A Proteomic Investigation to Discover Candidate Proteins Involved in Novel Mechanisms of 5-Fluorouracil Resistance in Colorectal Cancer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Chemosensitivity Assay
2.3. Development of 5-FU-Resistant Human CRC Sublines
2.4. SILAC Approach for Quantitative Proteomic Analysis
2.5. Protein Extraction and Protein Digestion
2.6. Shotgun Proteomics
2.7. Immunoblotting
2.8. Immunofluorescence
2.9. Statistical Analysis
3. Results
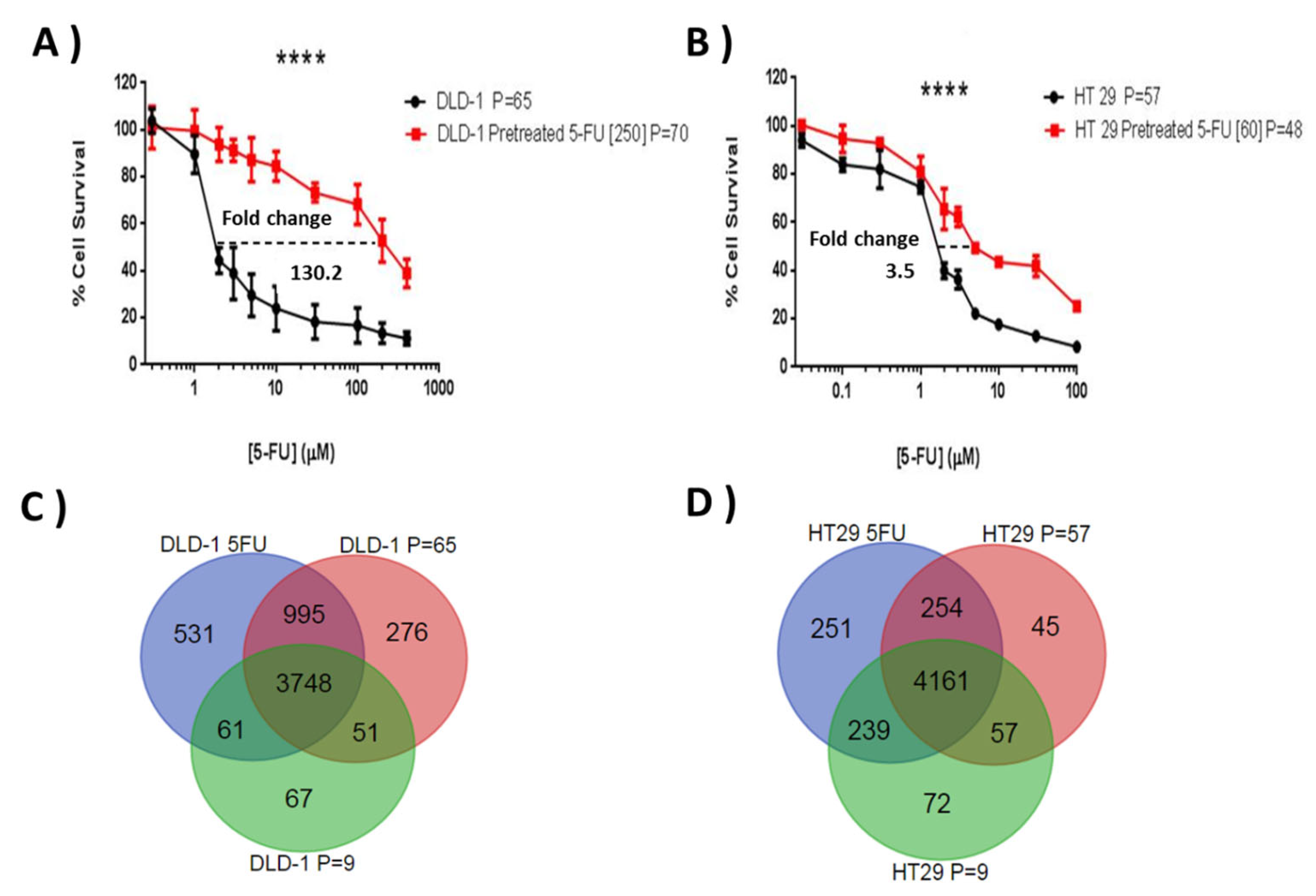
3.1. Establishment of DLD-1 and HT-29 Drug-Resistant Sublines to 5-FU
3.2. Identification of Proteome Changes in DLD-1- and HT-29-Resistant Sublines to 5-FU Using a Mass Spectrometry by SILAC Approach
3.3. Validation of the SILAC Proteomics Approach Using Immunodetection of CD44 and CD63
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Longley, D.B.; Harkin, D.P.; Johnston, P.G. 5-fluorouracil: Mechanisms of action and clinical strategies. Nat. Rev. Cancer 2003, 3, 330–338. [Google Scholar] [CrossRef]
- Peters, G.J.; Backus, H.H.; Freemantle, S.; van Triest, B.; Codacci-Pisanelli, G.; van der Wilt, C.L.; Smid, K.; Lunec, J.; Calvert, A.H.; Marsh, S.; et al. Induction of thymidylate synthase as a 5-fluorouracil resistance mechanism. Biochim. Biophys. Acta 2002, 1587, 194–205. [Google Scholar] [CrossRef]
- Gmeiner, W.H.; Okechukwu, C.C. Review of 5-FU resistance mechanisms in colorectal cancer: Clinical significance of attenuated on-target effects. Cancer Drug Resist. 2023, 6, 257–272. [Google Scholar] [CrossRef]
- Zhang, N.; Yin, Y.; Xu, S.J.; Chen, W.S. 5-Fluorouracil: Mechanisms of resistance and reversal strategies. Molecules 2008, 13, 1551–1569. [Google Scholar] [CrossRef] [PubMed]
- Blondy, S.; David, V.; Verdier, M.; Mathonnet, M.; Perraud, A.; Christou, N. 5-Fluorouracil resistance mechanisms in colorectal cancer: From classical pathways to promising processes. Cancer Sci. 2020, 111, 3142–3154. [Google Scholar] [CrossRef] [PubMed]
- Dallas, N.A.; Xia, L.; Fan, F.; Gray, M.J.; Gaur, P.; van Buren, G., 2nd; Samuel, S.; Kim, M.P.; Lim, S.J.; Ellis, L.M. Chemoresistant colorectal cancer cells, the cancer stem cell phenotype, and increased sensitivity to insulin-like growth factor-I receptor inhibition. Cancer Res. 2009, 69, 1951–1957. [Google Scholar] [CrossRef] [PubMed]
- Pylvas-Eerola, M.; Liakka, A.; Puistola, U.; Koivunen, J.; Karihtala, P. Cancer Stem Cell Properties as Factors Predictive of Chemoresistance in Neoadjuvantly-treated Patients with Ovarian Cancer. Anticancer Res. 2016, 36, 3425–3431. [Google Scholar]
- Xia, P.; Xu, X.Y. Prognostic significance of CD44 in human colon cancer and gastric cancer: Evidence from bioinformatic analyses. Oncotarget 2016, 7, 45538–45546. [Google Scholar] [CrossRef] [PubMed]
- Hammond, W.A.; Swaika, A.; Mody, K. Pharmacologic resistance in colorectal cancer: A review. Ther. Adv. Med. Oncol. 2016, 8, 57–84. [Google Scholar] [CrossRef] [PubMed]
- Williams, G.R.; Bethard, J.R.; Berkaw, M.N.; Nagel, A.K.; Luttrell, L.M.; Ball, L.E. Exploring G protein-coupled receptor signaling networks using SILAC-based phosphoproteomics. Methods 2016, 92, 36–50. [Google Scholar] [CrossRef]
- Ivanisevic, T.; Sewduth, R.N. Multi-Omics Integration for the Design of Novel Therapies and the Identification of Novel Biomarkers. Proteomes 2023, 11, 34. [Google Scholar] [CrossRef]
- Hekmat, O.; Munk, S.; Fogh, L.; Yadav, R.; Francavilla, C.; Horn, H.; Wurtz, S.O.; Schrohl, A.S.; Damsgaard, B.; Romer, M.U.; et al. TIMP-1 increases expression and phosphorylation of proteins associated with drug resistance in breast cancer cells. J. Proteome. Res. 2013, 12, 4136–4151. [Google Scholar] [CrossRef]
- Ong, S.E.; Blagoev, B.; Kratchmarova, I.; Kristensen, D.B.; Steen, H.; Pandey, A.; Mann, M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteom. 2002, 1, 376–386. [Google Scholar] [CrossRef] [PubMed]
- Stebbing, J.; Zhang, H.; Xu, Y.; Grothey, A.; Ajuh, P.; Angelopoulos, N.; Giamas, G. Characterization of the Tyrosine Kinase-Regulated Proteome in Breast Cancer by Combined use of RNA interference (RNAi) and Stable Isotope Labeling with Amino Acids in Cell Culture (SILAC) Quantitative Proteomics. Mol. Cell. Proteom. 2015, 14, 2479–2492. [Google Scholar] [CrossRef] [PubMed]
- Lindemann, C.; Thomanek, N.; Hundt, F.; Lerari, T.; Meyer, H.E.; Wolters, D.; Marcus, K. Strategies in relative and absolute quantitative mass spectrometry based proteomics. Biol. Chem. 2017, 398, 687–699. [Google Scholar] [CrossRef]
- Ong, S.E.; Foster, L.J.; Mann, M. Mass spectrometric-based approaches in quantitative proteomics. Methods 2003, 29, 124–130. [Google Scholar] [CrossRef]
- Shnyder, S.D.; Cooper, P.A.; Millington, N.J.; Pettit, G.R.; Bibby, M.C. Auristatin PYE, a novel synthetic derivative of dolastatin 10, is highly effective in human colon tumour models. Int. J. Oncol. 2007, 31, 353–360. [Google Scholar] [CrossRef]
- Van Hoof, D.; Pinkse, M.W.; Oostwaard, D.W.; Mummery, C.L.; Heck, A.J.; Krijgsveld, J. An experimental correction for arginine-to-proline conversion artifacts in SILAC-based quantitative proteomics. Nat. Methods 2007, 4, 677–678. [Google Scholar] [CrossRef]
- Ong, S.E.; Mann, M. A practical recipe for stable isotope labeling by amino acids in cell culture (SILAC). Nat. Protoc. 2006, 1, 2650–2660. [Google Scholar] [CrossRef] [PubMed]
- Waanders, L.F.; Hanke, S.; Mann, M. Top-down quantitation and characterization of SILAC-labeled proteins. J. Am. Soc. Mass. Spectrom. 2007, 18, 2058–2064. [Google Scholar] [CrossRef]
- Draffin, J.E.; McFarlane, S.; Hill, A.; Johnston, P.G.; Waugh, D.J. CD44 potentiates the adherence of metastatic prostate and breast cancer cells to bone marrow endothelial cells. Cancer Res. 2004, 64, 5702–5711. [Google Scholar] [CrossRef]
- Ritchie, M.E.; Phipson, B.; Wu, D.; Hu, Y.; Law, C.W.; Shi, W.; Smyth, G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015, 43, e47. [Google Scholar] [CrossRef]
- Glaab, E.; Baudot, A.; Krasnogor, N.; Schneider, R.; Valencia, A. EnrichNet: Network-based gene set enrichment analysis. Bioinformatics 2012, 28, i451–i457. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Kirsch, R.; Koutrouli, M.; Nastou, K.; Mehryary, F.; Hachilif, R.; Gable, A.L.; Fang, T.; Doncheva, N.T.; Pyysalo, S.; et al. The STRING database in 2023: Protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res 2023, 51, D638–D646. [Google Scholar] [CrossRef]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res 2022, 50, W216–W221. [Google Scholar] [CrossRef] [PubMed]
- Tam, S.Y.; Islam Khan, M.Z.; Chen, J.Y.; Yip, J.H.; Yan, H.Y.; Tam, T.Y.; Law, H.K. Proteomic Profiling of Chemotherapy Responses in FOLFOX-Resistant Colorectal Cancer Cells. Int. J. Mol. Sci. 2023, 24, 9899. [Google Scholar] [CrossRef] [PubMed]
- Nojadeh, J.N.; Behrouz Sharif, S.; Sakhinia, E. Microsatellite instability in colorectal cancer. EXCLI J. 2018, 17, 159–168. [Google Scholar] [CrossRef] [PubMed]
- Boland, C.R.; Goel, A. Microsatellite instability in colorectal cancer. Gastroenterology 2010, 138, 2073–2087.e3. [Google Scholar] [CrossRef] [PubMed]
- Sargent, D.J.; Marsoni, S.; Monges, G.; Thibodeau, S.N.; Labianca, R.; Hamilton, S.R.; French, A.J.; Kabat, B.; Foster, N.R.; Torri, V.; et al. Defective mismatch repair as a predictive marker for lack of efficacy of fluorouracil-based adjuvant therapy in colon cancer. J. Clin. Oncol. 2010, 28, 3219–3226. [Google Scholar] [CrossRef] [PubMed]
- Ribic, C.M.; Sargent, D.J.; Moore, M.J.; Thibodeau, S.N.; French, A.J.; Goldberg, R.M.; Hamilton, S.R.; Laurent-Puig, P.; Gryfe, R.; Shepherd, L.E.; et al. Tumor microsatellite-instability status as a predictor of benefit from fluorouracil-based adjuvant chemotherapy for colon cancer. N. Engl. J. Med. 2003, 349, 247–257. [Google Scholar] [CrossRef] [PubMed]
- Jover, R.; Zapater, P.; Castells, A.; Llor, X.; Andreu, M.; Cubiella, J.; Pinol, V.; Xicola, R.M.; Bujanda, L.; Rene, J.M.; et al. Mismatch repair status in the prediction of benefit from adjuvant fluorouracil chemotherapy in colorectal cancer. Gut 2006, 55, 848–855. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.Y.; Lee, J.S.; Min, H.Y.; Lee, H.Y. Acquired resistance to 5-fluorouracil via HSP90/Src-mediated increase in thymidylate synthase expression in colon cancer. Oncotarget 2015, 6, 32622–32633. [Google Scholar] [CrossRef] [PubMed]
- Zhijun, X.; Shulan, Z.; Zhuo, Z. Expression and significance of the protein and mRNA of metastasis suppressor gene ME491/CD63 and integrin alpha5 in ovarian cancer tissues. Eur. J. Gynaecol. Oncol. 2007, 28, 179–183. [Google Scholar] [PubMed]
- Toricelli, M.; Melo, F.H.; Peres, G.B.; Silva, D.C.; Jasiulionis, M.G. Timp1 interacts with beta-1 integrin and CD63 along melanoma genesis and confers anoikis resistance by activating PI3-K signaling pathway independently of Akt phosphorylation. Mol. Cancer 2013, 12, 22. [Google Scholar] [CrossRef] [PubMed]
- Sordat, I.; Decraene, C.; Silvestre, T.; Petermann, O.; Auffray, C.; Pietu, G.; Sordat, B. Complementary DNA arrays identify CD63 tetraspanin and alpha3 integrin chain as differentially expressed in low and high metastatic human colon carcinoma cells. Lab. Investig. 2002, 82, 1715–1724. [Google Scholar] [CrossRef]
- Kimhi, O.; Drucker, L.; Neumann, A.; Shapiro, H.; Shapira, J.; Yarkoni, S.; Lahav, M.; Radnay, J.; Lishner, M. Fluorouracil induces apoptosis and surface molecule modulation of peripheral blood leukocytes. Clin. Lab. Haematol. 2004, 26, 327–333. [Google Scholar] [CrossRef]
- Hiraga, T.; Ito, S.; Nakamura, H. Cancer stem-like cell marker CD44 promotes bone metastases by enhancing tumorigenicity, cell motility, and hyaluronan production. Cancer Res. 2013, 73, 4112–4122. [Google Scholar] [CrossRef]
- Jing, F.; Kim, H.J.; Kim, C.H.; Kim, Y.J.; Lee, J.H.; Kim, H.R. Colon cancer stem cell markers CD44 and CD133 in patients with colorectal cancer and synchronous hepatic metastases. Int. J. Oncol. 2015, 46, 1582–1588. [Google Scholar] [CrossRef]
- Sahlberg, S.H.; Spiegelberg, D.; Glimelius, B.; Stenerlow, B.; Nestor, M. Evaluation of cancer stem cell markers CD133, CD44, CD24: Association with AKT isoforms and radiation resistance in colon cancer cells. PLoS ONE 2014, 9, e94621. [Google Scholar] [CrossRef]
- Yu, Y.; Sarkar, F.H.; Majumdar, A.P. Down-regulation of miR-21 Induces Differentiation of Chemoresistant Colon Cancer Cells and Enhances Susceptibility to Therapeutic Regimens. Transl. Oncol. 2013, 6, 180–186. [Google Scholar] [CrossRef]
- Lei, X.; He, Q.; Li, Z.; Zou, Q.; Xu, P.; Yu, H.; Ding, Y.; Zhu, W. Cancer stem cells in colorectal cancer and the association with chemotherapy resistance. Med. Oncol. 2021, 38, 43. [Google Scholar] [CrossRef]
- Nautiyal, J.; Du, J.; Yu, Y.; Kanwar, S.S.; Levi, E.; Majumdar, A.P. EGFR regulation of colon cancer stem-like cells during aging and in response to the colonic carcinogen dimethylhydrazine. Am. J. Physiol. Gastrointest. Liver Physiol. 2012, 302, G655–G663. [Google Scholar] [CrossRef]
- Patel, B.B.; Yu, Y.; Du, J.; Levi, E.; Phillip, P.A.; Majumdar, A.P. Age-related increase in colorectal cancer stem cells in macroscopically normal mucosa of patients with adenomas: A risk factor for colon cancer. Biochem. Biophys. Res. Commun. 2009, 378, 344–347. [Google Scholar] [CrossRef]
- Yan, Y.; Zuo, X.; Wei, D. Concise Review: Emerging Role of CD44 in Cancer Stem Cells: A Promising Biomarker and Therapeutic Target. Stem Cells Transl. Med. 2015, 4, 1033–1043. [Google Scholar] [CrossRef]
- Muller, U.C.; Zheng, H. Physiological functions of APP family proteins. Cold Spring Harb Perspect. Med. 2012, 2, a006288. [Google Scholar] [CrossRef]
- Wu, X.L.; Chen, Y.; Kong, W.C.; Zhao, Z.Q. Amyloid precursor protein regulates 5-fluorouracil resistance in human hepatocellular carcinoma cells by inhibiting the mitochondrial apoptotic pathway. J. Zhejiang Univ. Sci. B 2020, 21, 234–245. [Google Scholar] [CrossRef]
- Tian, S.; Chu, Y.; Hu, J.; Ding, X.; Liu, Z.; Fu, D.; Yuan, Y.; Deng, Y.; Wang, G.; Wang, L.; et al. Tumour-associated neutrophils secrete AGR2 to promote colorectal cancer metastasis via its receptor CD98hc-xCT. Gut 2022, 71, 2489–2501. [Google Scholar] [CrossRef]
- Li, J.; Hu, J.; Luo, Z.; Zhou, C.; Huang, L.; Zhang, H.; Chi, J.; Chen, Z.; Li, Q.; Deng, M.; et al. AGR2 is controlled by DNMT3a-centered signaling module and mediates tumor resistance to 5-Aza in colorectal cancer. Exp. Cell Res. 2019, 385, 111644. [Google Scholar] [CrossRef]
- Xue, R.; Yang, K.; Xiao, F.; Yang, L.; Chen, G.; Li, Y.; Ye, Y.; Chen, K.; Smith, S.T.; Li, G.; et al. dNAGLU Extends Life Span and Promotes Fitness and Stress Resistance in Drosophila. Int. J. Mol. Sci. 2022, 23, 14433. [Google Scholar] [CrossRef]
- Choi, Y.; Kwon, C.H.; Lee, S.J.; Park, J.; Shin, J.Y.; Park, D.Y. Integrative analysis of oncogenic fusion genes and their functional impact in colorectal cancer. Br. J. Cancer 2018, 119, 230–240. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, K.; Swaminathan, K.; Peche, V.S.; Clemen, C.S.; Knyphausen, P.; Lammers, M.; Noegel, A.A.; Rastetter, R.H. Novel Coronin7 interactions with Cdc42 and N-WASP regulate actin organization and Golgi morphology. Sci. Rep. 2016, 6, 25411. [Google Scholar] [CrossRef]
- Barpanda, A.; Tuckley, C.; Ray, A.; Banerjee, A.; Duttagupta, S.P.; Kantharia, C.; Srivastava, S. A protein microarray-based serum proteomic investigation reveals distinct autoantibody signature in colorectal cancer. Proteom. Clin. Appl. 2023, 17, e2200062. [Google Scholar] [CrossRef] [PubMed]
- Dal Col, J.; Lamberti, M.J.; Nigro, A.; Casolaro, V.; Fratta, E.; Steffan, A.; Montico, B. Phospholipid scramblase 1: A protein with multiple functions via multiple molecular interactors. Cell Commun. Signal. 2022, 20, 78. [Google Scholar] [CrossRef]
- Kuo, Y.B.; Chan, C.C.; Chang, C.A.; Fan, C.W.; Hung, R.P.; Hung, Y.S.; Chen, K.T.; Yu, J.S.; Chang, Y.S.; Chan, E.C. Identification of phospholipid scramblase 1 as a biomarker and determination of its prognostic value for colorectal cancer. Mol. Med. 2011, 17, 41–47. [Google Scholar] [CrossRef]
- Menyhart, O.; Fekete, J.T.; Gyorffy, B. Gene expression-based biomarkers designating glioblastomas resistant to multiple treatment strategies. Carcinogenesis 2021, 42, 804–813. [Google Scholar] [CrossRef]
- Frey, L.; Zietara, N.; Lyszkiewicz, M.; Marquardt, B.; Mizoguchi, Y.; Linder, M.I.; Liu, Y.; Giesert, F.; Wurst, W.; Dahlhoff, M.; et al. Mammalian VPS45 orchestrates trafficking through the endosomal system. Blood 2021, 137, 1932–1944. [Google Scholar] [CrossRef]
- Hwang, G.W.; Murai, Y.; Takahashi, T.; Naganuma, A. The protein transportation pathway from Golgi to vacuoles via endosomes plays a role in enhancement of methylmercury toxicity. Sci. Rep. 2014, 4, 5888. [Google Scholar] [CrossRef]
- Sun, X.; Hu, Y.; Wu, J.; Shi, L.; Zhu, L.; Xi, P.W.; Wei, J.F.; Ding, Q. RBMS2 inhibits the proliferation by stabilizing P21 mRNA in breast cancer. J. Exp. Clin. Cancer Res. 2018, 37, 298. [Google Scholar] [CrossRef]
- Xu, F.; Xia, T.; Xu, Q.T.; Zhang, X.; Huang, Y.Z.; Sun, X.; Shi, L.; Zhou, X.J.; Wei, J.F.; Ding, Q. RBMS2 Chemosensitizes Breast Cancer Cells to Doxorubicin by Regulating BMF Expression. Int. J. Biol. Sci. 2022, 18, 1724–1736. [Google Scholar] [CrossRef]
- Widmann, B.; Wandrey, F.; Badertscher, L.; Wyler, E.; Pfannstiel, J.; Zemp, I.; Kutay, U. The kinase activity of human Rio1 is required for final steps of cytoplasmic maturation of 40S subunits. Mol. Biol. Cell 2012, 23, 22–35. [Google Scholar] [CrossRef]
- Weinberg, F.; Reischmann, N.; Fauth, L.; Taromi, S.; Mastroianni, J.; Kohler, M.; Halbach, S.; Becker, A.C.; Deng, N.; Schmitz, T.; et al. The Atypical Kinase RIOK1 Promotes Tumor Growth and Invasive Behavior. eBioMedicine 2017, 20, 79–97. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhou, S.; Wan, K.; Yu, L.; Zhao, C.; Deng, H.; Ou, Q.; Qin, J.; Hu, J.; Hou, Z. RIOK1 mediates p53 degradation and radioresistance in colorectal cancer through phosphorylation of G3BP2. Oncogene 2022, 41, 3433–3444. [Google Scholar] [CrossRef] [PubMed]
- Williams, C.L. The polybasic region of Ras and Rho family small GTPases: A regulator of protein interactions and membrane association and a site of nuclear localization signal sequences. Cell Signal. 2003, 15, 1071–1080. [Google Scholar] [CrossRef]
- Bowler, E.; Porazinski, S.; Uzor, S.; Thibault, P.; Durand, M.; Lapointe, E.; Rouschop, K.M.A.; Hancock, J.; Wilson, I.; Ladomery, M. Hypoxia leads to significant changes in alternative splicing and elevated expression of CLK splice factor kinases in PC3 prostate cancer cells. BMC Cancer 2018, 18, 355. [Google Scholar] [CrossRef]
- Lea, S. Interactions of CD55 with non-complement ligands. Biochem. Soc. Trans. 2002, 30, 1014–1019. [Google Scholar] [CrossRef]
- Durrant, L.G.; Chapman, M.A.; Buckley, D.J.; Spendlove, I.; Robins, R.A.; Armitage, N.C. Enhanced expression of the complement regulatory protein CD55 predicts a poor prognosis in colorectal cancer patients. Cancer Immunol. Immunother. 2003, 52, 638–642. [Google Scholar] [CrossRef]
- Dho, S.H.; Cho, E.H.; Lee, J.Y.; Lee, S.Y.; Jung, S.H.; Kim, L.K.; Lim, J.C. A novel therapeutic anti-CD55 monoclonal antibody inhibits the proliferation and metastasis of colorectal cancer cells. Oncol. Rep. 2019, 42, 2686–2693. [Google Scholar] [CrossRef]
- Miura, K.; Karasawa, H.; Sasaki, I. cIAP2 as a therapeutic target in colorectal cancer and other malignancies. Expert Opin. Ther. Targets 2009, 13, 1333–1345. [Google Scholar] [CrossRef] [PubMed]

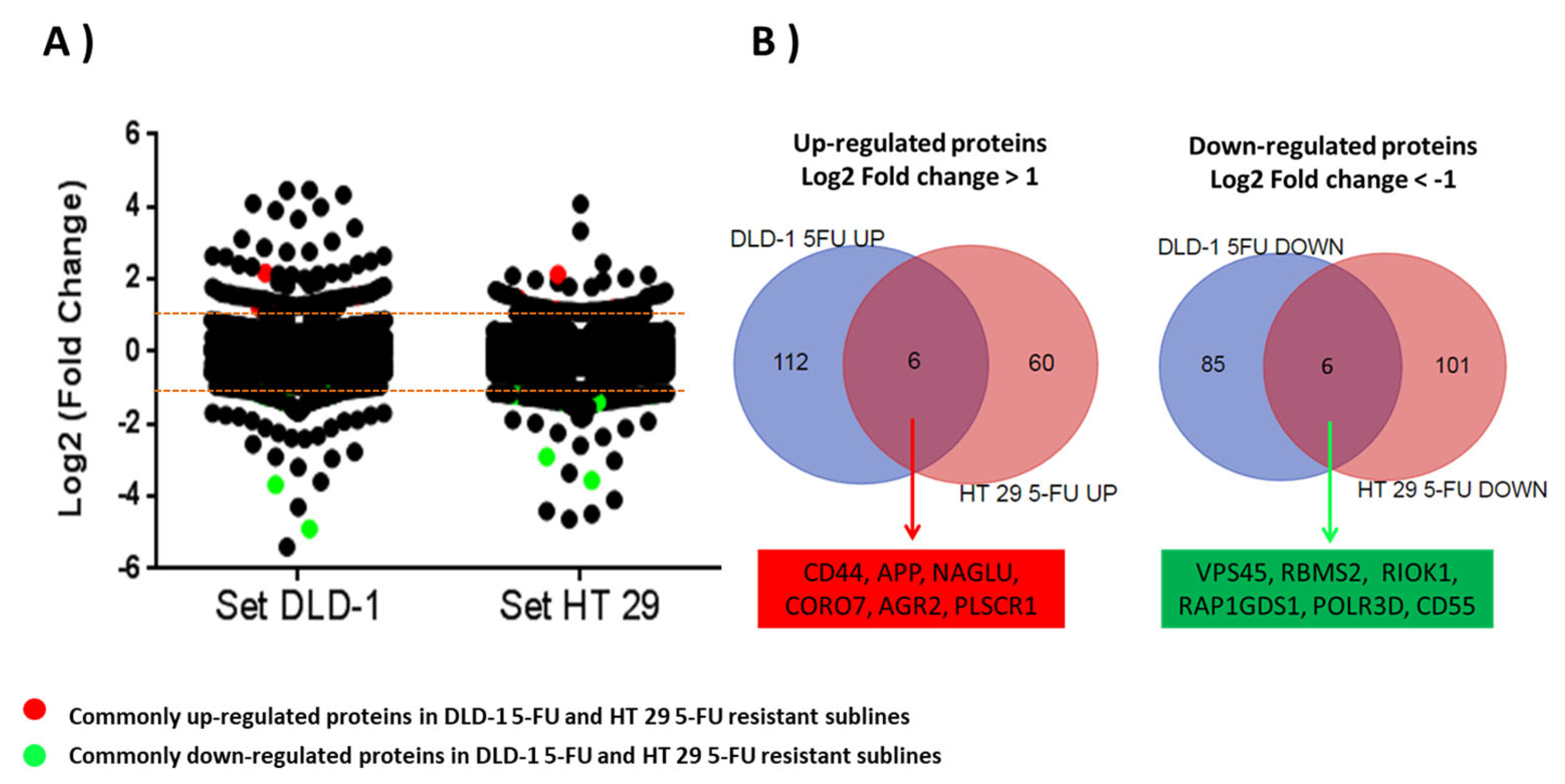

| Gene Name | UniProt Accession 1 | MW 2 [kDa] | Calc. pI 2 | Seq Coverage 3 | Peptides 4 | PSMs 5 | Mascot Score 6 | Abundance Ratio: Log2 R/LP 7 | ||
|---|---|---|---|---|---|---|---|---|---|---|
| Commonly up-regulated | DLD-1 | HT-29 | ||||||||
| CD44 antigen | CD44 | P16070 | 81.5 | 5.3 | 8.2 | 5 | 76 | 1261 | 0.97 | 2.23 |
| Amyloid-beta A4 precursor protein | APP | P05067 | 86.9 | 4.8 | 24.8 | 14 | 103 | 1122 | 1.39 | 1.04 |
| N-acetyl-alpha-glucosaminidase | NAGLU | P54802 | 82.2 | 6.7 | 26.9 | 12 | 61 | 561 | 1.45 | 1.43 |
| Coronin 7 | CORO7 | P57737 | 100.5 | 5.8 | 28.6 | 12 | 64 | 864 | 1.09 | 0.82 |
| Anterior gradient protein 2 homolog | AGR2 | O95994 | 20.0 | 9.0 | 57.1 | 10 | 488 | 7943 | 0.32 | 2.47 |
| Phospholipid scramblase 1 | PLSCR1 | O15162 | 35.0 | 4.9 | 9.4 | 3 | 27 | 515 | 0.74 | 1.30 |
| Commonly down-regulated | ||||||||||
| Vacuolar protein sorting-associated protein 45 | VPS45 | Q9NRW7 | 65.0 | 8.2 | 11.9 | 6 | 30 | 377 | −4.83 | −2.93 |
| RNA binding motif single stranded interacting protein 2 | RBMS2 | Q15434 | 43.9 | 9.1 | 27.5 | 6 | 27 | 533 | −0.87 | −2.00 |
| Serine/threonine-protein kinase RIO1 | RIOK1 | Q9BRS2 | 65.5 | 6.2 | 13.7 | 4 | 11 | 150 | −1.13 | −0.05 |
| Rap1 GTPase-GDP dissociation stimulator 1 | RAP1GSD1 | P52306 | 66.3 | 5.3 | 23.9 | 10 | 50 | 1010 | −1.75 | −1.46 |
| DNA-directed RNA polymerase III subunit RPC4 | POLR3D | P05423 | 44.4 | 7.0 | 4.5 | 1 | 7 | 159 | −0.33 | −0.09 |
| Complement decay-accelerating factor | CD55 | P08174 | 41.4 | 7.6 | 25.5 | 5 | 15 | 123 | −2.28 | −1.78 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ortega Duran, M.; Shaheed, S.u.; Sutton, C.W.; Shnyder, S.D. A Proteomic Investigation to Discover Candidate Proteins Involved in Novel Mechanisms of 5-Fluorouracil Resistance in Colorectal Cancer. Cells 2024, 13, 342. https://doi.org/10.3390/cells13040342
Ortega Duran M, Shaheed Su, Sutton CW, Shnyder SD. A Proteomic Investigation to Discover Candidate Proteins Involved in Novel Mechanisms of 5-Fluorouracil Resistance in Colorectal Cancer. Cells. 2024; 13(4):342. https://doi.org/10.3390/cells13040342
Chicago/Turabian StyleOrtega Duran, Mario, Sadr ul Shaheed, Christopher W. Sutton, and Steven D. Shnyder. 2024. "A Proteomic Investigation to Discover Candidate Proteins Involved in Novel Mechanisms of 5-Fluorouracil Resistance in Colorectal Cancer" Cells 13, no. 4: 342. https://doi.org/10.3390/cells13040342






