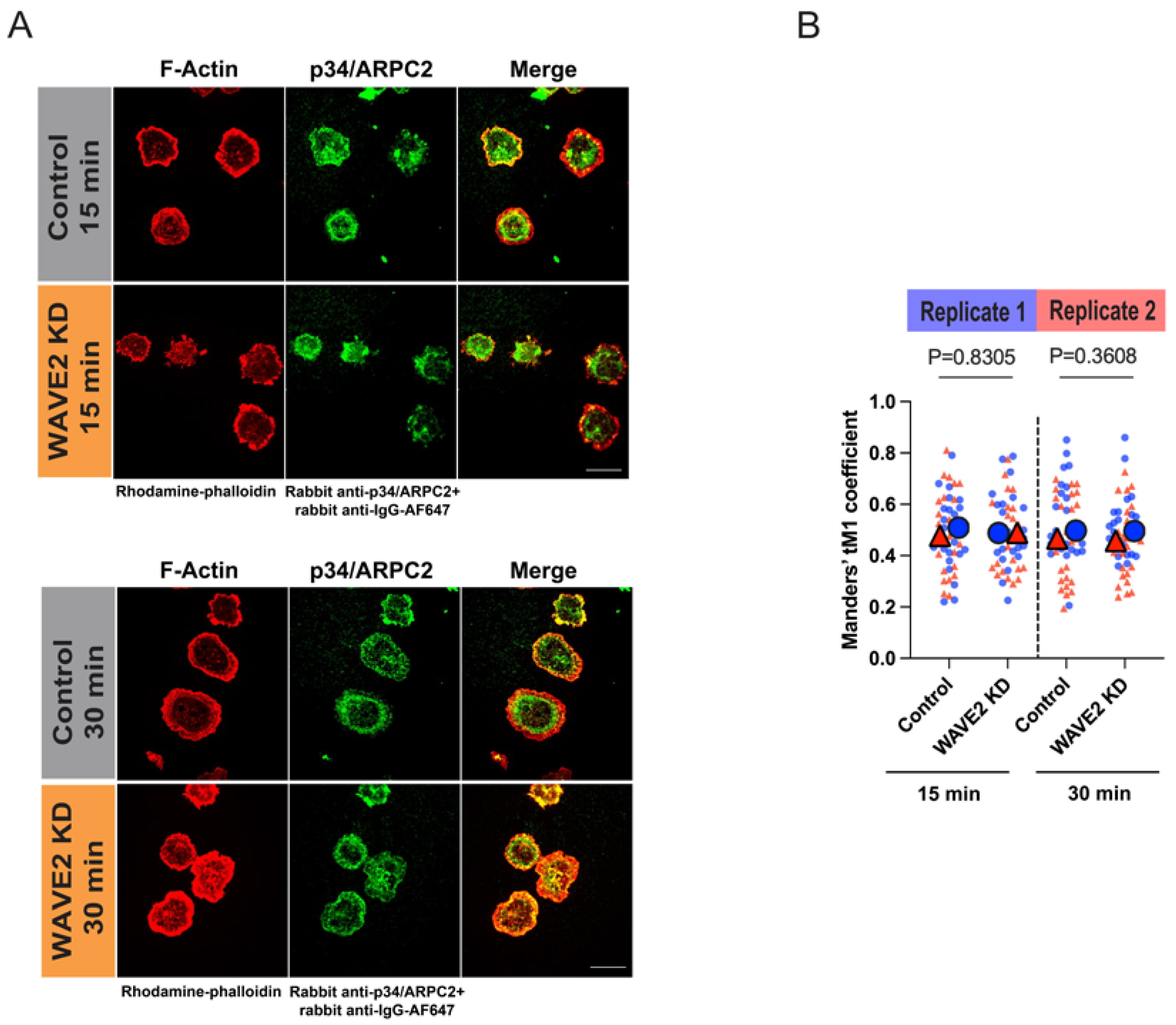
2.3. Co-Localization of WAVE2 or the Arp2/3 Complex with F-Actin
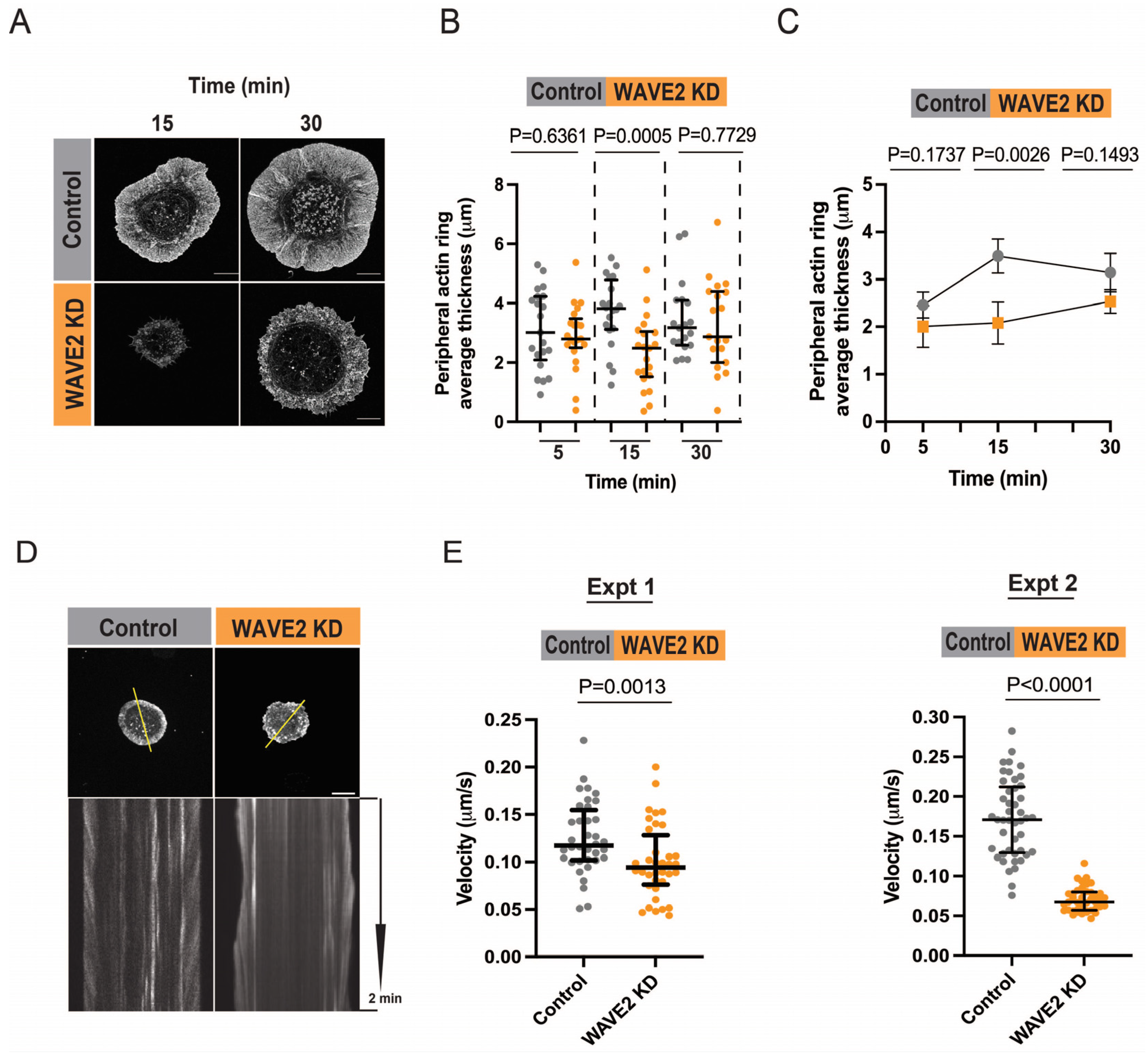
2.4. Actin Retrograde Flow
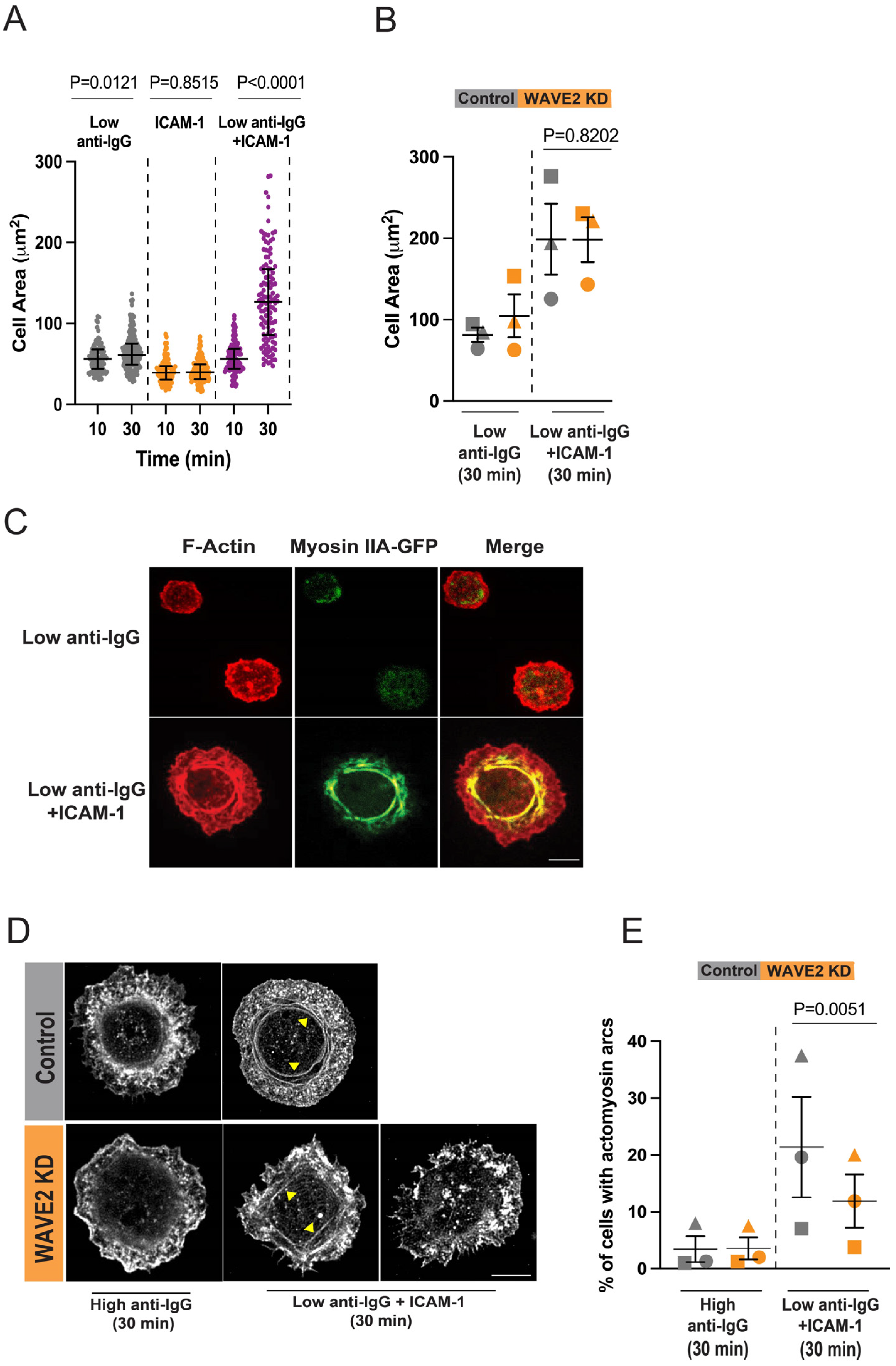
2.5. Visualization of Actomyosin Arcs
2.6. BCR Signaling in Response to Immobilized Anti-IgG
2.7. Immunoblotting
2.8. B Cell Responses to Antigen-Bearing Surrogate APCs
2.9. Statistical Analysis
3. Results
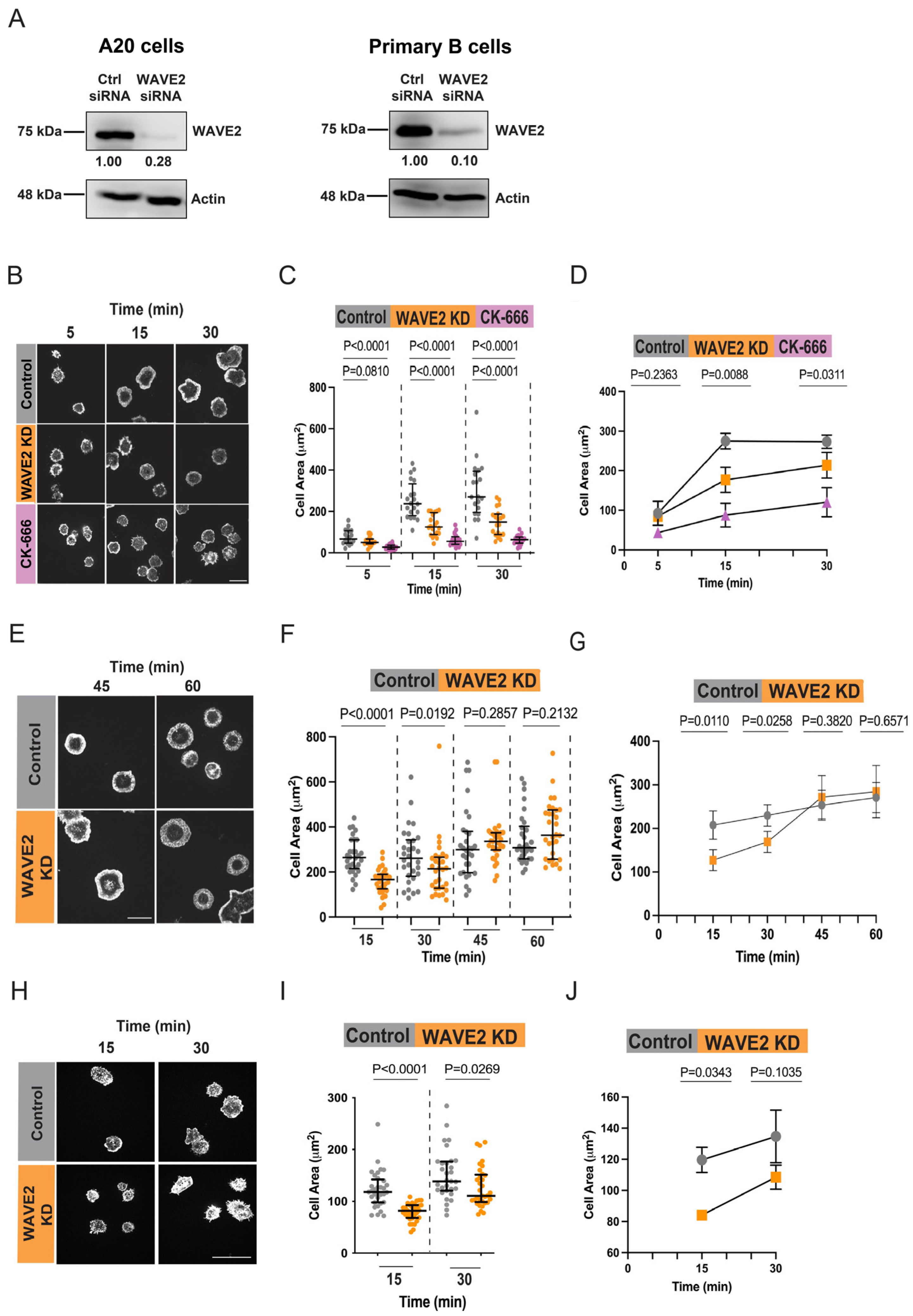
3.1. WAVE2 Regulates BCR-Induced B Cell Spreading
3.2. WAVE2 Regulates Peripheral Actin Dynamics
3.3. WAVE2 Regulates LFA-1-Induced Formation of Actomyosin Arcs
3.4. WAVE2 Regulates Integrin-Induced B Cell Spreading on Fibronectin
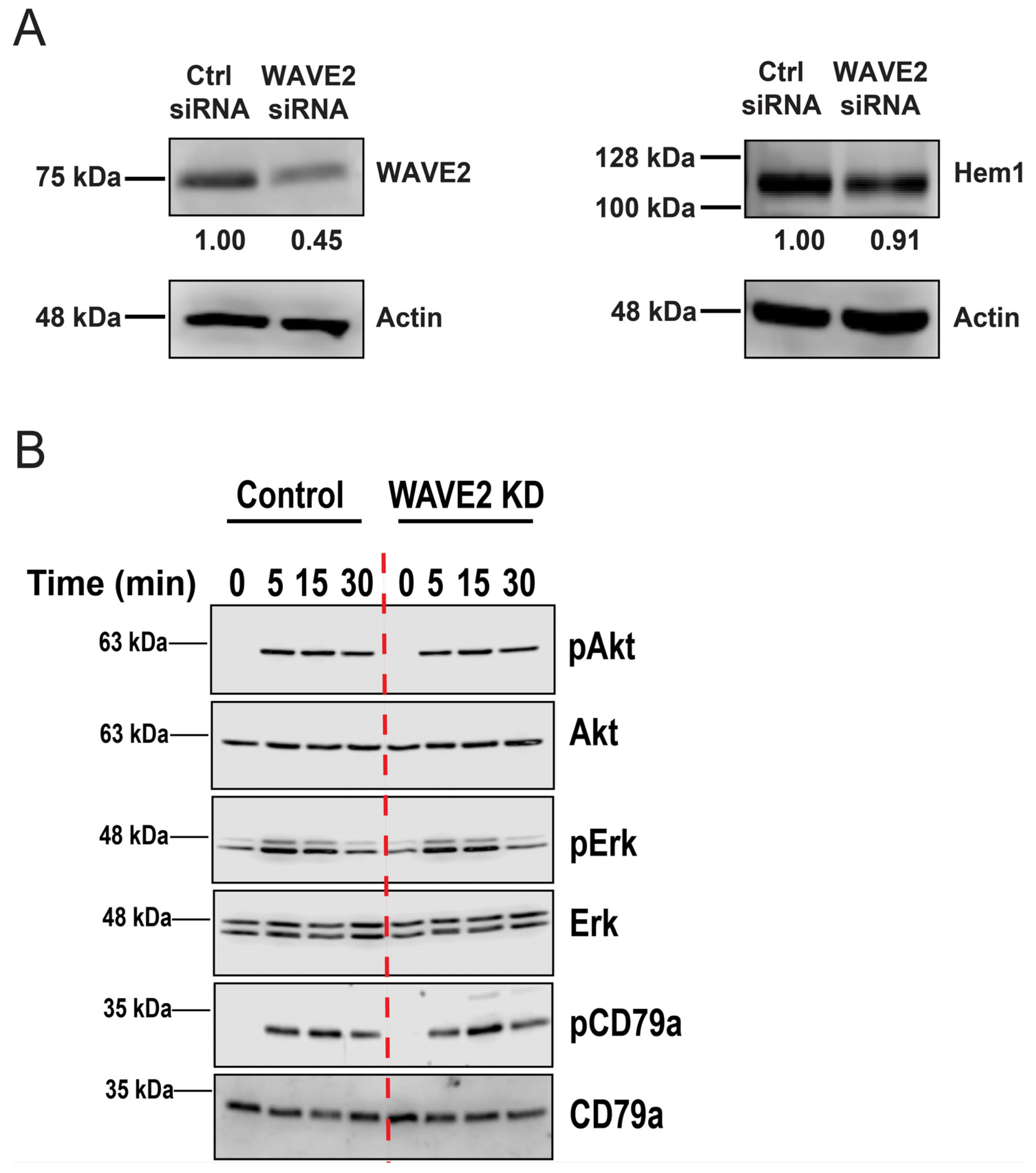
3.5. WAVE2 Enhances APC-Induced Proximal BCR Signaling and B Cell Activation
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abraham, L.; Wang, J.C.; Bolger-Munro, M.; Gold, M.R. Structure, function, and spatial organization of the B cell receptor. In Encyclopedia of Immunobiology; Ratcliffe, M.J.H., Ed.; Elsivier: Amsterdam, The Netherlands, 2016; Volume 2, pp. 40–54. [Google Scholar] [CrossRef]
- Shen, P.; Fillatreau, S. Antibody-independent functions of B cells: A focus on cytokines. Nat. Rev. Immunol. 2015, 15, 441–451. [Google Scholar] [CrossRef] [PubMed]
- Kwak, K.; Akkaya, M.; Pierce, S.K. B cell signaling in context. Nat. Immunol. 2019, 20, 963–969. [Google Scholar] [CrossRef] [PubMed]
- Kurosaki, T.; Shinohara, H.; Baba, Y. B cell signaling and fate decision. Annu. Rev. Immunol. 2010, 28, 21–55. [Google Scholar] [CrossRef] [PubMed]
- Fleire, S.J.; Goldman, J.P.; Carrasco, Y.R.; Weber, M.; Bray, D.; Batista, F.D. B cell ligand discrimination through a spreading and contraction response. Science 2006, 312, 738–741. [Google Scholar] [CrossRef] [PubMed]
- Harwood, N.E.; Batista, F.D. The cytoskeleton coordinates the early events of B-cell activation. Cold Spring Harb. Perspect. Biol. 2011, 3, a002360. [Google Scholar] [CrossRef] [PubMed]
- Song, W.; Liu, C.; Upadhyaya, A. The pivotal position of the actin cytoskeleton in the initiation and regulation of B cell receptor activation. Biochim. Biophys. Acta 2014, 1838, 569–578. [Google Scholar] [CrossRef] [PubMed]
- Bhanja, A.; Rey-Suarez, I.; Song, W.; Upadhyaya, A. Bidirectional feedback between BCR signaling and actin cytoskeletal dynamics. FEBS J. 2022, 289, 4430–4446. [Google Scholar] [CrossRef]
- Heesters, B.A.; van der Poel, C.E.; Das, A.; Carroll, M.C. Antigen presentation to B cells. Trends Immunol. 2016, 37, 844–854. [Google Scholar] [CrossRef]
- Batista, F.D.; Harwood, N.E. The who, how and where of antigen presentation to B cells. Nat. Rev. Immunol. 2009, 9, 15–27. [Google Scholar] [CrossRef]
- Bolger-Munro, M.; Choi, K.; Scurll, J.M.; Abraham, L.; Chappell, R.S.; Sheen, D.; Dang-Lawson, M.; Wu, X.; Priatel, J.J.; Coombs, D.; et al. Arp2/3 complex-driven spatial patterning of the BCR enhances immune synapse formation, BCR signaling and B cell activation. eLife 2019, 8, e44574. [Google Scholar] [CrossRef]
- Carrasco, Y.R.; Batista, F.D. B-cell activation by membrane-bound antigens is facilitated by the interaction of VLA-4 with VCAM-1. EMBO J. 2006, 25, 889–899. [Google Scholar] [CrossRef]
- Carrasco, Y.R.; Fleire, S.J.; Cameron, T.; Dustin, M.L.; Batista, F.D. LFA-1/ICAM-1 interaction lowers the threshold of B cell activation by facilitating B cell adhesion and synapse formation. Immunity 2004, 20, 589–599. [Google Scholar] [CrossRef]
- Liu, C.; Miller, H.; Orlowski, G.; Hang, H.; Upadhyaya, A.; Song, W. Actin reorganization is required for the formation of polarized B cell receptor signalosomes in response to both soluble and membrane-associated antigens. J. Immunol. 2012, 188, 3237–3246. [Google Scholar] [CrossRef]
- Tolar, P. Cytoskeletal control of B cell responses to antigens. Nat. Rev. Immunol. 2017, 17, 621–634. [Google Scholar] [CrossRef]
- Sprenkeler, E.G.G.; Webbers, S.D.S.; Kuijpers, T.W. When actin is not actin’ like it should: A new category of distinct primary immunodeficiency disorders. J. Innate Immun. 2021, 13, 3–25. [Google Scholar] [CrossRef]
- Tur-Gracia, S.; Martinez-Quiles, N. Emerging functions of cytoskeletal proteins in immune diseases. J. Cell Sci. 2021, 134, jcs253534. [Google Scholar] [CrossRef]
- Tangye, S.G.; Bucciol, G.; Casas-Martin, J.; Pillay, B.; Ma, C.S.; Moens, L.; Meyts, I. Human inborn errors of the actin cytoskeleton affecting immunity: Way beyond WAS and WIP. Immunol. Cell Biol. 2019, 97, 389–402. [Google Scholar] [CrossRef]
- Candotti, F. Clinical manifestations and pathophysiological mechanisms of the Wiskott-Aldrich Syndrome. J. Clin. Immunol. 2018, 38, 13–27. [Google Scholar] [CrossRef]
- Cook, S.; Lenardo, M.J.; Freeman, A.F. HEM1 actin immunodysregulatory disorder: Genotypes, phenotypes, and future directions. J. Clin. Immunol. 2022, 42, 1583–1592. [Google Scholar] [CrossRef]
- Janssen, E.; Geha, R.S. Primary immunodeficiencies caused by mutations in actin regulatory proteins. Immunol. Rev. 2019, 287, 121–134. [Google Scholar] [CrossRef]
- Leung, G.; Zhou, Y.; Ostrowski, P.; Mylvaganam, S.; Boroumand, P.; Mulder, D.J.; Guo, C.; Muise, A.M.; Freeman, S.A. ARPC1B binds WASP to control actin polymerization and curtail tonic signaling in B cells. JCI Insight 2021, 6, e149376. [Google Scholar] [CrossRef]
- Pfajfer, L.; Mair, N.K.; Jiménez-Heredia, R.; Genel, F.; Gulez, N.; Ardeniz, Ö.; Hoeger, B.; Bal, S.K.; Madritsch, C.; Kalinichenko, A.; et al. Mutations affecting the actin regulator WD repeat–containing protein 1 lead to aberrant lymphoid immunity. J. Allergy Clin. Immunol. 2018, 142, 1589–1604.e11. [Google Scholar] [CrossRef]
- Keppler, S.J.; Burbage, M.; Gasparrini, F.; Hartjes, L.; Aggarwal, S.; Massaad, M.J.; Geha, R.S.; Bruckbauer, A.; Batista, F.D. The lack of WIP binding to actin results in impaired B cell migration and altered humoral immune responses. Cell Rep. 2018, 24, 619–629. [Google Scholar] [CrossRef]
- Salzer, E.; Zoghi, S.; Kiss, M.G.; Kage, F.; Rashkova, C.; Stahnke, S.; Haimel, M.; Platzer, R.; Caldera, M.; Ardy, R.C.; et al. The cytoskeletal regulator HEM1 governs B-cell development and prevents autoimmunity. Sci. Immunol. 2020, 5, eabc3979. [Google Scholar] [CrossRef]
- Park, H.; Staehling-Hampton, K.; Appleby, M.W.; Brunkow, M.E.; Habib, T.; Zhang, Y.; Ramsdell, F.; Liggitt, H.D.; Freie, B.; Tsang, M.; et al. A point mutation in the murine Heml gene reveals an essential role for Hematopoietic Protein 1 in lymphopoiesis and innate immunity. J. Exp. Med. 2008, 205, 2899–2913. [Google Scholar] [CrossRef]
- Randall, K.L.; Lambe, T.; Johnson, A.; Treanor, B.; Kucharska, E.; Domaschenz, H.; Whittle, B.; Tze, L.E.; Enders, A.; Crockford, T.L.; et al. DOCK8 mutations cripple B cell immune synapse, germinal centers and long-lived antibody production. Nat. Immunol. 2009, 10, 1283–1291. [Google Scholar] [CrossRef]
- Sun, X.; Wang, J.; Qin, T.; Zhang, Y.; Huang, L.; Niu, L.; Bai, X.; Jing, Y.; Xuan, X.; Miller, H.; et al. Dock8 regulates BCR signaling and activation of memory B cells via WASP and CD19. Blood Adv. 2018, 2, 401–413. [Google Scholar] [CrossRef]
- Michelot, A.; Drubin, D.G. Building distinct actin filament networks in a common cytoplasm. Curr. Biol. 2011, 21, R560–R569. [Google Scholar] [CrossRef]
- Chhabra, E.S.; Higgs, H.N. The many faces of actin: Matching assembly factors with cellular structures. Nat. Cell Biol. 2007, 9, 1110–1121. [Google Scholar] [CrossRef]
- Goley, E.D.; Welch, M.D. The ARP2/3 complex: An actin nucleator comes of age. Nat. Rev. Mol. Cell Biol. 2006, 7, 713–726. [Google Scholar] [CrossRef]
- Pollard, T.D.; Borisy, G.G. Cellular motility driven by assembly and disassembly of actin filaments. Cell 2003, 112, 453–465. [Google Scholar] [CrossRef]
- Mogilner, A.; Oster, G. Cell motility driven by actin polymerization. Biophys. J. 1996, 71, 3030–3045. [Google Scholar] [CrossRef]
- Dimchev, G.; Steffen, A.; Kage, F.; Dimchev, V.; Pernier, J.; Carlier, M.F.; Rottner, K. Efficiency of lamellipodia protrusion is determined by the extent of cytosolic actin assembly. Mol. Biol. Cell 2017, 28, 1311–1325. [Google Scholar] [CrossRef]
- Pollitt, A.Y.; Insall, R.H. WASP and SCAR/WAVE proteins: The drivers of actin assembly. J. Cell Sci. 2009, 122, 2575–2578. [Google Scholar] [CrossRef]
- Pauker, M.H.; Reicher, B.; Joseph, N.; Wortzel, I.; Jakubowicz, S.; Noy, E.; Perl, O.; Barda-Saad, M. WASp family verprolin-homologous protein-2 (WAVE2) and Wiskott-Aldrich Syndrome protein (WASp) engage in distinct downstream signaling interactions at the T cell antigen receptor site. J. Biol. Chem. 2014, 289, 34503–34519. [Google Scholar] [CrossRef]
- Gaertner, F.; Reis-Rodrigues, P.; de Vries, I.; Hons, M.; Aguilera, J.; Riedl, M.; Leithner, A.; Tasciyan, S.; Kopf, A.; Merrin, J.; et al. WASp triggers mechanosensitive actin patches to facilitate immune cell migration in dense tissues. Dev. Cell 2022, 57, 47–62.e9. [Google Scholar] [CrossRef]
- Liu, C.; Miller, H.; Hui, K.L.; Grooman, B.; Bolland, S.; Upadhyaya, A.; Song, W. A balance of Bruton’s tyrosine kinase and SHIP activation regulates B cell receptor cluster formation by controlling actin remodeling. J. Immunol. 2011, 187, 230–239. [Google Scholar] [CrossRef]
- Westerberg, L.S.; Dahlberg, C.; Baptista, M.; Moran, C.J.; Detre, C.; Keszei, M.; Eston, M.A.; Alt, F.W.; Terhorst, C.; Notarangelo, L.D.; et al. Wiskott-Aldrich syndrome protein (WASP) and N-WASP are critical for peripheral B-cell development and function. Blood 2012, 119, 3966–3974. [Google Scholar] [CrossRef]
- Meyer-Bahlburg, A.; Becker-Herman, S.; Humblet-Baron, S.; Khim, S.; Weber, M.; Bouma, G.; Thrasher, A.J.; Batista, F.D.; Rawlings, D.J. Wiskott-Aldrich syndrome protein deficiency in B cells results in impaired peripheral homeostasis. Blood 2008, 112, 4158–4169. [Google Scholar] [CrossRef]
- Westerberg, L.; Larsson, M.; Hardy, S.J.; Fernández, C.; Thrasher, A.J.; Severinson, E. Wiskott-Aldrich syndrome protein deficiency leads to reduced B-cell adhesion, migration, and homing, and a delayed humoral immune response. Blood 2005, 105, 1144–1152. [Google Scholar] [CrossRef]
- Westerberg, L.S.; De La Fuente, M.A.; Wermeling, F.; Ochs, H.D.; Karlsson, M.C.I.; Snapper, S.B.; Notarangelo, L.D. WASP confers selective advantage for specific hematopoietic cell populations and serves a unique role in marginal zone B-cell homeostasis and function. Blood 2008, 112, 4139–4147. [Google Scholar] [CrossRef]
- Recher, M.; Burns, S.O.; De La Fuente, M.A.; Volpi, S.; Dahlberg, C.; Walter, J.E.; Moffitt, K.; Mathew, D.; Honke, N.; Lang, P.A.; et al. B cell-intrinsic deficiency of the Wiskott-Aldrich syndrome protein (WASp) causes severe abnormalities of the peripheral B-cell compartment in mice. Blood 2012, 119, 2819–2828. [Google Scholar] [CrossRef]
- Becker-Herman, S.; Meyer-Bahlburg, A.; Schwartz, M.A.; Jackson, S.W.; Hudkins, K.L.; Liu, C.; Sather, B.D.; Khim, S.; Liggitt, D.; Song, W.; et al. WASp-deficient B cells play a critical, cell-intrinsic role in triggering autoimmunity. J. Exp. Med. 2011, 208, 2033–2042. [Google Scholar] [CrossRef]
- Volpi, S.; Santori, E.; Abernethy, K.; Mizui, M.; Dahlberg, C.I.M.; Recher, M.; Capuder, K.; Csizmadia, E.; Ryan, D.; Mathew, D.; et al. N-WASP is required for B-cell–mediated autoimmunity in Wiskott-Aldrich syndrome. Blood 2016, 127, 216–220. [Google Scholar] [CrossRef]
- Vermi, W.; Blanzuoli, L.; Kraus, M.D.; Grigolato, P.; Donato, F.; Loffredo, G.; Marino, C.E.; Alberti, D.; Notarangelo, L.D.; Facchetti, F. The spleen in the Wiskott-Aldrich syndrome: Histopathologic abnormalities of the white pulp correlate with the clinical phenotype of the disease. Am. J. Surg. Pathol. 1999, 23, 182–191. [Google Scholar] [CrossRef]
- Takenawa, T.; Suetsugu, S. The WASP–WAVE protein network: Connecting the membrane to the cytoskeleton. Nat. Rev. Mol. Cell Biol. 2007, 8, 37–48. [Google Scholar] [CrossRef]
- Gene Skyline. Available online: http://rstats.immgen.org/Skyline/skyline.html (accessed on 11 June 2023).
- COVID-19 Expression Data. Available online: https://www.immgen.org/Databrowser19/HumanExpressionData.html (accessed on 11 June 2023).
- Chen, Z.; Borek, D.; Padrick, S.B.; Gomez, T.S.; Metlagel, Z.; Ismail, A.M.; Umetani, J.; Billadeau, D.D.; Otwinowski, Z.; Rosen, M.K. Structure and control of the actin regulatory WAVE complex. Nature 2010, 468, 533–538. [Google Scholar] [CrossRef]
- Rottner, K.; Stradal, T.E.B.; Chen, B. WAVE regulatory complex. Curr. Biol. 2021, 31, R512–R517. [Google Scholar] [CrossRef]
- Avalos, A.; Tietsort, J.T.; Suwankitwat, N.; Woods, J.D.; Jackson, S.W.; Christodoulou, A.; Morrill, C.; Liggitt, H.D.; Zhu, C.; Li, Q.Z.; et al. Hem-1 regulates protective humoral immunity and limits autoantibody production in a B cell–specific manner. JCI Insight 2022, 7, e153597. [Google Scholar] [CrossRef]
- Cook, S.A.; Comrie, W.A.; Poli, M.C.; Similuk, M.; Oler, A.J.; Faruqi, A.J.; Kuhns, D.B.; Yang, S.; Vargas-Hernández, A.; Carisey, A.F.; et al. HEM1 deficiency disrupts mTORC2 and F-actin control in inherited immunodysregulatory disease. Science 2020, 369, 202–207. [Google Scholar] [CrossRef]
- Weiner, O.D.; Rentel, M.C.; Ott, A.; Brown, G.E.; Jedrychowski, M.; Yaffe, M.B.; Gygi, S.P.; Cantley, L.C.; Bourne, H.R.; Kirschner, M.W. Hem-1 complexes are essential for rac activation, actin polymerization, and myosin regulation during neutrophil chemotaxis. PLoS Biol. 2006, 4, e38. [Google Scholar] [CrossRef]
- Sarbassov, D.D.; Guertin, D.A.; Ali, S.M.; Sabatini, D.M. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 2005, 307, 1098–1101. [Google Scholar] [CrossRef]
- Jacinto, E.; Loewith, R.; Schmidt, A.; Lin, S.; Rüegg, M.A.; Hall, A.; Hall, M.N. Mammalian TOR complex 2 controls the actin cytoskeleton and is rapamycin insensitive. Nat. Cell Biol. 2004, 6, 1122–1128. [Google Scholar] [CrossRef]
- Dos, D.S.; Ali, S.M.; Kim, D.H.; Guertin, D.A.; Latek, R.R.; Erdjument-Bromage, H.; Tempst, P.; Sabatini, D.M. Rictor, a novel binding partner of mTOR, defines a rapamycin-insensitive and raptor-independent pathway that regulates the cytoskeleton. Curr. Biol. 2004, 14, 1296–1302. [Google Scholar] [CrossRef]
- Wu, D.; Yu, D.; Wang, X.; Yu, B. F-actin rearrangement is regulated by mTORC2/Akt/Girdin in mouse fertilized eggs. Cell Prolif. 2016, 49, 740–750. [Google Scholar] [CrossRef]
- Diz-Muñoz, A.; Thurley, K.; Chintamen, S.; Altschuler, S.J.; Wu, L.F.; Fletcher, D.A.; Weiner, O.D. Membrane tension acts through PLD2 and mTORC2 to limit actin network assembly during neutrophil migration. PLoS Biol. 2016, 14, e1002474. [Google Scholar] [CrossRef]
- Kim, K.J.; Kanellopoulos-Langevin, C.; Merwin, R.M.; Sachs, D.H.; Asofsky, R. Establishment and characterization of BALB/c lymphoma lines with B cell properties. J. Immunol. 1979, 122, 549–554. [Google Scholar] [CrossRef]
- Wang, J.C.; Bolger-Munro, M.; Gold, M.R. Imaging the interactions between B cells and antigen-presenting cells. Methods Mol. Biol. 2018, 1707, 131–161. [Google Scholar] [CrossRef]
- Wang, J.C.; Bolger-Munro, M.; Gold, M.R. Visualizing the actin and microtubule cytoskeletons at the B-cell immune synapse using stimulated emission depletion (STED) microscopy. J. Vis. Exp. 2018, 134, e57028. [Google Scholar] [CrossRef]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef]
- Johnson, H.W.; Schell, M.J. Neuronal IP3 3-kinase is an F-actin-bundling protein: Role in dendritic targeting and regulation of spine morphology. Mol. Biol. Cell 2009, 20, 5166–5180. [Google Scholar] [CrossRef]
- Jacobelli, J.; Chmura, S.A.; Buxton, D.B.; Davis, M.M.; Krummel, M.F. A single class II myosin modulates T cell motility and stopping, but not synapse formation. Nat. Immunol. 2004, 5, 531–538. [Google Scholar] [CrossRef] [PubMed]
- Gold, M.R.; Matsuuchi, L.; Kelly, R.B.; DeFranco, A.L. Tyrosine phosphorylation of components of the B-cell antigen receptors following receptor crosslinking. Proc. Natl. Acad. Sci. USA 1991, 88, 3436–3440. [Google Scholar] [CrossRef] [PubMed]
- Freeman, S.A.; Lei, V.; Dang-Lawson, M.; Mizuno, K.; Roskelley, C.D.; Gold, M.R. Cofilin-mediated F-actin severing Is regulated by the Rap GTPase and controls the cytoskeletal dynamics that drive lymphocyte spreading and BCR microcluster formation. J. Immunol. 2011, 187, 5887–9000. [Google Scholar] [CrossRef] [PubMed]
- Ait-Azzouzene, D.; Verkoczy, L.; Peters, J.; Gavin, A.; Skog, P.; Vela, J.L.; Nemazee, D. An immunoglobulin Cκ-reactive single chain antibody fusion protein induces tolerance through receptor editing in a normal polyclonal immune system. J. Exp. Med. 2005, 201, 817–828. [Google Scholar] [CrossRef] [PubMed]
- FIJImacros/APC_Analyser_MBM.ijm at Master Mbolgermunro/FIJImacros GitHub. Available online: https://github.com/mbolgermunro/FIJImacros/blob/master/APC_analyser_MBM.ijm (accessed on 30 January 2023).
- Bolger-Munro, M.; Choi, K.; Cheung, F.; Liu, Y.T.; Dang-Lawson, M.; Deretic, N.; Keane, C.; Gold, M.R. The Wdr1-LIMK-cofilin axis controls B cell antigen receptor-induced actin remodeling and signaling at the immune synapse. Front. Cell Dev. Biol. 2021, 9, 649433. [Google Scholar] [CrossRef]
- Motulsky, H.J.; Brown, R.E. Detecting outliers when fitting data with nonlinear regression—A new method based on robust nonlinear regression and the false discovery rate. BMC Bioinform. 2006, 7, 123. [Google Scholar] [CrossRef]
- Smith, B.A.; Daugherty-Clarke, K.; Goode, B.L.; Gelles, J. Pathway of actin filament branch formation by Arp2/3 complex revealed by single-molecule imaging. Proc. Natl. Acad. Sci. USA 2013, 110, 1285–1290. [Google Scholar] [CrossRef]
- Gautreau, A.M.; Fregoso, F.E.; Simanov, G.; Dominguez, R. Nucleation, stabilization, and disassembly of branched actin networks. Trends Cell Biol. 2022, 32, 421–432. [Google Scholar] [CrossRef]
- Carrasco, Y.R. Molecular cues involved in the regulation of B cell dynamics: Assistants of antigen hunting. J. Leukoc. Biol. 2020, 107, 1107–1113. [Google Scholar] [CrossRef]
- Mace, E.M.; Zhang, J.; Siminovitch, K.A.; Takei, F. Elucidation of the integrin LFA-1–mediated signaling pathway of actin polarization in natural killer cells. Blood 2010, 116, 1272–1279. [Google Scholar] [CrossRef]
- Wang, J.C.; Yim, Y.-I.; Wu, X.; Jaumouille, V.; Cameron, A.; Waterman, C.M.; Kehrl, J.H.; Hammer, J.A. A B-cell actomyosin arc network couples integrin co-stimulation to mechanical force-dependent immune synapse formation. eLife 2022, 11, e72805. [Google Scholar] [CrossRef]
- Stupack, D.G.; Stewart, S.; Carter, W.G.; Wayner, E.A.; Wilkins, J.A. B lymphocyte fibronectin receptors: Expression and utilization. Scand. J. Immunol. 1991, 34, 761–769. [Google Scholar] [CrossRef]
- Ketchum, C.; Miller, H.; Song, W.; Upadhyaya, A. Ligand mobility regulates B cell receptor clustering and signaling activation. Biophys. J. 2014, 106, 26–36. [Google Scholar] [CrossRef]
- BioRender. Available online: https://www.biorender.com (accessed on 4 February 2023).
- Dolati, S.; Kage, F.; Mueller, J.; Müsken, M.; Kirchner, M.; Dittmar, G.; Sixt, M.; Rottner, K.; Falckea, M. On the relation between filament density, force generation, and protrusion rate in mesenchymal cell motility. Mol. Biol. Cell 2018, 29, 2674–2886. [Google Scholar] [CrossRef]
- Liu, C.; Bai, X.; Wu, J.; Sharma, S.; Upadhyaya, A.; Dahlberg, C.I.M.; Westerberg, L.S.; Snapper, S.B.; Zhao, X.; Song, W. N-WASP is essential for the negative regulation of B cell receptor signaling. PLoS Biol. 2013, 11, e1001704. [Google Scholar] [CrossRef]
- Zimmermann, J.; Brunner, C.; Enculescu, M.; Goegler, M.; Ehrlicher, A.; Käs, J.; Falcke, M. Actin filament elasticity and retrograde flow shape the force-velocity relation of motile cells. Biophys. J. 2012, 102, 287–295. [Google Scholar] [CrossRef]
- Bieling, P.; Hansen, S.D.; Akin, O.; Li, T.; Hayden, C.C.; Fletcher, D.A.; Mullins, R.D. WH2 and proline-rich domains of WASP-family proteins collaborate to accelerate actin filament elongation. EMBO J. 2018, 37, 102–121. [Google Scholar] [CrossRef]
- Tang, Q.; Schaks, M.; Koundinya, N.; Yang, C.; Pollard, L.W.; Svitkina, T.M.; Rottner, K.; Goode, B.L. WAVE1 and WAVE2 have distinct and overlapping roles in controlling actin assembly at the leading edge. Mol. Biol. Cell 2020, 31, 2168–2178. [Google Scholar] [CrossRef]
- Buracco, S.; Singh, S.; Claydon, S.; Paschke, P.; Tweedy, L.; Whitelaw, J.; McGarry, L.; Thomason, P.A.; Insall, R.H. The Scar/WAVE complex drives normal actin protrusions without the Arp2/3 complex, but proline-rich domains are required. bioRxiv 2022, 491902. [Google Scholar] [CrossRef]
- Pipathsouk, A.; Brunetti, R.M.; Town, J.P.; Graziano, B.R.; Breuer, A.; Pellett, P.A.; Marchuk, K.; Tran, N.H.T.; Krummel, M.F.; Stamou, D.; et al. The WAVE complex associates with sites of saddle membrane curvature. J. Cell Biol. 2021, 220, e202003086. [Google Scholar] [CrossRef] [PubMed]
- Murugesan, S.; Hong, J.; Yi, J.; Li, D.; Beach, J.R.; Shao, L.; Meinhardt, J.; Madison, G.; Wu, X.; Betzig, E.; et al. Formin-generated actomyosin arcs propel T cell receptor microcluster movement at the immune synapse. J. Cell Biol. 2016, 215, 383–399. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Martín, L.; Sánchez-Sánchez, N.; Gutiérrez-López, M.D.; Rojo, A.I.; Vicente-Manzanares, M.; Pérez-Alvarez, M.J.; Sánchez-Mateos, P.; Bustelo, X.R.; Cuadrado, A.; Sánchez-Madrid, F.; et al. Signaling through the leukocyte integrin LFA-1 in T cells induces a transient activation of Rac-1 that is regulated by Vav and PI3K/Akt-1. J. Biol. Chem. 2004, 279, 16194–16205. [Google Scholar] [CrossRef] [PubMed]
- Lomakin, A.J.; Lee, K.C.; Han, S.J.; Bui, D.A.; Davidson, M.; Mogilner, A.; Danuser, G. Competition for actin between two distinct F-actin networks defines a bistable switch for cell polarization. Nat. Cell Biol. 2015, 17, 1435–1445. [Google Scholar] [CrossRef] [PubMed]
- Das, M.; Subbayya Ithychanda, S.; Qin, J.; Plow, E.F. Mechanisms of talin-dependent integrin signaling and crosstalk. Biochim. Biophys. Acta 2014, 1838, 579–588. [Google Scholar] [CrossRef]
- Härzchel, A.; Zucchetto, A.; Gattei, V.; Hartmann, T.N. VLA-4 expression and activation in B cell malignancies: Functional and clinical aspects. Int. J. Mol. Sci. 2020, 21, 2206. [Google Scholar] [CrossRef]
- Lin, K.B.L.; Tan, P.; Freeman, S.A.; Lam, M.; McNagny, K.M.; Gold, M.R. The Rap GTPases regulate the migration, invasiveness and in vivo dissemination of B-cell lymphomas. Oncogene 2010, 29, 608–615. [Google Scholar] [CrossRef]
- Yan, C.; Martinez-Quiles, N.; Eden, S.; Shibata, T.; Takeshima, F.; Shinkura, R.; Fujiwara, Y.; Bronson, R.; Snapper, S.B.; Kirschner, M.W.; et al. WAVE2 deficiency reveals distinct roles in embryogenesis and Rac-mediated actin-based motility. EMBO J. 2003, 22, 3602–3612. [Google Scholar] [CrossRef]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bedi, A.; Choi, K.; Keane, C.; Bolger-Munro, M.; Ambrose, A.R.; Gold, M.R. WAVE2 Regulates Actin-Dependent Processes Induced by the B Cell Antigen Receptor and Integrins. Cells 2023, 12, 2704. https://doi.org/10.3390/cells12232704
Bedi A, Choi K, Keane C, Bolger-Munro M, Ambrose AR, Gold MR. WAVE2 Regulates Actin-Dependent Processes Induced by the B Cell Antigen Receptor and Integrins. Cells. 2023; 12(23):2704. https://doi.org/10.3390/cells12232704
Chicago/Turabian StyleBedi, Abhishek, Kate Choi, Connor Keane, Madison Bolger-Munro, Ashley R. Ambrose, and Michael R. Gold. 2023. "WAVE2 Regulates Actin-Dependent Processes Induced by the B Cell Antigen Receptor and Integrins" Cells 12, no. 23: 2704. https://doi.org/10.3390/cells12232704







