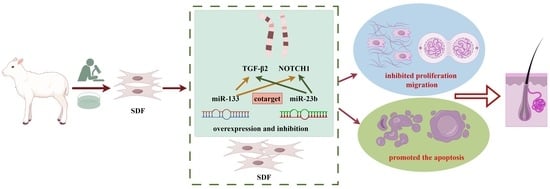
MiR-23b and miR-133 Cotarget TGFβ2/NOTCH1 in Sheep Dermal Fibroblasts, Affecting Hair Follicle Development
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Selection and Skin Tissue Preparation
2.2. Cell Culture and Transfection
2.3. RT‒qPCR
2.4. Western Blotting Analysis
2.5. Prediction and Enrichment Analysis of Target Genes
2.6. Luciferase Reporter Assay
2.7. Cell Transfection
2.8. Cell Proliferation Assay
2.9. Cell Apoptosis Analysis by Annexin V-FITC Staining
2.10. Transwell Cell Migration Assay
2.11. Flow Cytometry Analysis
2.12. Statistical Analysis
3. Results
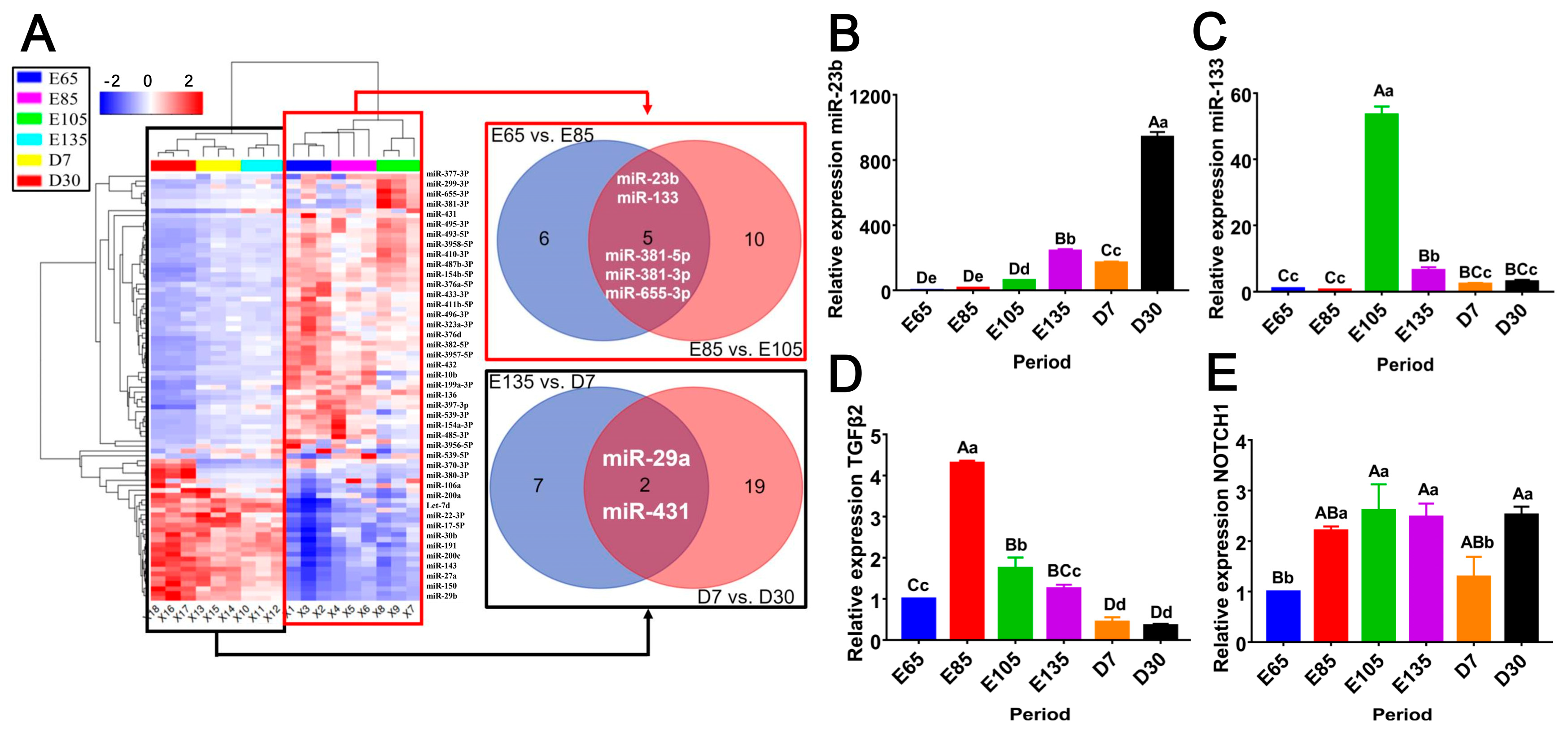
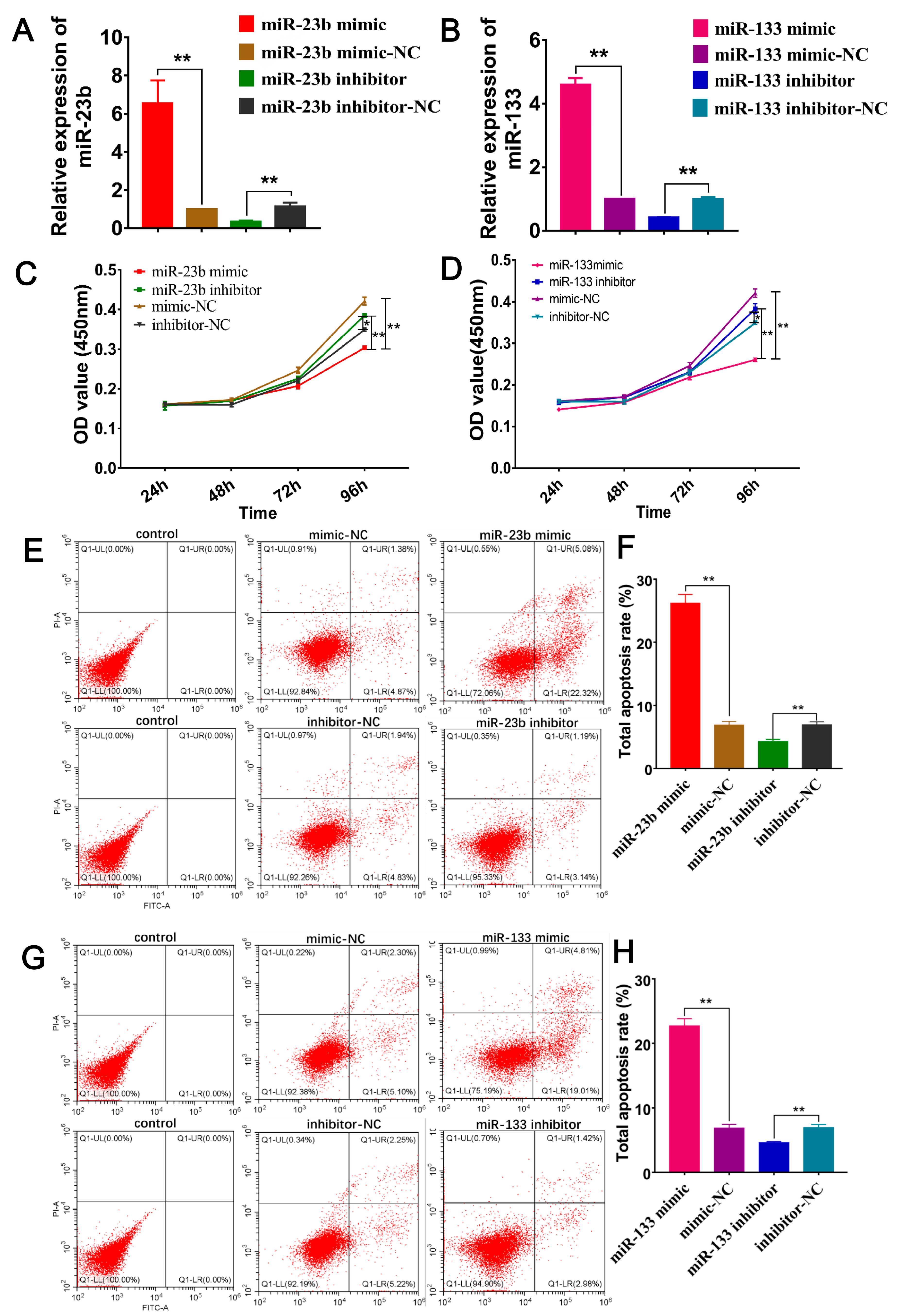
3.1. DE-miRNA Clustering and RT‒qPCR Analysis
3.2. Functional Enrichment Analysis of Oar-miR-23b and Oar-miR-133 Target Genes
3.3. Oar-miR-23b and Oar-miR-133 Cotarget NOTCH1/TGFβ2
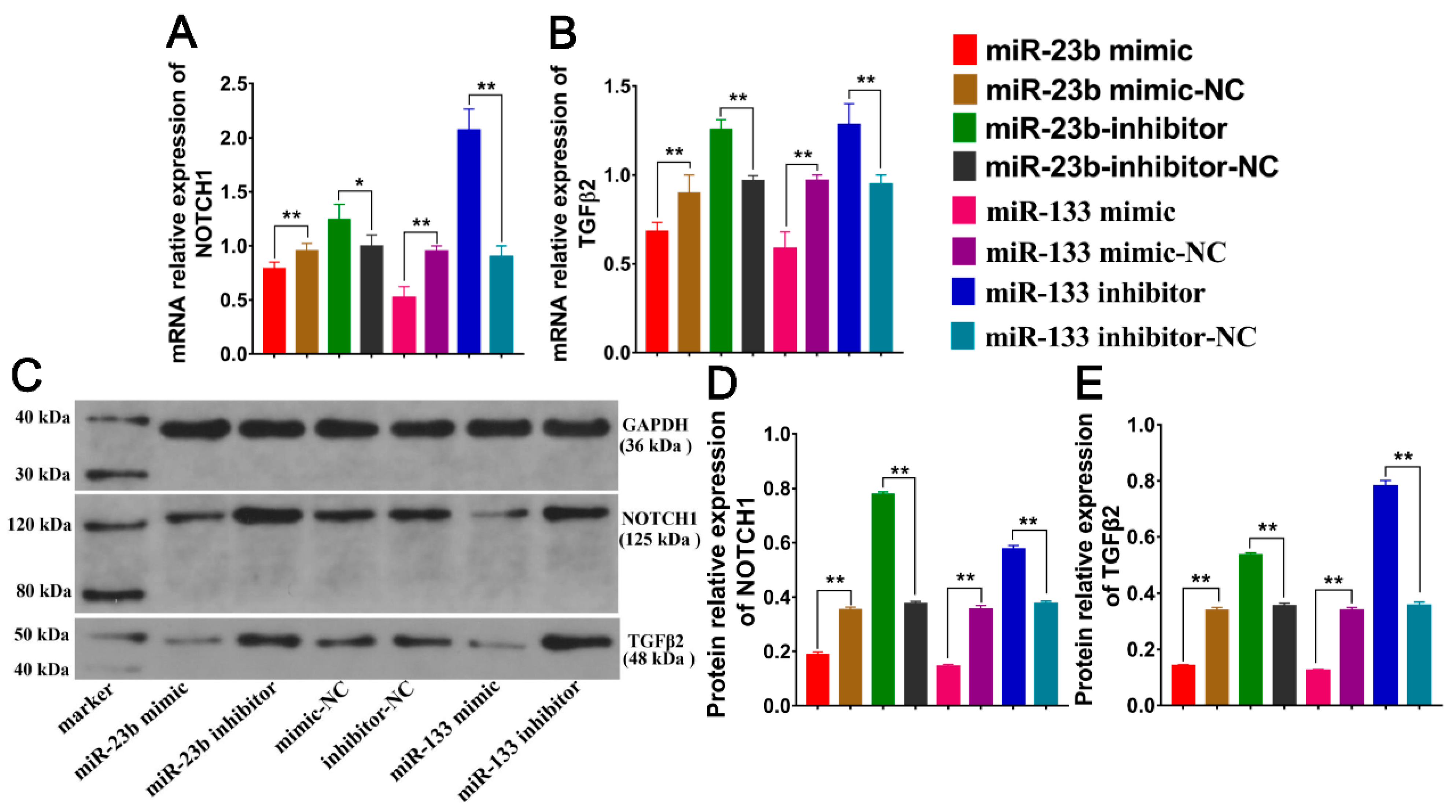
3.4. Oar-miR-23 and Oar-miR-133 Inhibit NOTCH1/TGFβ2 Expression
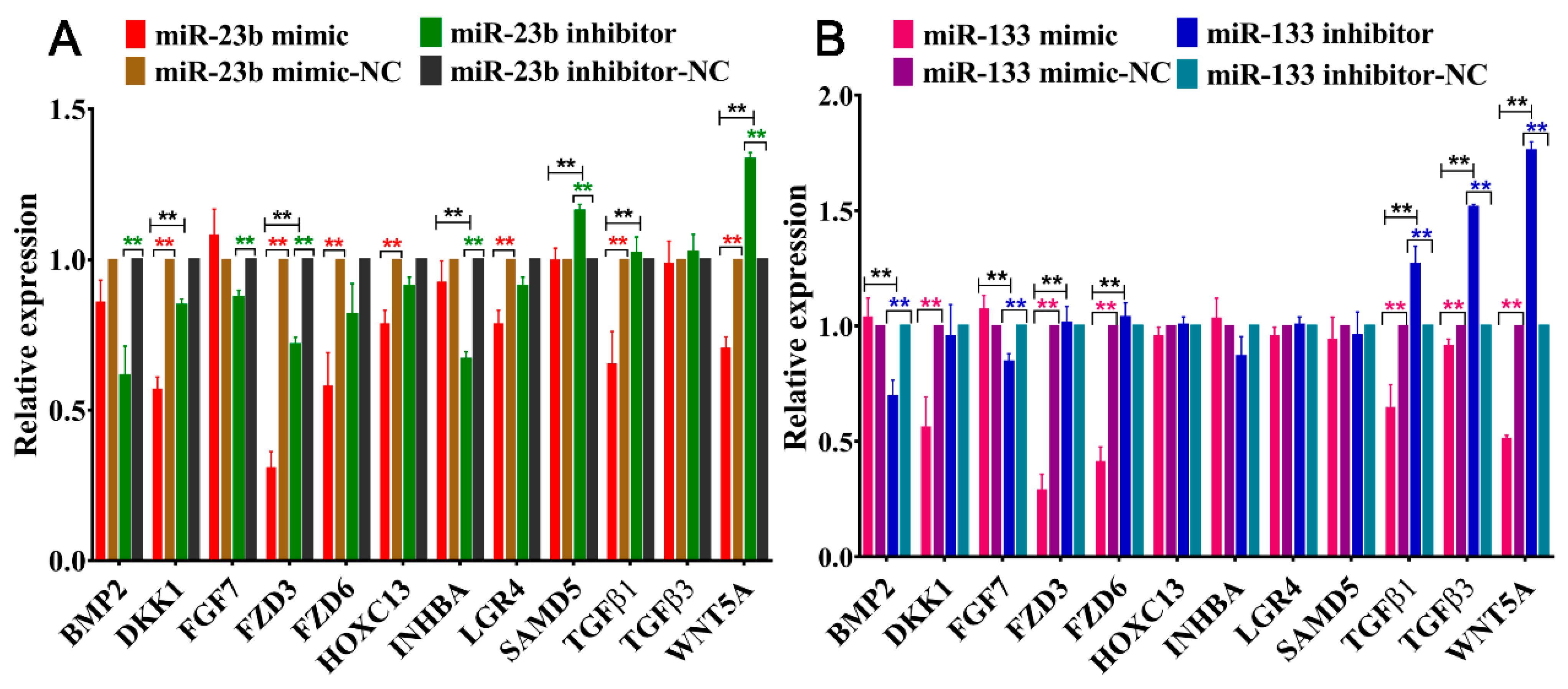
3.5. Effects of Oar-miR-23b and Oar-miR-133 on Pathway-Related Gene Expression
3.6. Oar-miR-23b and Oar-miR-133 Enhance Apoptosis and Inhibit Proliferation of SDFs
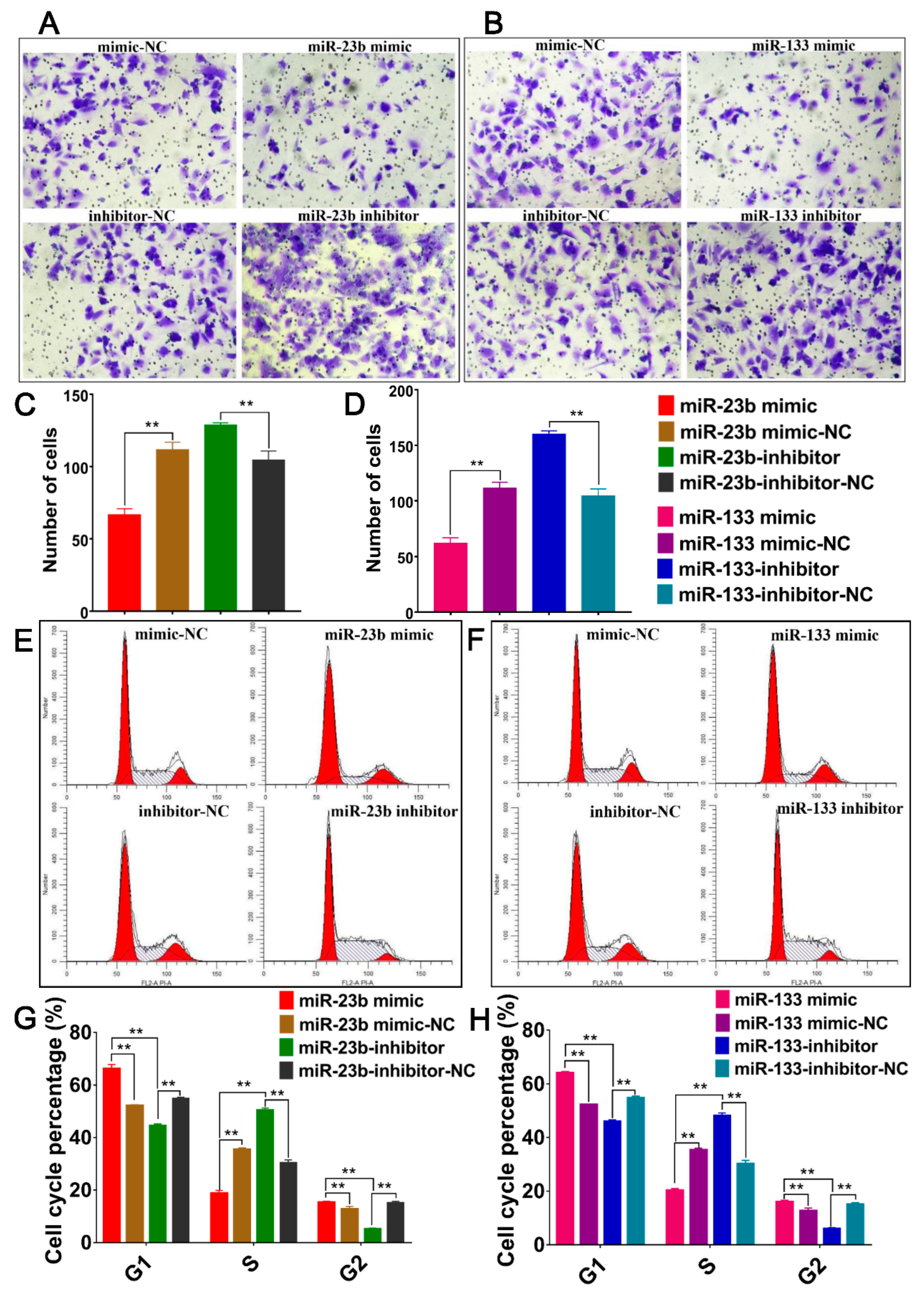
3.7. Oar-miR-23b and Oar-miR-133 Inhibit Cell Migration and Alter Cell Cycle Progression of SDFs
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Paus, R.; Muller, R.S.; Van Der, V.C.; Maurer, M.; Eichmuller, S.; Ling, G.; Hofmann, U.; Foitzik, K.; Mecklenburg, L.; Handjiski, B. A Comprehensive Guide for the Recognition and Classification of Distinct Stages of Hair Follicle Morphogenesis. J. Investig. Dermatol. 1999, 113, 523–532. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Luo, H.; He, J.; Huang, X.; Chen, S.; Fu, X.; Zeng, W.; Tian, Y.; Liu, S.; Li, C.; et al. Comprehensive Transcriptome and Methylome Analysis Delineates the Biological Basis of Hair Follicle Development and Wool-Related Traits in Merino Sheep. BMC Biol. 2021, 19, 197. [Google Scholar] [CrossRef] [PubMed]
- Hardy, M.; Lyne, A.G. The Pre-Natal Development of Wool Follicles in Merino Sheep. Aust. J. Biol. Sci. 1956, 9, 423–441. [Google Scholar] [CrossRef]
- Chapman, R.E.; Hopkins, P.S.; Thorburn, G.D. The Effects of Fetal Thyroidectomy and Thyroxine Administration on the Development of Skin and Wool Follicles of Sheep Fetuses. J. Anat. 1974, 117, 419–432. [Google Scholar]
- Brook, A.H.; Short, B.F.; Lyne, A.G. Formation of New Wool Follicles in the Adult Sheep. Nature 1960, 185, 51. [Google Scholar] [CrossRef]
- Hui, T.; Zhu, Y.; Shen, J.; Bai, M.; Fan, Y.; Feng, S.; Wang, Z.; Zhao, J.; Zhang, Q.; Liu, X.; et al. Identification and Molecular Analysis of M6a-CircRNAs from Cashmere Goat Reveal their Integrated Regulatory Network and Putative Functions in Secondary Hair Follicle during Anagen Stage. Animals 2022, 12, 694. [Google Scholar] [CrossRef]
- Lindner, G.; Botchkarev, V.A.; Botchkareva, N.V.; Ling, G.; van der Veen, C.; Paus, R. Analysis of Apoptosis during Hair Follicle Regression (Catagen). Am. J. Path. 1997, 151, 1601–1617. [Google Scholar]
- Zhang, Y.; Andl, T.; Yang, S.H.; Teta, M.; Liu, F.; Seykora, J.T.; Tobias, J.W.; Piccolo, S.; Schmidt-Ullrich, R.; Nagy, A.; et al. Activation of Beta-Catenin Signaling Programs Embryonic Epidermis to Hair Follicle Fate. Development 2008, 135, 2161–2172. [Google Scholar] [CrossRef]
- Gentile, P.; Garcovich, S. Advances in Regenerative Stem Cell Therapy in Androgenic Alopecia and Hair Loss: Wnt Pathway, Growth-Factor, and Mesenchymal Stem Cell Signaling Impact Analysis on Cell Growth and Hair Follicle Development. Cells 2019, 8, 466. [Google Scholar] [CrossRef]
- Foitzik, K.; Lindner, G.; Mueller, R.S.; Maurer, M.; Botchkareva, N.; Botchkarev, V.; Handjiski, B.; Metz, M.; Hibino, T.; Soma, T.; et al. Control of Murine Hair Follicle Regression (Catagen) by TGF-β1 In Vivo. FASEB J. 2000, 14, 752–760. [Google Scholar] [CrossRef]
- Telerman, S.B.; Rognoni, E.; Sequeira, I.; Pisco, A.O.; Lichtenberger, B.M.; Culley, O.J.; Viswanathan, P.; Driskell, R.R.; Watt, F.M. Dermal Blimp1 Acts Downstream of Epidermal TGF beta and Wnt/Beta-Catenin to Regulate Hair Follicle Formation and Growth. J. Investig. Dermatol. 2017, 137, 2270–2281. [Google Scholar] [CrossRef]
- Rendl, M.; Polak, L.; Fuchs, E. BMP signaling in dermal papilla cells is required for their hair follicle-inductive properties. Genes Dev. 2008, 22, 543–557. [Google Scholar] [CrossRef]
- Tian, Y.; Yang, X.; Du, J.; Zeng, W.; Wu, W.; Di, J.; Huang, X.; Tian, K. Differential Methylation and Transcriptome Integration Analysis Identified Differential Methylation Annotation Genes and Functional Research Related to Hair Follicle Development in Sheep. Front. Genet. 2021, 12, 735827. [Google Scholar] [CrossRef]
- Botchkareva, N.V. MicroRNA/mRNA Regulatory Networks in the Control of Skin Development and Regeneration. Cell Cycle 2012, 11, 468–474. [Google Scholar] [CrossRef]
- He, L.; Hannon, G.J. MicroRNAs: Small RNAs with a Big Role in Gene Regulation. Nat. Rev. Genet. 2004, 5, 522–531. [Google Scholar] [CrossRef]
- Fan, J.; An, X.; Yang, Y.; Xu, H.; Fan, L.; Deng, L.; Li, T.; Weng, X.; Zhang, J.; Zhao, R.C. MiR-1292 Targets FZD4 to Regulate Senescence and Osteogenic Differentiation of Stem Cells in TE/SJ/Mesenchymal Tissue System via the Wnt/β-Catenin Pathway. Aging Dis. 2018, 9, 1103–1121. [Google Scholar] [CrossRef]
- Wang, Z.; Pang, L.; Zhao, H.; Song, L.; Wang, Y.; Sun, Q.; Guo, C.; Wang, B.; Qin, X.; Pan, A. MiR-128 Regulates Differentiation of Hair Follicle Mesenchymal Stem Cells into Smooth Muscle Cells by Targeting SMAD2. Acta Histochem. 2016, 118, 393–400. [Google Scholar] [CrossRef]
- Qu, H.; Wu, S.; Li, J.; Ma, T.; Li, J.; Xiang, B.; Jiang, H.; Zhang, Q. MiR-125B Regulates the Differentiation of Hair Follicles in Fine-Wool Sheep and Cashmere Goats by Targeting MXD4 and FGFR2. Anim. Biotechnol. 2023, 34, 357–364. [Google Scholar] [CrossRef]
- Zhao, B.; Chen, Y.; Yang, N.; Chen, Q.; Bao, Z.; Liu, M.; Hu, S.; Li, J.; Wu, X. MiR-218-5P Regulates Skin and Hair Follicle Development through Wnt/β-Catenin Signaling Pathway by Targeting SFRP2. J. Cell Physiol. 2019, 234, 20329–20341. [Google Scholar] [CrossRef]
- Ahmed, M.I.; Mardaryev, A.N.; Lewis, C.J.; Sharov, A.A.; Botchkareva, N.V. MicroRNA-21 is an Important Downstream Component of BMP Signaling in Epidermal Keratinocytes. J. Cell Sci. 2011, 124, 3399–3404. [Google Scholar] [CrossRef]
- Mardaryev, A.N.; Ahmed, M.I.; Vlahov, N.V.; Fessing, M.Y.; Gill, J.H.; Sharov, A.A.; Botchkareva, N.V. Micro-RNA-31 Controls Hair Cycle-Associated Changes in Gene Expression Programs of the Skin and Hair Follicle. FASEB J. 2010, 24, 3869–3881. [Google Scholar] [CrossRef]
- Ahmed, M.I.; Alam, M.; Emelianov, V.U.; Poterlowicz, K.; Patel, A.; Sharov, A.A.; Mardaryev, A.N.; Botchkareva, N.V. MicroRNA-214 Controls Skin and Hair Follicle Development by Modulating the Activity of the Wnt Pathway. J. Cell Biol. 2014, 207, 549–567. [Google Scholar] [CrossRef]
- Du, K.T.; Deng, J.Q.; He, X.G.; Liu, Z.P.; Peng, C.; Zhang, M.S. MiR-214 Regulates the Human Hair Follicle Stem Cell Proliferation and Differentiation by Targeting EZH2 and Wnt/Beta-Catenin Signaling Way In Vitro. Tissue Eng. Regen. Med. 2018, 15, 341–350. [Google Scholar] [CrossRef]
- Zhai, B.; Zhang, L.; Wang, C.; Zhao, Z.; Zhang, M.; Li, X. Identification of microRNA-21 Target Genes Associated with Hair Follicle Development in Sheep. PeerJ 2019, 7, e7167. [Google Scholar] [CrossRef]
- Yin, R.H.; Zhao, S.J.; Jiao, Q.; Wang, Z.Y.; Bai, M.; Fan, Y.X.; Zhu, Y.B.; Bai, W.L. CircRNA-1926 Promotes the Differentiation of Goat SHF Stem Cells into Hair Follicle Lineage by miR-148a/b-3p/CDK19 Axis. Animals 2020, 10, 1552. [Google Scholar] [CrossRef] [PubMed]
- Lv, X.; Gao, W.; Jin, C.; Wang, L.; Wang, Y.; Chen, W.; Zou, S.; Huang, S.; Li, Z.; Wang, J.; et al. Preliminary Study on microR-148a and microR-10a in Dermal Papilla Cells of Hu Sheep. BMC Genet. 2019, 20, 70. [Google Scholar] [CrossRef]
- Zhu, N.; Huang, K.; Liu, Y.; Zhang, H.; Lin, E.; Zeng, Y.; Li, H.; Xu, Y.; Cai, B.; Yuan, Y.; et al. MiR-195-5P Regulates Hair Follicle Inductivity of Dermal Papilla Cells by Suppressing Wnt/Beta-Catenin Activation. Biomed. Res. Int. 2018, 2018, 4924356. [Google Scholar] [CrossRef]
- Ma, T.; Li, J.; Jiang, Q.; Wu, S.; Jiang, H.; Zhang, Q. Differential Expression of miR-let7a in Hair Follicle Cycle of Liaoning Cashmere Goats and Identification of its Targets. Funct. Integr. Genom. 2018, 18, 701–707. [Google Scholar] [CrossRef]
- Cai, B.; Li, M.; Zheng, Y.; Yin, Y.; Jin, F.; Li, X.; Dong, J.; Jiao, X.; Liu, X.; Zhang, K.; et al. MicroRNA-149-Mediated MAPK1/ERK2 Suppression Attenuates Hair Follicle Stem Cell Differentiation. Hum. Gene Ther. 2022, 33, 625–637. [Google Scholar] [CrossRef]
- Yi, R.; O’Carroll, D.; Pasolli, H.A.; Zhang, Z.; Dietrich, F.S.; Tarakhovsky, A.; Fuchs, E. Morphogenesis in Skin is Governed by Discrete Sets of Differentially Expressed microRNAs. Nat. Genet. 2006, 38, 356–362. [Google Scholar] [CrossRef]
- Yi, R.; Poy, M.N.; Stoffel, M.; Fuchs, E. A Skin microRNA Promotes Differentiation by Repressing ‘Stemness’. Nature 2008, 452, 225–229. [Google Scholar] [CrossRef]
- Zhang, L.; Stokes, N.; Polak, L.; Fuchs, E. Specific microRNAs are Preferentially Expressed by Skin Stem Cells to Balance Self-Renewal and Early Lineage Commitment. Cell Stem Cell 2011, 8, 294–308. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Tredget, E.E.; Wu, Y. Dynamic Signals for Hair Follicle Development and Regeneration. Stem Cells Dev. 2012, 21, 7–18. [Google Scholar] [CrossRef] [PubMed]
- Ito, M.; Yang, Z.; Andl, T.; Cui, C.; Kim, N.; Millar, S.E.; Cotsarelis, G. Wnt-Dependent De Novo Hair Follicle Regeneration in Adult Mouse Skin after Wounding. Nature 2007, 447, 316–320. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; Zeisberg, M. Fibroblasts in Cancer. Nat. Rev. Cancer 2006, 6, 392–401. [Google Scholar] [CrossRef] [PubMed]
- Rosenquist, T.A.; Martin, G.R. Fibroblast Growth Factor Signaling in the Hair Growth Cycle: Expression of the Fibroblast Growth Factor Receptor and Ligand Genes in the Murine Hair Follicle. Dev. Dynam. 1996, 205, 379–386. [Google Scholar] [CrossRef]
- Thangapazham, R.; Darling, T.; Meyerle, J. Alteration of Skin Properties with Autologous Dermal Fibroblasts. Int. J. Mol. Sci. 2014, 15, 8407–8427. [Google Scholar] [CrossRef]
- Kulessa, H.; Turk, G.; Hogan, B.L. Inhibition of BMP Signaling Affects Growth and Differentiation in the Anagen Hair Follicle. Embo J. 2000, 19, 6664–6674. [Google Scholar] [CrossRef]
- Botchkarev, V.A. Bone Morphogenetic Proteins and their Antagonists in Skin and Hair Follicle Biology. J. Investig. Dermatol. 2003, 120, 36–47. [Google Scholar] [CrossRef]
- Sorrell, J.M.; Caplan, A.I. Fibroblast Heterogeneity: More than Skin Deep. J. Cell Sci. 2004, 117, 667–675. [Google Scholar] [CrossRef] [PubMed]
- Woodley, D.T. Distinct Fibroblasts in the Papillary and Reticular Dermis. Dermatol. Clin. 2017, 35, 95–100. [Google Scholar] [CrossRef] [PubMed]
- Driskell, R.R.; Lichtenberger, B.M.; Hoste, E.; Kretzschmar, K.; Simons, B.D.; Charalambous, M.; Ferron, S.R.; Herault, Y.; Pavlovic, G.; Ferguson-Smith, A.C.; et al. Distinct Fibroblast Lineages Determine Dermal Architecture in Skin Development and Repair. Nature 2013, 504, 277–281. [Google Scholar] [CrossRef] [PubMed]
- Kasper, M.; Jaks, V.; Are, A.; Bergstrom, A.; Schwager, A.; Svard, J.; Teglund, S.; Barker, N.; Toftgård, R. Wounding Enhances Epidermal Tumorigenesis by Recruiting Hair Follicle Keratinocytes. Proc. Natl. Acad. Sci. USA 2011, 108, 4099–4104. [Google Scholar] [CrossRef] [PubMed]
- Martin, P. Wound Healing—Aiming for Perfect Skin Regeneration. Science 1997, 276, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Shah, M.; Foreman, D.M.; Ferguson, M.W. Neutralising Antibody to TGF-Beta 1, 2 Reduces Cutaneous Scarring in Adult Rodents. J. Cell Sci. 1994, 107, 1137–1157. [Google Scholar] [CrossRef] [PubMed]
- Huelsken, J.; Vogel, R.; Erdmann, B.; Cotsarelis, G.; Birchmeier, W. Beta-Catenin Controls Hair Follicle Morphogenesis and Stem Cell Differentiation in the Skin. Cell 2001, 105, 533–545. [Google Scholar] [CrossRef] [PubMed]
- Collins, C.A.; Kretzschmar, K.; Watt, F.M. Reprogramming Adult Dermis to a Neonatal State through Epidermal Activation of β-Catenin. Development 2011, 138, 5189–5199. [Google Scholar] [CrossRef]
- Gong, J.; Lin, Y.; Zhang, H.; Liu, C.; Cheng, Z.; Yang, X.; Zhang, J.; Xiao, Y.; Sang, N.; Qian, X.; et al. Reprogramming of Lipid Metabolism in Cancer-Associated Fibroblasts Potentiates Migration of Colorectal Cancer Cells. Cell Death Dis. 2020, 11, 267. [Google Scholar] [CrossRef]
- He, J.; Huang, X.; Zhao, B.; Liu, G.; Tian, Y.; Zhang, G.; Wei, C.; Mao, J.; Tian, K. Integrated Analysis of miRNAs and mRNA Profiling Reveals the Potential Roles of miRNAs in Sheep Hair Follicle Development. BMC Genom. 2022, 23, 722. [Google Scholar] [CrossRef]
- He, J.; Zhao, B.; Huang, X.; Fu, X.; Liu, G.; Tian, Y.; Wu, C.; Mao, J.; Liu, J.; Gun, S.; et al. Gene Network Analysis Reveals Candidate Genes Related with the Hair Follicle Development in Sheep. BMC Genom. 2022, 23, 428. [Google Scholar] [CrossRef]
- Zeng, J.; Zeng, Z.; Zhang, K.; Zhao, Y.; Liu, Y.; Chen, J.; Tong, H.; Wei, D.; Jiang, Z.; Wang, Z. MiR-23b-3p and miR-125b-5p Downregulate Apo(a) Expression by Targeting ETS1 in HEPG2 Cells. Cell Biol. Int. 2018, 42, 313–323. [Google Scholar] [CrossRef]
- Xian, X.; Tang, L.; Wu, C.; Huang, L. MiR-23b-3p and miR-130a-5p Affect Cell Growth; Migration and Invasion by Targeting CB1R via the Wnt/Beta-Catenin Signaling Pathway in Gastric Carcinoma. Oncotargets Ther. 2018, 11, 7503–7512. [Google Scholar] [CrossRef]
- Wang, J.; Zhao, B. MiR-23b-3p Functions as a Positive Factor for Osteoporosis Progression by Targeting CCND1 in MC3T3-E1 Cells. Vitr. Cell. Dev. Biol.-Anim. 2021, 57, 324–331. [Google Scholar] [CrossRef] [PubMed]
- Smola, H.; Thiekotter, G.; Baur, M.; Stark, H.J.; Breitkreutz, D.; Fusenig, N.E. Organotypic and Epidermal-Dermal Co-Cultures of Normal Human Keratinocytes and Dermal Cells: Regulation of Transforming Growth Factor Alpha, Beta1 and Beta2 mRNA Levels. Toxicol. Vitr. 1994, 8, 641–650. [Google Scholar] [CrossRef]
- Hu, H.; Tang, J.; Liu, C.; Cen, Y. MiR-23b Promotes the Migration of Keratinocytes through Downregulating TIMP3. J. Surg. Res. 2020, 254, 102–109. [Google Scholar] [CrossRef]
- Yang, Z.; Xiao, Z.; Guo, H.; Fang, X.; Liang, J.; Zhu, J.; Yang, J.; Li, H.; Pan, R.; Yuan, S.; et al. Novel Role of the Clustered miR-23b-3p and miR-27b-3p in Enhanced Expression of Fibrosis-Associated Genes by Targeting TGEBR3 in Atrial Fibroblasts. J. Cell Mol. Med. 2019, 23, 3246–3256. [Google Scholar] [CrossRef] [PubMed]
- Mamalis, A.; Koo, E.; Tepper, C.; Jagdeo, J. MicroRNA Expression Analysis of Human Skin Fibroblasts Treated with High-Fluence Light-Emitting Diode-Red Light. J. Biophotonics 2019, 12, e201800207. [Google Scholar] [CrossRef]
- Chen, Z.; Liang, Y.; Lu, Q.; Nazar, M.; Mao, Y.; Aboragah, A.; Yang, Z.; Loor, J.J. Cadmium Promotes Apoptosis and Inflammation via the Circ08409/miR-133a/TGFβ2 Axis in Bovine Mammary Epithelial Cells and Mouse Mammary Gland. Ecotox. Environ. Safe 2021, 222, 112477. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Zhou, H.; Qian, C.; Huang, N.; Gu, Y.; Huang, Y.; Li, W.; Wei, B.; Mao, S.; Li, S.; et al. Effect of miR-133 on Myocardial Cell Apoptosis in Rats with Myocardial Infarction through the Notch1 Signaling Pathway. Minerva Med. 2021, 112, 303–305. [Google Scholar] [CrossRef]
- Shao, X.; Kong, W.; Li, Y. MiR-133 Inhibits Kidney Injury in Rats with Diabetic Nephropathy via MAPK/ERK Pathway. Eur. Rev. Med. Pharmaco. 2019, 23, 10957–10963. [Google Scholar] [CrossRef]
- Xu, F.; Li, F.; Zhang, W.; Jia, P. Growth of Glioblastoma is Inhibited by miR-133-Mediated EGFR Suppression. Tumour Biol. 2015, 36, 9553–9558. [Google Scholar] [CrossRef]
- Mina, S.Z.; Fatemeh, K.; Masoud, S.; Amir, A.; Tasharrofi, N. Overexpression of miR-133 Decrease Primary Endothelial Cells Proliferation and Migration via FGFR1 Targeting. Exp. Cell Res. 2018, 369, 11–16. [Google Scholar]
- Nan, W.; Li, G.; Si, H.; Lou, Y.; Wang, D.; Guo, R.; Zhang, H. All-Trans-Retinoic Acid Inhibits Mink Hair Follicle Growth via Inhibiting Proliferation and Inducing Apoptosis of Dermal Papilla Cells through TGF-β2/SMAD2/3 Pathway. Acta Histochem. 2020, 122, 151603. [Google Scholar] [CrossRef]
- Lu, G.; Wu, Z.; Chu, X.; Bi, Z.; Fan, W. An Investigation of Crosstalk Between Wnt/β-Catenin and Transforming Growth Factor-Β Signaling in Androgenetic Alopecia. Med. 2016, 95, e4297. [Google Scholar] [CrossRef]
- Foitzik, K.; Paus, R.; Doetschman, T.; Paolo, D.G. The TGF-β2 Isoform is Both a Required and Sufficient Inducer of Murine Hair Follicle Morphogenesis. Dev. Biol. 1999, 212, 278–289. [Google Scholar] [CrossRef] [PubMed]
- Woo, W.; Zhen, H.H.; Oro, A.E. Shh Maintains Dermal Papilla Identity and Hair Morphogenesis via a Noggin-Shh Regulatory Loop. Gene Dev. 2012, 26, 1235–1246. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Massagué, J. Mechanisms of TGF-β Signaling from Cell Membrane to the Nucleus. Cell 2003, 113, 685–700. [Google Scholar] [CrossRef]
- Hai, E.; Han, W.; Wu, Z.; Ma, R.; Shang, F.; Wang, M.; Liang, L.; Rong, Y.; Pan, J.; Wang, Z.; et al. Chi-miR-370-3P Regulates Hair Follicle Morphogenesis of Inner Mongolian Cashmere Goats. G3 2021, 11, jkab091. [Google Scholar] [CrossRef] [PubMed]
- Han, W.; Yang, F.; Wu, Z.; Guo, F.; Zhang, J.; Hai, E.; Shang, F.; Su, R.; Wang, R.; Wang, Z.; et al. Inner Mongolian Cashmere Goat Secondary Follicle Development Regulation Research Based on mRNA-miRNA Co-Analysis. Sci. Rep. 2020, 10, 4519. [Google Scholar] [CrossRef]
- Aubin, H.G. Notch Signaling and the Developing Hair Follicle. Adv. Exp. Med. Biol. 2012, 727, 142–160. [Google Scholar] [CrossRef]
- Powell, B.C.; Passmore, E.A.; Nesci, A.; Dunn, S.M. The Notch Signaling Pathway in Hair Growth. Mech. Dev. 1998, 78, 189–192. [Google Scholar] [CrossRef]
- Blanpain, C.; Lowry, W.E.; Pasolli, H.A.; Fuchs, E. Canonical Notch Signaling Functions as a Commitment Switch in the Epidermal Lineage. Gene Dev. 2006, 20, 3022–3035. [Google Scholar] [CrossRef]
- Kopan, R.; Weintraub, H. Mouse Notch: Expression in Hair Follicles Correlates with Cell Fate Determination. J. Cell Biol. 1993, 121, 631–641. [Google Scholar] [CrossRef]
- Xavier, S.P.; Gordon, T.C.; Wynn, P.C.; McCullagh, P.; Thomson, P.C.; Tomkins, L.; Mason, R.S.; Moore, G.P.M. Evidence that Notch and Delta Expressions Have a Role in Dermal Condensate Aggregation during Wool Follicle Initiation. Exp. Dermatol. 2013, 22, 659–662. [Google Scholar] [CrossRef]
- Cai, J.; Lee, J.; Kopan, R.; Ma, L. Genetic Interplays Between MSX2 and FOXN1 are Required for NOTCH1 Expression and Hair Shaft Differentiation. Dev. Biol. 2009, 326, 420–430. [Google Scholar] [CrossRef]
- Pan, Y.; Lin, M.H.; Tian, X.; Cheng, H.T.; Gridley, T.; Shen, J.; Kopan, R. Gamma-Secretase Functions through Notch Signaling to Maintain Skin Appendages but is Not Required for their Patterning or Initial Morphogenesis. Dev. Cell 2004, 7, 731–743. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
He, J.; Wei, C.; Huang, X.; Zhang, G.; Mao, J.; Li, X.; Yang, C.; Zhang, W.; Tian, K.; Liu, G. MiR-23b and miR-133 Cotarget TGFβ2/NOTCH1 in Sheep Dermal Fibroblasts, Affecting Hair Follicle Development. Cells 2024, 13, 557. https://doi.org/10.3390/cells13060557
He J, Wei C, Huang X, Zhang G, Mao J, Li X, Yang C, Zhang W, Tian K, Liu G. MiR-23b and miR-133 Cotarget TGFβ2/NOTCH1 in Sheep Dermal Fibroblasts, Affecting Hair Follicle Development. Cells. 2024; 13(6):557. https://doi.org/10.3390/cells13060557
Chicago/Turabian StyleHe, Junmin, Chen Wei, Xixia Huang, Guoping Zhang, Jingyi Mao, Xue Li, Cunming Yang, Wenjing Zhang, Kechuan Tian, and Guifen Liu. 2024. "MiR-23b and miR-133 Cotarget TGFβ2/NOTCH1 in Sheep Dermal Fibroblasts, Affecting Hair Follicle Development" Cells 13, no. 6: 557. https://doi.org/10.3390/cells13060557