Drosophila Models Reveal Properties of Mutant Lamins That Give Rise to Distinct Diseases
Abstract
:1. Introduction
2. Materials and Methods
2.1. Drosophila Stocks
2.2. Western Analysis
2.3. Immunohistochemistry of Drosophila Larval Tissues
2.4. Oil Red O Staining of Adipose Tissue
2.5. Larval Body Wall Muscle and Adipose Cell Size Measurements
2.6. Larval Motility Assays
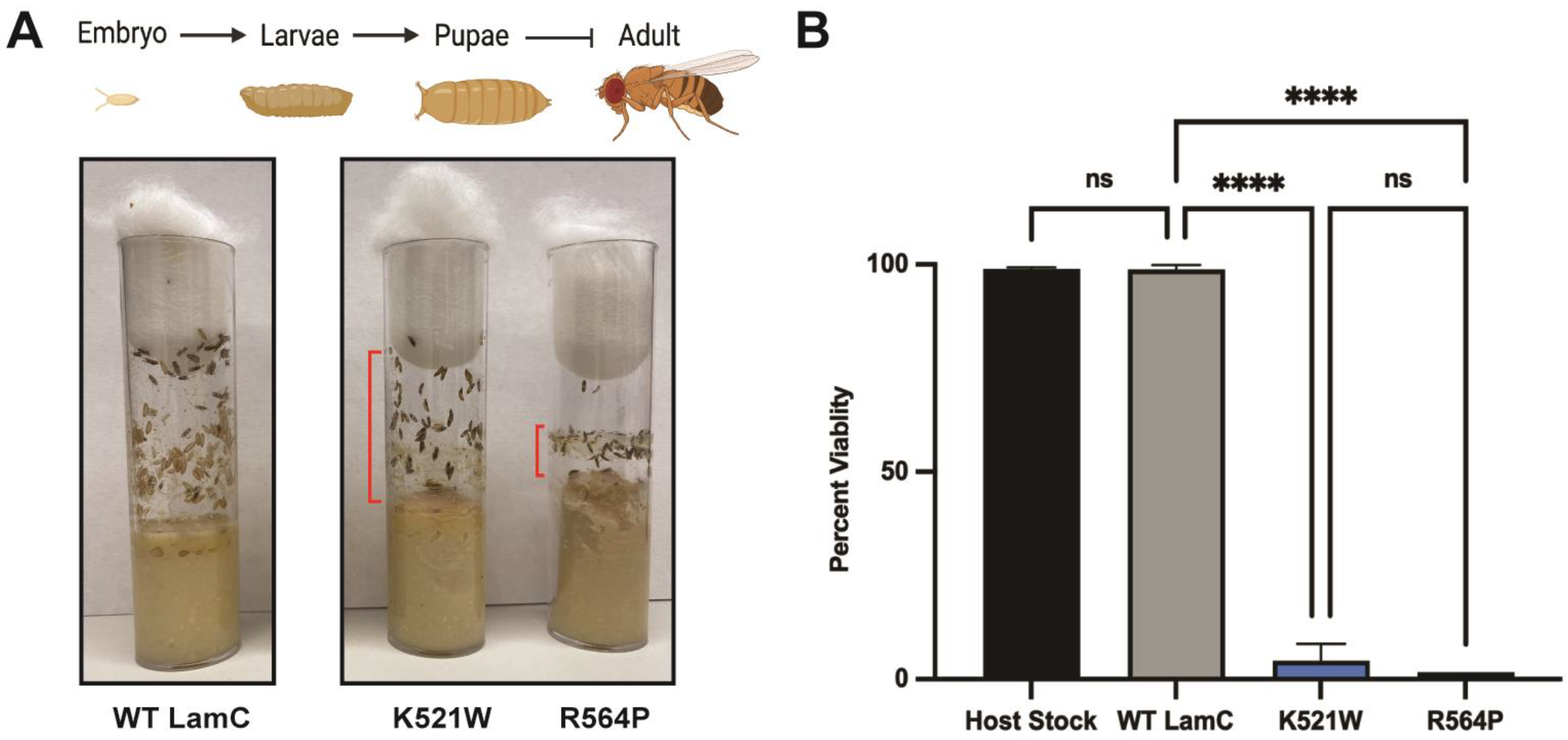
2.7. Adult Viability Measurements
2.8. Statistical Analysis
3. Results
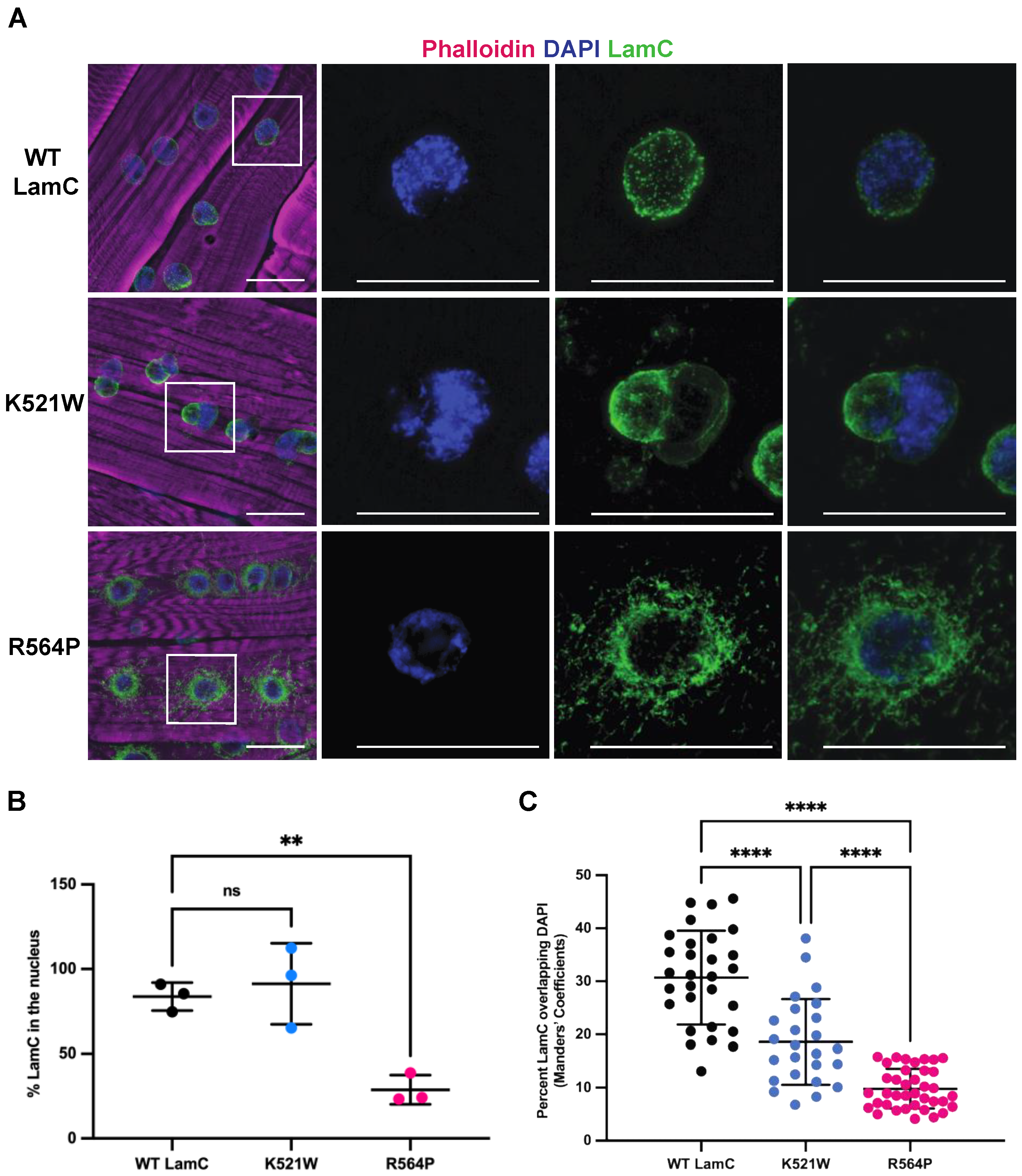
3.1. Mutant Lamins Have Different Patterns of Localization and Effects on Myonuclear Morphology
3.2. Mutant Lamins Have Different Effects on Nuclear Envelope Protein Localization
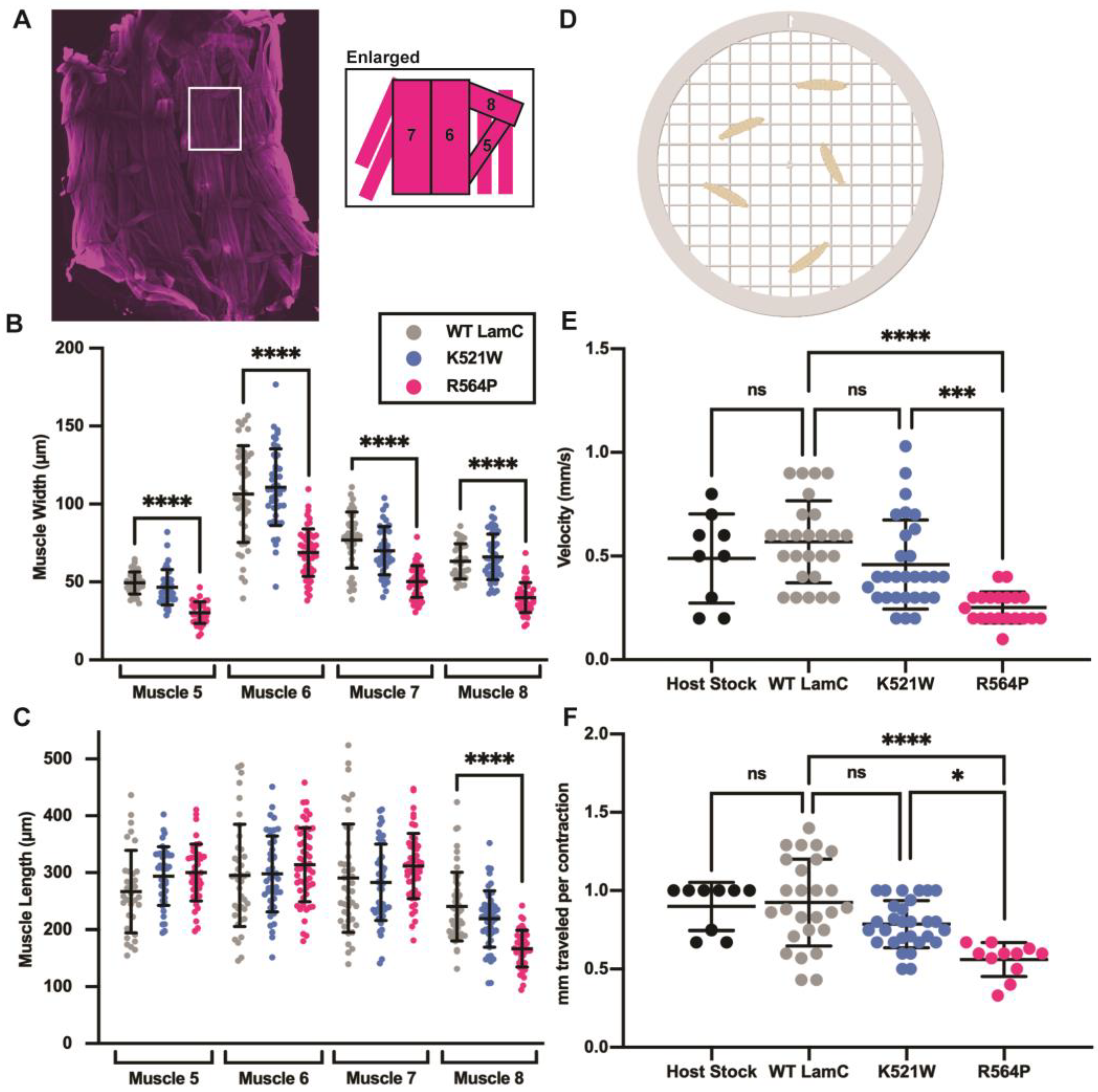
3.3. Mutant Lamins Have Different Effects on Larval Motility and Body Wall Size
3.4. Mutant Lamins Have Different Effects on Adipose Tissue
3.5. Mutant Lamins Have Different Effects on Lifespan When Expressed in Cardiac Tissue
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Verbrugge, S.A.J.; Schonfelder, M.; Becker, L.; Yaghoob Nezhad, F.; Hrabe de Angelis, M.; Wackerhage, H. Genes Whose Gain or Loss-Of-Function Increases Skeletal Muscle Mass in Mice: A Systematic Literature Review. Front. Physiol. 2018, 9, 553. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salari, N.; Fatahi, B.; Valipour, E.; Kazeminia, M.; Fatahian, R.; Kiaei, A.; Shohaimi, S.; Mohammadi, M. Global prevalence of Duchenne and Becker muscular dystrophy: A systematic review and meta-analysis. J. Orthop. Surg. Res. 2022, 17, 96. [Google Scholar] [CrossRef] [PubMed]
- Moser, H. Duchenne muscular dystrophy: Pathogenetic aspects and genetic prevention. Hum. Genet. 1984, 66, 17–40. [Google Scholar] [CrossRef]
- Emery, A.E. Emery-Dreifuss syndrome. J. Med. Genet. 1989, 26, 637–641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zatz, M.; Vainzof, M.; Passos-Bueno, M.R. Limb-girdle muscular dystrophy: One gene with different phenotypes, one phenotype with different genes. Curr. Opin. Neurol. 2000, 13, 511–517. [Google Scholar] [CrossRef]
- Aiello, D.; Patel, K.; Lasagna, E. The myostatin gene: An overview of mechanisms of action and its relevance to livestock animals. Anim. Genet. 2018, 49, 505–519. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Esposito, P.; Picciotto, D.; Battaglia, Y.; Costigliolo, F.; Viazzi, F.; Verzola, D. Myostatin: Basic biology to clinical application. Adv. Clin. Chem. 2022, 106, 181–234. [Google Scholar] [CrossRef]
- Youn, B.Y.; Ko, S.G.; Kim, J.Y. Genetic basis of elite combat sports athletes: A systematic review. Biol. Sport 2021, 38, 667–675. [Google Scholar] [CrossRef]
- McNally, E.M. Powerful genes—Myostatin regulation of human muscle mass. N. Engl. J. Med. 2004, 350, 2642–2644. [Google Scholar] [CrossRef]
- Joulia-Ekaza, D.; Cabello, G. The myostatin gene: Physiology and pharmacological relevance. Curr. Opin. Pharmacol. 2007, 7, 310–315. [Google Scholar] [CrossRef]
- Chen, M.M.; Zhao, Y.P.; Zhao, Y.; Deng, S.L.; Yu, K. Regulation of Myostatin on the Growth and Development of Skeletal Muscle. Front. Cell Dev. Biol. 2021, 9, 785712. [Google Scholar] [CrossRef] [PubMed]
- Qian, L.; Tang, M.; Yang, J.; Wang, Q.; Cai, C.; Jiang, S.; Li, H.; Jiang, K.; Gao, P.; Ma, D.; et al. Targeted mutations in myostatin by zinc-finger nucleases result in double-muscled phenotype in Meishan pigs. Sci. Rep. 2015, 5, 14435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, J.; Song, Z.; Yu, S.; Cui, D.; Wang, B.; Ding, F.; Li, S.; Dai, Y.; Li, N. Efficient generation of myostatin (MSTN) biallelic mutations in cattle using zinc finger nucleases. PLoS ONE 2014, 9, e95225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frezarim, G.B.; Fonseca, L.F.S.; Salatta, B.M.; Silva, D.B.S.; Bresolin, T.; Oliveira Seno, L.; Barufatti, A.; Ferro, J.A.; Albuquerque, L.G. Genes and proteins associated with ribeye area and meat tenderness in a commercial Nellore cattle population. Genome 2021, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Petersen, B. More abundant and healthier meat: Will the MSTN editing epitome empower the commercialization of gene editing in livestock? Sci. China Life Sci. 2022, 65, 448–450. [Google Scholar] [CrossRef]
- Zimmers, T.A.; Davies, M.V.; Koniaris, L.G.; Haynes, P.; Esquela, A.F.; Tomkinson, K.N.; McPherron, A.C.; Wolfman, N.M.; Lee, S.J. Induction of cachexia in mice by systemically administered myostatin. Science 2002, 296, 1486–1488. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Epstein, D. Do These Genes Make Me Look Fatless? “Something Only I Can See”; This American Life: New York, NY, USA, 2016. [Google Scholar]
- Ahn, J.; Jo, I.; Kang, S.M.; Hong, S.; Kim, S.; Jeong, S.; Kim, Y.H.; Park, B.J.; Ha, N.C. Structural basis for lamin assembly at the molecular level. Nat. Commun. 2019, 10, 3757. [Google Scholar] [CrossRef] [Green Version]
- Tenga, R.; Medalia, O. Structure and unique mechanical aspects of nuclear lamin filaments. Curr. Opin. Struct. Biol. 2020, 64, 152–159. [Google Scholar] [CrossRef]
- Turgay, Y.; Medalia, O. The structure of lamin filaments in somatic cells as revealed by cryo-electron tomography. Nucleus 2017, 8, 475–481. [Google Scholar] [CrossRef] [Green Version]
- Gruenbaum, Y.; Foisner, R. Lamins: Nuclear intermediate filament proteins with fundamental functions in nuclear mechanics and genome regulation. Annu. Rev. Biochem. 2015, 84, 131–164. [Google Scholar] [CrossRef]
- Heller, S.A.; Shih, R.; Kalra, R.; Kang, P.B. Emery-Dreifuss muscular dystrophy. Muscle Nerve 2020, 61, 436–448. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, A.T.; Brull, A.; Azibani, F.; Benarroch, L.; Chikhaoui, K.; Stewart, C.L.; Medalia, O.; Ben Yaou, R.; Bonne, G. Lamin A/C Assembly Defects in LMNA-Congenital Muscular Dystrophy Is Responsible for the Increased Severity of the Disease Compared with Emery-Dreifuss Muscular Dystrophy. Cells 2020, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muchir, A.; Worman, H.J. Emery-Dreifuss muscular dystrophy: Focal point nuclear envelope. Curr. Opin. Neurol. 2019, 32, 728–734. [Google Scholar] [CrossRef] [PubMed]
- Bagias, C.; Xiarchou, A.; Bargiota, A.; Tigas, S. Familial Partial Lipodystrophy (FPLD): Recent Insights. Diabetes Metab. Syndr. Obes. 2020, 13, 1531–1544. [Google Scholar] [CrossRef] [PubMed]
- Zammouri, J.; Vatier, C.; Capel, E.; Auclair, M.; Storey-London, C.; Bismuth, E.; Mosbah, H.; Donadille, B.; Janmaat, S.; Feve, B.; et al. Molecular and Cellular Bases of Lipodystrophy Syndromes. Front. Endocrinol. 2021, 12, 803189. [Google Scholar] [CrossRef] [PubMed]
- Garg, A. Lipodystrophies. Am. J. Med. 2000, 108, 143–152. [Google Scholar] [CrossRef] [PubMed]
- Worman, H.J.; Ostlund, C.; Wang, Y. Diseases of the nuclear envelope. Cold Spring Harb. Perspect. Biol. 2010, 2, a000760. [Google Scholar] [CrossRef]
- Worman, H.J.; Courvalin, J.C. How do mutations in lamins A and C cause disease? J. Clin. Investig. 2004, 113, 349–351. [Google Scholar] [CrossRef]
- Worman, H.J. Nuclear lamins and laminopathies. J. Pathol. 2012, 226, 316–325. [Google Scholar] [CrossRef]
- Hinz, B.E.; Walker, S.G.; Xiong, A.; Gogal, R.A.; Schnieders, M.J.; Wallrath, L.L. In Silico and In Vivo Analysis of Amino Acid Substitutions That Cause Laminopathies. Int. J. Mol. Sci. 2021, 22. [Google Scholar] [CrossRef]
- Lin, E.W.; Brady, G.F.; Kwan, R.; Nesvizhskii, A.I.; Omary, M.B. Genotype-phenotype analysis of LMNA-related diseases predicts phenotype-selective alterations in lamin phosphorylation. FASEB J. 2020, 34, 9051–9073. [Google Scholar] [CrossRef] [PubMed]
- Anderson, C.L.; Langer, E.R.; Routes, T.C.; McWilliams, S.F.; Bereslavskyy, I.; Kamp, T.J.; Eckhardt, L.L. Most myopathic lamin variants aggregate: A functional genomics approach for assessing variants of uncertain significance. NPJ Genom. Med. 2021, 6, 103. [Google Scholar] [CrossRef] [PubMed]
- Crasto, S.; My, I.; Di Pasquale, E. The Broad Spectrum of LMNA Cardiac Diseases: From Molecular Mechanisms to Clinical Phenotype. Front. Physiol. 2020, 11, 761. [Google Scholar] [CrossRef] [PubMed]
- Lazarte, J.; Jurgens, S.J.; Choi, S.H.; Khurshid, S.; Morrill, V.N.; Weng, L.C.; Nauffal, V.; Pirruccello, J.P.; Halford, J.L.; Hegele, R.A.; et al. LMNA Variants and Risk of Adult-Onset Cardiac Disease. J. Am. Coll Cardiol. 2022, 80, 50–59. [Google Scholar] [CrossRef]
- Scharner, J.; Lu, H.C.; Fraternali, F.; Ellis, J.A.; Zammit, P.S. Mapping disease-related missense mutations in the immunoglobulin-like fold domain of lamin A/C reveals novel genotype-phenotype associations for laminopathies. Proteins 2014, 82, 904–915. [Google Scholar] [CrossRef]
- Storey, E.C.; Fuller, H.R. Genotype-Phenotype Correlations in Human Diseases Caused by Mutations of LINC Complex-Associated Genes: A Systematic Review and Meta-Summary. Cells 2022, 11. [Google Scholar] [CrossRef]
- Petillo, R.; D’Ambrosio, P.; Torella, A.; Taglia, A.; Picillo, E.; Testori, A.; Ergoli, M.; Nigro, G.; Piluso, G.; Nigro, V.; et al. Novel mutations in LMNA A/C gene and associated phenotypes. Acta Myol. 2015, 34, 116–119. [Google Scholar]
- Bonne, G.; Di Barletta, M.R.; Varnous, S.; Becane, H.M.; Hammouda, E.H.; Merlini, L.; Muntoni, F.; Greenberg, C.R.; Gary, F.; Urtizberea, J.A.; et al. Mutations in the gene encoding lamin A/C cause autosomal dominant Emery-Dreifuss muscular dystrophy. Nat. Genet. 1999, 21, 285–288. [Google Scholar] [CrossRef]
- Van der Kooi, A.J.; Bonne, G.; Eymard, B.; Duboc, D.; Talim, B.; Van der Valk, M.; Reiss, P.; Richard, P.; Demay, L.; Merlini, L.; et al. Lamin A/C mutations with lipodystrophy, cardiac abnormalities, and muscular dystrophy. Neurology 2002, 59, 620–623. [Google Scholar] [CrossRef]
- Vantyghem, M.C.; Pigny, P.; Maurage, C.A.; Rouaix-Emery, N.; Stojkovic, T.; Cuisset, J.M.; Millaire, A.; Lascols, O.; Vermersch, P.; Wemeau, J.L.; et al. Patients with familial partial lipodystrophy of the Dunnigan type due to a LMNA R482W mutation show muscular and cardiac abnormalities. J. Clin. Endocrinol. Metab. 2004, 89, 5337–5346. [Google Scholar] [CrossRef] [Green Version]
- Plantie, E.; Migocka-Patrzalek, M.; Daczewska, M.; Jagla, K. Model organisms in the fight against muscular dystrophy: Lessons from drosophila and Zebrafish. Molecules 2015, 20, 6237–6253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Potikanond, S.; Nimlamool, W.; Noordermeer, J.; Fradkin, L.G. Muscular Dystrophy Model. Adv Exp Med Biol 2018, 1076, 147–172. [Google Scholar] [CrossRef]
- Palka, M.; Tomczak, A.; Grabowska, K.; Machowska, M.; Piekarowicz, K.; Rzepecka, D.; Rzepecki, R. Laminopathies: What can humans learn from fruit flies. Cell Mol. Biol. Lett. 2018, 23, 32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rzepecki, R.; Gruenbaum, Y. Invertebrate models of lamin diseases. Nucleus 2018, 9, 227–234. [Google Scholar] [CrossRef] [Green Version]
- Yamaguchi, M.; Yoshida, H. Drosophila as a Model Organism. In Drosopihla Models for Human Diseases; Springer: Berlin, Germany, 2018; Volume 1076, pp. 1–10. [Google Scholar] [CrossRef]
- Bawa, S.; Gameros, S.; Baumann, K.; Brooks, D.S.; Kollhoff, J.A.; Zolkiewski, M.; Re Cecconi, A.D.; Panini, N.; Russo, M.; Piccirillo, R.; et al. Costameric integrin and sarcoglycan protein levels are altered in a Drosophila model for Limb-girdle muscular dystrophy type 2H. Mol. Biol. Cell 2021, 32, 260–273. [Google Scholar] [CrossRef]
- Tsuda, L.; Lim, Y.M. Alzheimer’s Disease Model System Using Drosophila. Adv. Exp. Med. Biol. 2018, 1076, 25–40. [Google Scholar] [CrossRef] [PubMed]
- Dialynas, G.; Shrestha, O.K.; Ponce, J.M.; Zwerger, M.; Thiemann, D.A.; Young, G.H.; Moore, S.A.; Yu, L.; Lammerding, J.; Wallrath, L.L. Myopathic lamin mutations cause reductive stress and activate the nrf2/keap-1 pathway. PLoS Genet. 2015, 11, e1005231. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bhide, S.; Trujillo, A.S.; O’Connor, M.T.; Young, G.H.; Cryderman, D.E.; Chandran, S.; Nikravesh, M.; Wallrath, L.L.; Melkani, G.C. Increasing autophagy and blocking Nrf2 suppress laminopathy-induced age-dependent cardiac dysfunction and shortened lifespan. Aging Cell 2018, 17, e12747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishimura, M.; Ocorr, K.; Bodmer, R.; Cartry, J. Drosophila as a model to study cardiac aging. Exp Gerontol 2011, 46, 326–330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aryal, B.; Lee, Y. Disease model organism for Parkinson disease: Drosophila melanogaster. BMB Rep. 2019, 52, 250–258. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shaffer, C.D.; Wuller, J.M.; Elgin, S.C. Raising large quantities of Drosophila for biochemical experiments. Methods Cell Biol. 1994, 44, 99–108. [Google Scholar] [CrossRef] [PubMed]
- Rubin, G.M.; Spradling, A.C. Genetic transformation of Drosophila with transposable element vectors. Science 1982, 218, 348–353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barton, L.J.; Wilmington, S.R.; Martin, M.J.; Skopec, H.M.; Lovander, K.E.; Pinto, B.S.; Geyer, P.K. Unique and shared functions of nuclear lamina LEM domain proteins in Drosophila. Genetics 2014, 197, 653–665. [Google Scholar] [CrossRef] [Green Version]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [Green Version]
- Bolte, S.; Cordelieres, F.P. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 2006, 224, 213–232. [Google Scholar] [CrossRef]
- Alayari, N.N.; Vogler, G.; Taghli-Lamallem, O.; Ocorr, K.; Bodmer, R.; Cammarato, A. Fluorescent labeling of Drosophila heart structures. J. Vis. Exp. 2009. [Google Scholar] [CrossRef]
- Banerjee, K.K.; Ayyub, C.; Sengupta, S.; Kolthur-Seetharam, U. dSir2 deficiency in the fatbody, but not muscles, affects systemic insulin signaling, fat mobilization and starvation survival in flies. Aging 2012, 4, 206–223. [Google Scholar] [CrossRef] [Green Version]
- Fauzi, A.; Zubaidah, S.; Susanto, H. The Study of Larva and Adult Behavior of Drosophila melanogaster: Do Strains Affect Behavior? Aip. Conf. Proc. 2020, 2231. [Google Scholar]
- Koh, Y.H.; Popova, E.; Thomas, U.; Griffith, L.C.; Budnik, V. Regulation of DLG localization at synapses by CaMKII-dependent phosphorylation. Cell 1999, 98, 353–363. [Google Scholar] [CrossRef] [Green Version]
- Han, Z.; Olson, E.N. Hand is a direct target of Tinman and GATA factors during Drosophila cardiogenesis and hematopoiesis. Development 2005, 132, 3525–3536. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krimm, I.; Ostlund, C.; Gilquin, B.; Couprie, J.; Hossenlopp, P.; Mornon, J.P.; Bonne, G.; Courvalin, J.C.; Worman, H.J.; Zinn-Justin, S. The Ig-like structure of the C-terminal domain of lamin A/C, mutated in muscular dystrophies, cardiomyopathy, and partial lipodystrophy. Structure 2002, 10, 811–823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Coombs, G.S.; Rios-Monterrosa, J.L.; Lai, S.; Dai, Q.; Goll, A.C.; Ketterer, M.R.; Valdes, M.F.; Uche, N.; Benjamin, I.J.; Wallrath, L.L. Modulation of muscle redox and protein aggregation rescues lethality caused by mutant lamins. Redox Biol. 2021, 48, 102196. [Google Scholar] [CrossRef]
- Duffy, J.B. GAL4 system in Drosophila: A fly geneticist’s Swiss army knife. Genesis 2002, 34, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Caygill, E.E.; Brand, A.H. The GAL4 System: A Versatile System for the Manipulation and Analysis of Gene Expression. Methods Mol. Biol. 2016, 1478, 33–52. [Google Scholar] [CrossRef]
- Brand, A.H.; Perrimon, N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 1993, 118, 401–415. [Google Scholar] [CrossRef]
- Broers, J.L.; Machiels, B.M.; van Eys, G.J.; Kuijpers, H.J.; Manders, E.M.; van Driel, R.; Ramaekers, F.C. Dynamics of the nuclear lamina as monitored by GFP-tagged A-type lamins. J. Cell Sci. 1999, 112, 3463–3475. [Google Scholar] [CrossRef]
- Morin, X.; Daneman, R.; Zavortink, M.; Chia, W. A protein trap strategy to detect GFP-tagged proteins expressed from their endogenous loci in Drosophila. Proc. Natl. Acad. Sci. USA 2001, 98, 15050–15055. [Google Scholar] [CrossRef] [Green Version]
- Piekarowicz, K.; Machowska, M.; Dratkiewicz, E.; Lorek, D.; Madej-Pilarczyk, A.; Rzepecki, R. The effect of the lamin A and its mutants on nuclear structure, cell proliferation, protein stability, and mobility in embryonic cells. Chromosoma 2017, 126, 501–517. [Google Scholar] [CrossRef] [Green Version]
- Schejter, E.D.; Baylies, M.K. Born to run: Creating the muscle fiber. Curr. Opin Cell Biol. 2010, 22, 566–574. [Google Scholar] [CrossRef] [Green Version]
- Hartenstein, V. The Muscle Pattern of Drosophila. In Muscle Development in Drosophila; Springer New York: New York, NY, USA, 2006; pp. 8–27. [Google Scholar]
- Beckett, K.; Baylies, M.K. The development of the Drosophila larval body wall muscles. Int. Rev. Neurobiol. 2006, 75, 55–70. [Google Scholar] [CrossRef] [PubMed]
- Schulman, V.K.; Dobi, K.C.; Baylies, M.K. Morphogenesis of the somatic musculature in Drosophila melanogaster. Wiley Interdiscip Rev. Dev. Biol. 2015, 4, 313–334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fortier, T.M.; Vasa, P.P.; Woodard, C.T. Orphan nuclear receptor betaFTZ-F1 is required for muscle-driven morphogenetic events at the prepupal-pupal transition in Drosophila melanogaster. Dev. Biol. 2003, 257, 153–165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dialynas, G.; Speese, S.; Budnik, V.; Geyer, P.K.; Wallrath, L.L. The role of Drosophila Lamin C in muscle function and gene expression. Development 2010, 137, 3067–3077. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guelen, L.; Pagie, L.; Brasset, E.; Meuleman, W.; Faza, M.B.; Talhout, W.; Eussen, B.H.; de Klein, A.; Wessels, L.; de Laat, W.; et al. Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 2008, 453, 948–951. [Google Scholar] [CrossRef] [PubMed]
- Guerreiro, I.; Kind, J. Spatial chromatin organization and gene regulation at the nuclear lamina. Curr. Opin. Genet. Dev. 2019, 55, 19–25. [Google Scholar] [CrossRef]
- Paulsen, J.; Collas, P. Modeling the 3D Genome Using Hi-C and Nuclear Lamin-Genome Contacts. Methods Mol. Biol. 2022, 2301, 337–352. [Google Scholar] [CrossRef]
- Paulsen, J.; Liyakat Ali, T.M.; Collas, P. Computational 3D genome modeling using Chrom3D. Nat. Protoc. 2018, 13, 1137–1152. [Google Scholar] [CrossRef]
- Briand, N.; Collas, P. Lamina-associated domains: Peripheral matters and internal affairs. Genome Biol. 2020, 21, 85. [Google Scholar] [CrossRef] [Green Version]
- Pickersgill, H.; Kalverda, B.; de Wit, E.; Talhout, W.; Fornerod, M.; van Steensel, B. Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat. Genet. 2006, 38, 1005–1014. [Google Scholar] [CrossRef]
- Van Steensel, B.; Henikoff, S. Identification of in vivo DNA targets of chromatin proteins using tethered dam methyltransferase. Nat. Biotechnol. 2000, 18, 424–428. [Google Scholar] [CrossRef] [PubMed]
- Lieberman-Aiden, E.; van Berkum, N.L.; Williams, L.; Imakaev, M.; Ragoczy, T.; Telling, A.; Amit, I.; Lajoie, B.R.; Sabo, P.J.; Dorschner, M.O.; et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 2009, 326, 289–293. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Steensel, B.; Belmont, A.S. Lamina-Associated Domains: Links with Chromosome Architecture, Heterochromatin, and Gene Repression. Cell 2017, 169, 780–791. [Google Scholar] [CrossRef] [Green Version]
- Dittmer, T.A.; Misteli, T. The lamin protein family. Genome Biol. 2011, 12, 222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nmezi, B.; Xu, J.; Fu, R.; Armiger, T.J.; Rodriguez-Bey, G.; Powell, J.S.; Ma, H.; Sullivan, M.; Tu, Y.; Chen, N.Y.; et al. Concentric organization of A- and B-type lamins predicts their distinct roles in the spatial organization and stability of the nuclear lamina. Proc. Natl. Acad. Sci. USA 2019, 116, 4307–4315. [Google Scholar] [CrossRef] [Green Version]
- Shimi, T.; Kittisopikul, M.; Tran, J.; Goldman, A.E.; Adam, S.A.; Zheng, Y.; Jaqaman, K.; Goldman, R.D. Structural organization of nuclear lamins A, C, B1, and B2 revealed by superresolution microscopy. Mol. Biol. Cell 2015, 26, 4075–4086. [Google Scholar] [CrossRef]
- Al-Haboubi, T.; Shumaker, D.K.; Koser, J.; Wehnert, M.; Fahrenkrog, B. Distinct association of the nuclear pore protein Nup153 with A- and B-type lamins. Nucleus 2011, 2, 500–509. [Google Scholar] [CrossRef] [Green Version]
- Lussi, Y.C.; Hugi, I.; Laurell, E.; Kutay, U.; Fahrenkrog, B. The nucleoporin Nup88 is interacting with nuclear lamin A. Mol. Biol. Cell 2011, 22, 1080–1090. [Google Scholar] [CrossRef]
- Dialynas, G.; Flannery, K.M.; Zirbel, L.N.; Nagy, P.L.; Mathews, K.D.; Moore, S.A.; Wallrath, L.L. LMNA variants cause cytoplasmic distribution of nuclear pore proteins in Drosophila and human muscle. Hum. Mol. Genet. 2012, 21, 1544–1556. [Google Scholar] [CrossRef] [Green Version]
- Han, L.; Mich-Basso, J.D.; Li, Y.; Ammanamanchi, N.; Xu, J.; Bargaje, A.P.; Liu, H.; Wu, L.; Jeong, J.H.; Franks, J.; et al. Changes in nuclear pore numbers control nuclear import and stress response of mouse hearts. Dev. Cell 2022, 57, 2397–2411. [Google Scholar] [CrossRef]
- Ostlund, C.; Bonne, G.; Schwartz, K.; Worman, H.J. Properties of lamin A mutants found in Emery-Dreifuss muscular dystrophy, cardiomyopathy and Dunnigan-type partial lipodystrophy. J. Cell Sci. 2001, 114, 4435–4445. [Google Scholar] [CrossRef] [PubMed]
- Muchir, A.; Worman, H.J. Emery-Dreifuss muscular dystrophy. Curr. Neurol. Neurosci. Rep. 2007, 7, 78–83. [Google Scholar] [CrossRef] [PubMed]
- Padan, R.; Nainudel-Epszteyn, S.; Goitein, R.; Fainsod, A.; Gruenbaum, Y. Isolation and characterization of the Drosophila nuclear envelope otefin cDNA. J. Biol. Chem. 1990, 265, 7808–7813. [Google Scholar] [CrossRef] [PubMed]
- Bengtsson, L.; Otto, H. LUMA interacts with emerin and influences its distribution at the inner nuclear membrane. J. Cell Sci. 2008, 121, 536–548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, W.C.; Mitsuhashi, H.; Keduka, E.; Nonaka, I.; Noguchi, S.; Nishino, I.; Hayashi, Y.K. TMEM43 mutations in Emery-Dreifuss muscular dystrophy-related myopathy. Ann. Neurol. 2011, 69, 1005–1013. [Google Scholar] [CrossRef] [PubMed]
- Mukai, T.; Mori-Yoshimura, M.; Nishikawa, A.; Hokkoku, K.; Sonoo, M.; Nishino, I.; Takahashi, Y. Emery-Dreifuss muscular dystrophy-related myopathy with TMEM43 mutations. Muscle Nerve 2019, 59, E5–E7. [Google Scholar] [CrossRef]
- Klinke, N.; Meyer, H.; Ratnavadivel, S.; Reinhardt, M.; Heinisch, J.J.; Malmendal, A.; Milting, H.; Paululat, A. A Drosophila melanogaster model for TMEM43-related arrhythmogenic right ventricular cardiomyopathy type 5. Cell Mol. Life Sci. 2022, 79, 444. [Google Scholar] [CrossRef]
- Zhang, B.; Stewart, B. Electrophysiological recording from Drosophila larval body-wall muscles. Cold Spring Harb Protoc. 2010, 2010, pdb-prot5487. [Google Scholar] [CrossRef]
- Kimura, K.I.; Truman, J.W. Postmetamorphic cell death in the nervous and muscular systems of Drosophila melanogaster. J. Neurosci. 1990, 10, 403-1. [Google Scholar] [CrossRef]
- Von Kalm, L.; Fristrom, D.; Fristrom, J. The making of a fly leg: A model for epithelial morphogenesis. Bioessays 1995, 17, 693–702. [Google Scholar] [CrossRef]
- Brown, C.A.; Lanning, R.W.; McKinney, K.Q.; Salvino, A.R.; Cherniske, E.; Crowe, C.A.; Darras, B.T.; Gominak, S.; Greenberg, C.R.; Grosmann, C.; et al. Novel and recurrent mutations in lamin A/C in patients with Emery-Dreifuss muscular dystrophy. Am. J. Med. Genet. 2001, 102, 359–367. [Google Scholar] [CrossRef] [PubMed]
- Allikian, M.J.; Bhabha, G.; Dospoy, P.; Heydemann, A.; Ryder, P.; Earley, J.U.; Wolf, M.J.; Rockman, H.A.; McNally, E.M. Reduced life span with heart and muscle dysfunction in Drosophila sarcoglycan mutants. Hum. Mol. Genet. 2007, 16, 2933–2943. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paternostro, G.; Vignola, C.; Bartsch, D.U.; Omens, J.H.; McCulloch, A.D.; Reed, J.C. Age-associated cardiac dysfunction in Drosophila melanogaster. Circ Res. 2001, 88, 1053–1058. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blice-Baum, A.C.; Guida, M.C.; Hartley, P.S.; Adams, P.D.; Bodmer, R.; Cammarato, A. As time flies by: Investigating cardiac aging in the short-lived Drosophila model. Biochim. Biophys. Acta Mol. Basis Dis. 2019, 1865, 1831–1844. [Google Scholar] [CrossRef] [PubMed]
- Al-Sabri, M.H.; Behare, N.; Alsehli, A.M.; Berkins, S.; Arora, A.; Antoniou, E.; Moysiadou, E.I.; Anantha-Krishnan, S.; Cosmen, P.D.; Vikner, J.; et al. Statins Induce Locomotion and Muscular Phenotypes in Drosophila melanogaster That Are Reminiscent of Human Myopathy: Evidence for the Role of the Chloride Channel Inhibition in the Muscular Phenotypes. Cells 2022, 11. [Google Scholar] [CrossRef] [PubMed]
- Grice, S.J.; Liu, J.L. Motor defects in a Drosophila model for spinal muscular atrophy result from SMN depletion during early neurogenesis. PLoS Genet. 2022, 18, e1010325. [Google Scholar] [CrossRef]
- Lloyd, T.E.; Taylor, J.P. Flightless flies: Drosophila models of neuromuscular disease. Ann. NY Acad. Sci. 2010, 1184, e1-20. [Google Scholar] [CrossRef] [Green Version]
- Piazza, N.; Wessells, R.J. Drosophila models of cardiac disease. Prog. Mol. Biol. Transl. Sci. 2011, 100, 155–210. [Google Scholar] [CrossRef] [Green Version]
- Ribot, C.; Soler, C.; Chartier, A.; Al Hayek, S.; Nait-Saidi, R.; Barbezier, N.; Coux, O.; Simonelig, M. Activation of the ubiquitin-proteasome system contributes to oculopharyngeal muscular dystrophy through muscle atrophy. PLoS Genet. 2022, 18, e1010015. [Google Scholar] [CrossRef]
- Saoji, M.; Petersen, C.E.; Sen, A.; Tripoli, B.A.; Smyth, J.T.; Cox, R.T. Reduction of Drosophila Mitochondrial RNase P in Skeletal and Heart Muscle Causes Muscle Degeneration, Cardiomyopathy, and Heart Arrhythmia. Front. Cell Dev. Biol. 2022, 10, 788516. [Google Scholar] [CrossRef]
- Trujillo, A.S.; Hsu, K.H.; Puthawala, J.; Viswanathan, M.C.; Loya, A.; Irving, T.C.; Cammarato, A.; Swank, D.M.; Bernstein, S.I. Myosin dilated cardiomyopathy mutation S532P disrupts actomyosin interactions, leading to altered muscle kinetics, reduced locomotion, and cardiac dilation in Drosophila. Mol. Biol. Cell 2021, 32, 1690–1706. [Google Scholar] [CrossRef] [PubMed]
- Blazquez-Bernal, A.; Fernandez-Costa, J.M.; Bargiela, A.; Artero, R. Inhibition of autophagy rescues muscle atrophy in a LGMDD2 Drosophila model. FASEB J. 2021, 35, e21914. [Google Scholar] [CrossRef] [PubMed]
- Deng, J.; Guan, X.X.; Zhu, Y.B.; Deng, H.T.; Li, G.X.; Guo, Y.C.; Jin, P.; Duan, R.H.; Huang, W. Reducing the Excess Activin Signaling Rescues Muscle Degeneration in Myotonic Dystrophy Type 2 Drosophila Model. J. Pers. Med. 2022, 12. [Google Scholar] [CrossRef]
- Shaw, N.M.; Rios-Monterrosa, J.L.; Fedorchak, G.R.; Ketterer, M.R.; Coombs, G.S.; Lammerding, J.; Wallrath, L.L. Effects of mutant lamins on nucleo-cytoskeletal coupling in Drosophila models of LMNA muscular dystrophy. Front. Cell Dev. Biol. 2022, 10, 934586. [Google Scholar] [CrossRef] [PubMed]
- Thomas, J.; Smith, H.; Smith, C.A.; Coward, L.; Gorman, G.; De Luca, M.; Jumbo-Lucioni, P. The Angiotensin-Converting Enzyme Inhibitor Lisinopril Mitigates Memory and Motor Deficits in a Drosophila Model of Alzheimer’s Disease. Pathophysiology 2021, 28, 307–319. [Google Scholar] [CrossRef]
- Trujillo, A.S.; Ramos, R.; Bodmer, R.; Bernstein, S.I.; Ocorr, K.; Melkani, G.C. Drosophila as a potential model to ameliorate mutant Huntington-mediated cardiac amyloidosis. Rare Dis. 2014, 2, e968003. [Google Scholar] [CrossRef] [Green Version]
- Ahn, J.; Lee, J.; Jeong, S.; Kang, S.M.; Park, B.J.; Ha, N.C. Beta-strand-mediated dimeric formation of the Ig-like domains of human lamin A/C and B1. Biochem. Biophys. Res. Commun. 2021, 550, 191–196. [Google Scholar] [CrossRef]
- Earle, A.J.; Kirby, T.J.; Fedorchak, G.R.; Isermann, P.; Patel, J.; Iruvanti, S.; Moore, S.A.; Bonne, G.; Wallrath, L.L.; Lammerding, J. Mutant lamins cause nuclear envelope rupture and DNA damage in skeletal muscle cells. Nat. Mater. 2020, 19, 464–473. [Google Scholar] [CrossRef]
- Goldman, R.D.; Shumaker, D.K.; Erdos, M.R.; Eriksson, M.; Goldman, A.E.; Gordon, L.B.; Gruenbaum, Y.; Khuon, S.; Mendez, M.; Varga, R.; et al. Accumulation of mutant lamin A causes progressive changes in nuclear architecture in Hutchinson-Gilford progeria syndrome. Proc. Natl. Acad. Sci. USA 2004, 101, 8963–8968. [Google Scholar] [CrossRef] [Green Version]
- Heald, R.; McKeon, F. Mutations of phosphorylation sites in lamin A that prevent nuclear lamina disassembly in mitosis. Cell 1990, 61, 579–589. [Google Scholar] [CrossRef]
- Ward, G.E.; Kirschner, M.W. Identification of cell cycle-regulated phosphorylation sites on nuclear lamin C. Cell 1990, 61, 561–577. [Google Scholar] [CrossRef] [PubMed]
- Eggert, M.; Radomski, N.; Linder, D.; Tripier, D.; Traub, P.; Jost, E. Identification of novel phosphorylation sites in murine A-type lamins. Eur. J. Biochem. 1993, 213, 659–671. [Google Scholar] [CrossRef] [PubMed]
- Murray-Nerger, L.A.; Cristea, I.M. Lamin post-translational modifications: Emerging toggles of nuclear organization and function. Trends Biochem. Sci. 2021, 46, 832–847. [Google Scholar] [CrossRef] [PubMed]
- Simon, D.N.; Wilson, K.L. Partners and post-translational modifications of nuclear lamins. Chromosoma 2013, 122, 13–31. [Google Scholar] [CrossRef] [Green Version]
- Zheng, M.; Jin, G.; Zhou, Z. Post-Translational Modification of Lamins: Mechanisms and Functions. Front. Cell Dev. Biol. 2022, 10, 864191. [Google Scholar] [CrossRef]
- Kim, P.H.; Chen, N.Y.; Heizer, P.J.; Tu, Y.; Weston, T.A.; Fong, J.L.; Gill, N.K.; Rowat, A.C.; Young, S.G.; Fong, L.G. Nuclear membrane ruptures underlie the vascular pathology in a mouse model of Hutchinson-Gilford progeria syndrome. JCI Insight 2021, 6. [Google Scholar] [CrossRef]
- Balaji, A.K.; Saha, S.; Deshpande, S.; Poola, D.; Sengupta, K. Nuclear envelope, chromatin organizers, histones, and DNA: The many achilles heels exploited across cancers. Front. Cell Dev. Biol. 2022, 10, 1068347. [Google Scholar] [CrossRef]
- Starr, D.A. A network of nuclear envelope proteins and cytoskeletal force generators mediates movements of and within nuclei throughout Caenorhabditis elegans development. Exp. Biol. Med. 2019, 244, 1323–1332. [Google Scholar] [CrossRef]
- Fokkema, I.F.; Taschner, P.E.; Schaafsma, G.C.; Celli, J.; Laros, J.F.; den Dunnen, J.T. LOVD v.2.0: The next generation in gene variant databases. Hum. Mutat. 2011, 32, 557–563. [Google Scholar] [CrossRef]
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alfoldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P.; et al. The mutational constraint spectrum quantified from variation in 141,456 humans. Nature 2020, 581, 434–443. [Google Scholar] [CrossRef]
- Landrum, M.J.; Lee, J.M.; Benson, M.; Brown, G.R.; Chao, C.; Chitipiralla, S.; Gu, B.; Hart, J.; Hoffman, D.; Jang, W.; et al. ClinVar: Improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018, 46, D1062–D1067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montanucci, L.; Capriotti, E.; Birolo, G.; Benevenuta, S.; Pancotti, C.; Lal, D.; Fariselli, P. DDGun: An untrained predictor of protein stability changes upon amino acid variants. Nucleic Acids Res. 2022, 50, W222–W227. [Google Scholar] [CrossRef] [PubMed]
- Wilson, K.L.; Foisner, R. Lamin-binding Proteins. Cold Spring Harb. Perspect. Biol. 2010, 2, a000554. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, D.K.; Pande, S.; Kumar, J.; Yesodharan, D.; Nampoothiri, S.; Radhakrishnan, P.; Reddy, C.G.; Ranjan, A.; Girisha, K.M. The E262K mutation in Lamin A links nuclear proteostasis imbalance to laminopathy-associated premature aging. Aging Cell 2022, 21, e13688. [Google Scholar] [CrossRef] [PubMed]
- Liao, C.Y.; Anderson, S.S.; Chicoine, N.H.; Mayfield, J.R.; Academia, E.C.; Wilson, J.A.; Pongkietisak, C.; Thompson, M.A.; Lagmay, E.P.; Miller, D.M.; et al. Rapamycin Reverses Metabolic Deficits in Lamin A/C-Deficient Mice. Cell Rep. 2016, 17, 2542–2552. [Google Scholar] [CrossRef] [PubMed]
- Ramos, F.J.; Chen, S.C.; Garelick, M.G.; Dai, D.F.; Liao, C.Y.; Schreiber, K.H.; MacKay, V.L.; An, E.H.; Strong, R.; Ladiges, W.C.; et al. Rapamycin reverses elevated mTORC1 signaling in lamin A/C-deficient mice, rescues cardiac and skeletal muscle function, and extends survival. Sci. Transl. Med. 2012, 4, 144ra103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friesen, M.; Cowan, C.A. FPLD2 LMNA mutation R482W dysregulates iPSC-derived adipocyte function and lipid metabolism. Biochem. Biophys. Res. Commun. 2018, 495, 254–260. [Google Scholar] [CrossRef]
- Peng, Y.; Tang, Q.; Xiao, F.; Fu, N. Regulation of Lipid Metabolism by Lamin in Mutation-Related Diseases. Front. Pharmacol. 2022, 13, 820857. [Google Scholar] [CrossRef]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Walker, S.G.; Langland, C.J.; Viles, J.; Hecker, L.A.; Wallrath, L.L. Drosophila Models Reveal Properties of Mutant Lamins That Give Rise to Distinct Diseases. Cells 2023, 12, 1142. https://doi.org/10.3390/cells12081142
Walker SG, Langland CJ, Viles J, Hecker LA, Wallrath LL. Drosophila Models Reveal Properties of Mutant Lamins That Give Rise to Distinct Diseases. Cells. 2023; 12(8):1142. https://doi.org/10.3390/cells12081142
Chicago/Turabian StyleWalker, Sydney G., Christopher J. Langland, Jill Viles, Laura A. Hecker, and Lori L. Wallrath. 2023. "Drosophila Models Reveal Properties of Mutant Lamins That Give Rise to Distinct Diseases" Cells 12, no. 8: 1142. https://doi.org/10.3390/cells12081142




