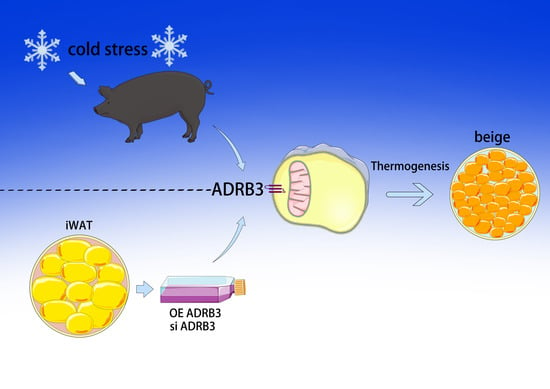
The Role of β3-Adrenergic Receptors in Cold-Induced Beige Adipocyte Production in Pigs
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals and Treatments
2.2. Histology
2.3. Quantitative RT-PCR Analysis
2.4. Mitochondrial DNA Copy Number
2.5. Isolation, Culture, and Differentiation of Porcine SVF Cells
2.6. Overexpression of Porcine ADRB1, ADRB2 and ADRB3
2.7. siRNA Knockdown of ADRB1, ADRB2, and ADRB3
2.8. Seahorse Metabolic Assays
2.9. Western Blotting
2.10. Fluorescence Microscopy
2.11. mRNA Sequencing, RNA-seq Data Analysis, and Functional Analysis
2.12. Statistical Analysis
3. Results
3.1. Cold Stimulation Induces Browning of Subcutaneous Adipose Tissue in Min Pigs through Adrenoceptors
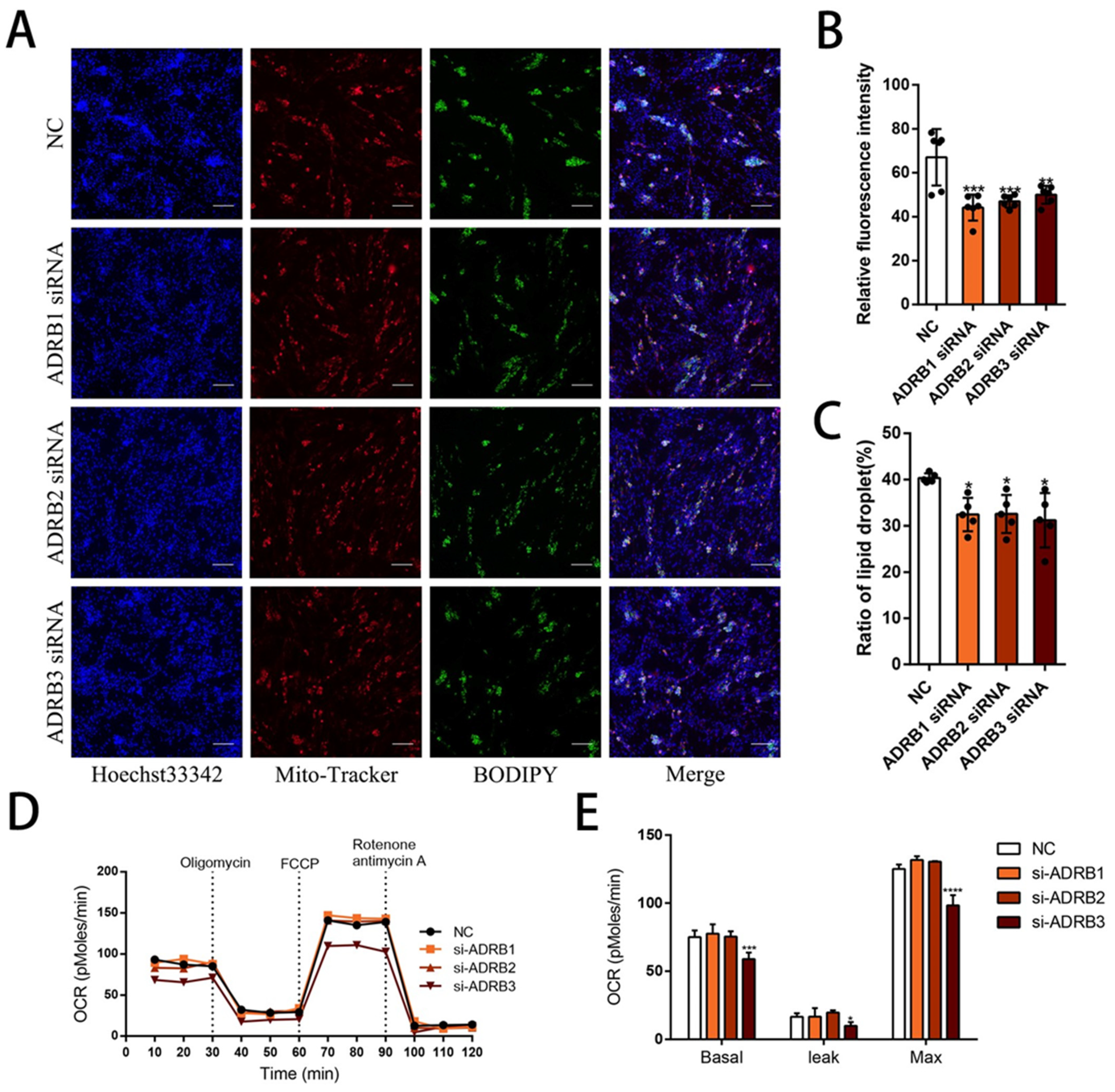
3.2. The Effect of Knocking down β-Adrenergic Receptors on Adipogenic Differentiation of Preadipocytes
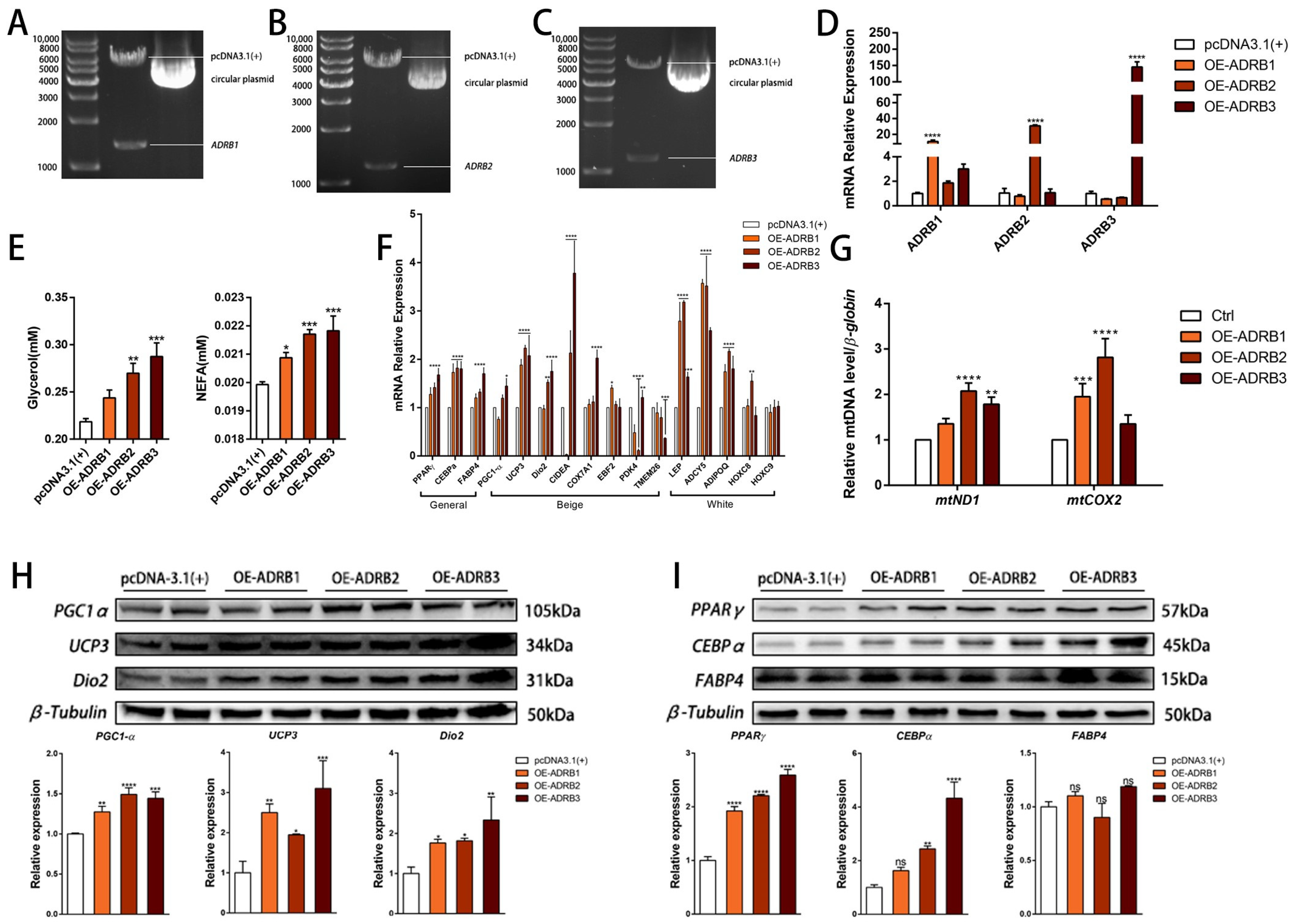
3.3. The Effect of Overexpression of β-Adrenergic Receptors on Adipogenic Differentiation of Preadipocytes
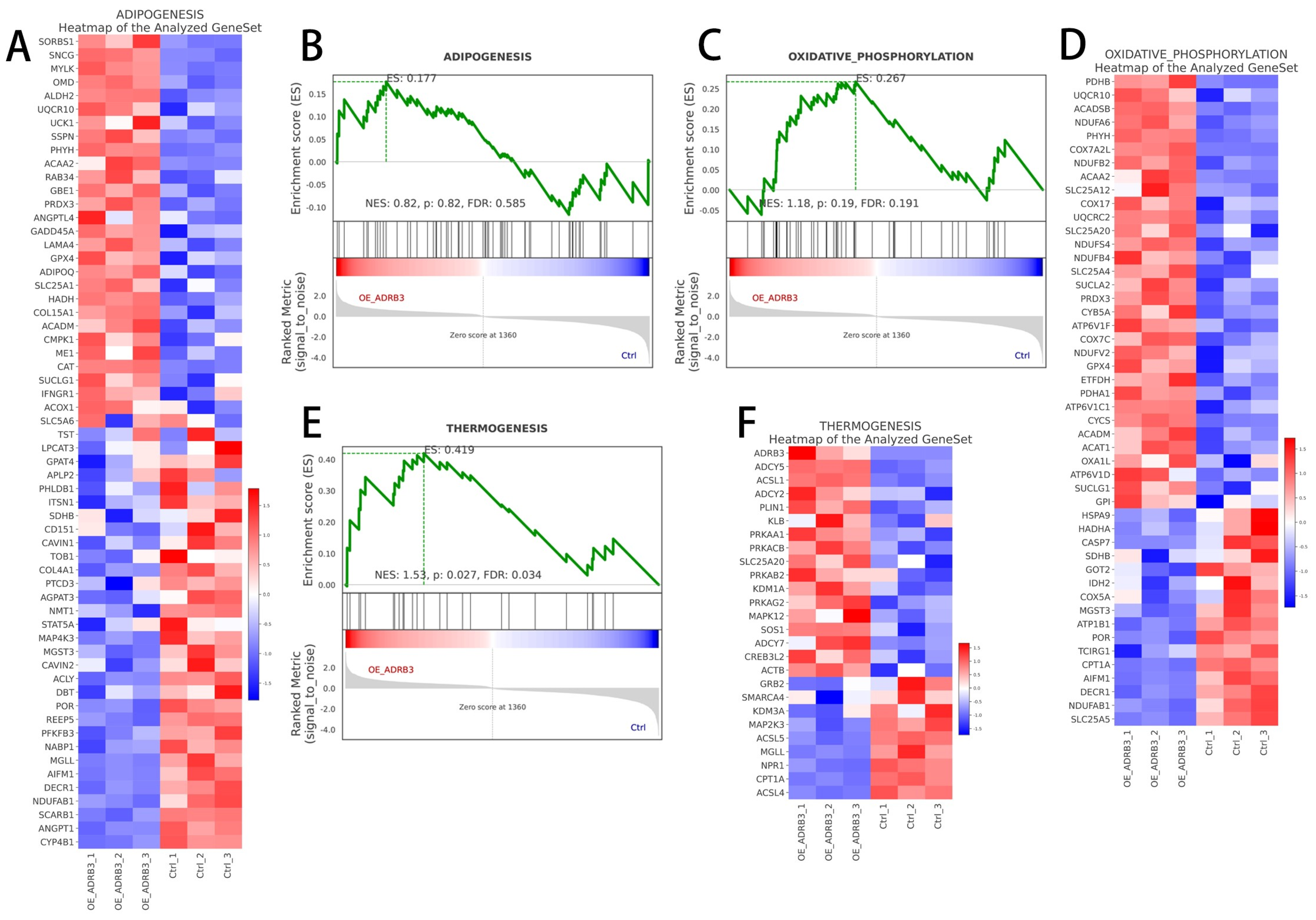
3.4. ADRB3 Plays a Role in Preadipocyte Differentiation at the Transcriptome Level
3.5. Regulatory Network of ADRB3 on Preadipocyte Differentiation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Cannon, B.; Nedergaard, J. Brown adipose tissue: Function and physiological significance. Physiol. Rev. 2004, 84, 277–359. [Google Scholar] [CrossRef] [PubMed]
- Spiegelman, B.M.; Flier, J.S. Obesity and the regulation of energy balance. Cell 2001, 104, 531–543. [Google Scholar] [CrossRef] [PubMed]
- Hafner, A.-L.; Dani, C. Human induced pluripotent stem cells: A new source for brown and white adipocytes. World J. Stem Cells 2014, 6, 467–472. [Google Scholar] [CrossRef]
- Yao, X.; Dani, V.; Dani, C. Human Pluripotent Stem Cells: A Relevant Model to Identify Pathways Governing Thermogenic Adipocyte Generation. Front. Endocrinol. 2020, 10, 932. [Google Scholar] [CrossRef] [PubMed]
- Pearson, S.; Loft, A.; Rajbhandari, P.; Simcox, J.; Lee, S.; Tontonoz, P.; Mandrup, S.; Villanueva, C.J. Loss of TLE3 promotes the mitochondrial program in beige adipocytes and improves glucose metabolism. Genes. Dev. 2019, 33, 747–762. [Google Scholar] [CrossRef]
- Kajimura, S.; Spiegelman, B.M.; Seale, P. Brown and Beige Fat: Physiological Roles beyond Heat Generation. Cell Metab. 2015, 22, 546–559. [Google Scholar] [CrossRef] [PubMed]
- Harms, M.; Seale, P. Brown and beige fat: Development, function and therapeutic potential. Nat. Med. 2013, 19, 1252–1263. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.A.; Tao, C.; Gupta, R.K.; Scherer, P.E. Tracking adipogenesis during white adipose tissue development, expansion and regeneration. Nat. Med. 2013, 19, 1338–1344. [Google Scholar] [CrossRef] [PubMed]
- Barbatelli, G.; Murano, I.; Madsen, L.; Hao, Q.; Jimenez, M.; Kristiansen, K.; Giacobino, J.P.; De Matteis, R.; Cinti, S. The emergence of cold-induced brown adipocytes in mouse white fat depots is determined predominantly by white to brown adipocyte transdifferentiation. Am. J. Physiol.-Endocrinol. Metab. 2010, 298, E1244–E1253. [Google Scholar] [CrossRef] [PubMed]
- Himms-Hagen, J.; Melnyk, A.; Zingaretti, M.C.; Ceresi, E.; Barbatelli, G.; Cinti, S. Multilocular fat cells in WAT of CL-316243-treated rats derive directly from white adipocytes. Am. J. Physiol.-Cell Physiol. 2000, 279, C670–C681. [Google Scholar] [CrossRef]
- Zhang, D.; Ma, S.; Wang, L.; Ma, H.; Wang, W.; Xia, J.; Liu, D. Min pig skeletal muscle response to cold stress. PLoS ONE 2022, 17, e0274184. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.Z.; Yang, X.Q.; Jing, X.Y.; He, X.M.; Wang, L.; Liu, Y.; Liu, D. Transcriptomics Analysis on Excellent Meat Quality Traits of Skeletal Muscles of the Chinese Indigenous Min Pig Compared with the Large White Breed. Int. J. Mol. Sci. 2018, 19, 21. [Google Scholar] [CrossRef] [PubMed]
- Hao, W.J.; Yang, Z.W.; Sun, Y.L.; Li, J.X.; Zhang, D.J.; Liu, D.; Yang, X.Q. Characterization of Alternative Splicing Events in Porcine Skeletal Muscles with Different Intramuscular Fat Contents. Biomolecules 2022, 12, 154. [Google Scholar] [CrossRef]
- Lin, J.; Cao, C.W.; Tao, C.; Ye, R.C.; Dong, M.; Zheng, Q.T.; Wang, C.; Jiang, X.X.; Qin, G.S.; Yan, C.G.; et al. Cold adaptation in pigs depends on UCP3 in beige adipocytes. J. Mol. Cell Biol. 2017, 9, 364–375. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.L.; Hu, S.L.; Cao, C.W.; Chen, C.H.; Liu, J.L.; Wang, Y.; Liu, J.F.; Zhao, J.G.; Tao, C.; Wang, Y.F. Functional and Genetic Characterization of Porcine Beige Adipocytes. Cells 2022, 11, 751. [Google Scholar] [CrossRef]
- Lu, M.; He, Y.; Gong, M.; Li, Q.; Tang, Q.; Wang, X.; Wang, Y.; Yuan, M.; Yu, Z.; Xu, B. Role of Neuro-Immune Cross-Talk in the Anti-obesity Effect of Electro-Acupuncture. Front. Neurosci. 2020, 14, 151. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, R.K.; Ruiz de Azua, I.; Conrad, A.; Purrio, M.; Lutz, B. Cannabinoid CB1 Receptor Deletion from Catecholaminergic Neurons Protects from Diet-Induced Obesity. Int. J. Mol. Sci. 2022, 23, 12635. [Google Scholar] [CrossRef] [PubMed]
- Kouidhi, M.; Villageois, P.; Mounier, C.M.; Ménigot, C.; Rival, Y.; Piwnica, D.; Aubert, J.; Chignon-Sicard, B.; Dani, C. Characterization of human knee and chin adipose-derived stromal cells. Stem Cells Int. 2015, 2015, 592090. [Google Scholar] [CrossRef] [PubMed]
- Kurylowicz, A.; Jonas, M.; Lisik, W.; Jonas, M.; Wicik, Z.A.; Wierzbicki, Z.; Chmura, A.; Puzianowska-Kuznicka, M. Obesity is associated with a decrease in expression but not with the hypermethylation of thermogenesis-related genes in adipose tissues. J. Transl. Med. 2015, 13, 31. [Google Scholar] [CrossRef]
- Collins, S.; Surwit, R.S. The beta-adrenergic receptors and the control of adipose tissue metabolism and thermogenesis. Recent Prog. Horm. Res. 2001, 56, 309–328. [Google Scholar] [CrossRef]
- Shin, S.; Ajuwon, K.M. Divergent Response of Murine and Porcine Adipocytes to Stimulation of Browning Genes by 18-Carbon Polyunsaturated Fatty Acids and Beta-Receptor Agonists. Lipids 2018, 53, 65–75. [Google Scholar] [CrossRef] [PubMed]
- Shin, S.; Ajuwon, K.M. Activation of Adrenergic Receptor Subtypes Differentially Regulate Expression of Metabolic Genes in Porcine Adipocytes. FASEB J. 2017, 31, 792.16. [Google Scholar] [CrossRef]
- Mills, S. Beta-adrenergic receptor subtypes mediating lipolysis in porcine adipocytes. Studies with BRL-37344, a putative beta3-adrenergic agonist. Comp. Biochem. Physiol. Toxicol. Pharmacol. CBP 2000, 126, 11–20. [Google Scholar]
- Berretz, G.; Packheiser, J.; Wolf, O.T.; Ocklenburg, S. Acute stress increases left hemispheric activity measured via changes in frontal alpha asymmetries. iScience 2022, 25, 103841. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.L.; Lin, X.; Zhang, Q.; Pang, Y.; Zhang, X.H.; Zhao, X.L.; Liu, D.; Yang, X.Q. Genome-wide characterization of lncRNAs and mRNAs in muscles with differential intramuscular fat contents. Front. Vet. Sci. 2022, 9, 982258. [Google Scholar] [CrossRef]
- Yang, Y.; Sun, C.C.; Li, F.; Shan, A.S.; Shi, B.M. Characteristics of faecal bacterial flora and volatile fatty acids in Min pig, Landrace pig, and Yorkshire pig. Electron. J. Biotechnol. 2021, 53, 33–43. [Google Scholar] [CrossRef]
- Teng, T.; Sun, G.D.; Ding, H.W.; Song, X.; Bai, G.D.; Shi, B.M.; Shang, T.T. Characteristics of glucose and lipid metabolism and the interaction between gut microbiota and colonic mucosal immunity in pigs during cold exposure. J. Anim. Sci. Biotechnol. 2023, 14, 84. [Google Scholar] [CrossRef] [PubMed]
- Gaudry, M.J.; Jastroch, M.; Treberg, J.R.; Hofreiter, M.; Paijmans, J.L.A.; Starrett, J.; Wales, N.; Signore, A.V.; Springer, M.S.; Campbell, K.L. Inactivation of thermogenic UCP1 as a historical contingency in multiple placental mammal clades. Sci. Adv. 2017, 3, e1602878. [Google Scholar] [CrossRef] [PubMed]
- Berg, F.; Gustafson, U.; Andersson, L. The uncoupling protein 1 gene (UCP1) is disrupted in the pig lineage: A genetic explanation for poor thermoregulation in piglets. PLoS Genet. 2006, 2, e129. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Xu, Z.; Wang, L.; Ling, D.; Nong, Q.; Xie, J.; Zhu, X.; Shan, T. Cold Exposure Induces Depot-Specific Alterations in Fatty Acid Composition and Transcriptional Profile in Adipose Tissues of Pigs. Front. Endocrinol. 2022, 13, 827523. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.W.; Berryit, D.C.; Grain, J.M. Distinct cellular and molecular mechanisms for β3 adrenergic receptor-induced beige adipocyte formation. eLife 2017, 6, e30329. [Google Scholar] [CrossRef] [PubMed]
- Heazlewood, J.L.; Tonti-Filippini, J.S.; Gout, A.M.; Day, D.A.; Whelan, J.; Millar, A.H. Experimental analysis of the Arabidopsis mitochondrial proteome highlights signaling and regulatory components, provides assessment of targeting prediction programs, and indicates plant-specific mitochondrial proteins. Plant Cell 2004, 16, 241–256. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Sánchez, R.; Marín-Hernández, A.; Saavedra, E.; Pardo, J.P.; Ralph, S.J.; Rodríguez-Enríquez, S. Who controls the ATP supply in cancer cells? Biochemistry lessons to understand cancer energy metabolism. Int. J. Biochem. Cell Biol. 2014, 50, 10–23. [Google Scholar] [CrossRef] [PubMed]
- Colson, C.; Batrow, P.L.; Gautier, N.; Rochet, N.; Ailhaud, G.; Peiretti, F.; Amri, E.Z. The Rosmarinus Bioactive Compound Carnosic Acid Is a Novel PPAR Antagonist That Inhibits the Browning of White Adipocytes. Cells 2020, 9, 2433. [Google Scholar] [CrossRef] [PubMed]
- Woodall, B.P.; Gresham, K.S.; Woodall, M.A.; Valenti, M.C.; Cannavo, A.; Pfleger, J.; Chuprun, J.K.; Drosatos, K.; Koch, W.J. Alteration of myocardial GRK2 produces a global metabolic phenotype. JCI Insight 2019, 5, e123848. [Google Scholar] [CrossRef] [PubMed]
- Kahoul, Y.; Yao, X.; Oger, F.; Moreno, M.; Amanzougarene, S.; Derhourhi, M.; Durand, E.; Boutry, R.; Bonnefond, A.; Froguel, P.; et al. Knocking Down CDKN2A in 3D hiPSC-Derived Brown Adipose Progenitors Potentiates Differentiation, Oxidative Metabolism and Browning Process. Cells 2023, 12, 870. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Zhu, S.; Li, C.; Lyu, L.; Yu, J.; Wang, D.; Xu, Z.; Ni, J.; Gao, B.; Lu, J.; et al. Gene essentiality profiling reveals a novel determinant of stresses preventing protein aggregation in Salmonella. Emerg. Microbes Infect. 2022, 11, 1554–1571. [Google Scholar] [CrossRef] [PubMed]
- Fenzl, A.; Kiefer, F.W. Brown adipose tissue and thermogenesis. Horm. Mol. Biol. Clin. Investig. 2014, 19, 25–37. [Google Scholar] [CrossRef] [PubMed]
- Ladoux, A.; Peraldi, P.; Chignon-Sicard, B.; Dani, C. Distinct Shades of Adipocytes Control the Metabolic Roles of Adipose Tissues: From Their Origins to Their Relevance for Medical Applications. Biomedicines 2021, 9, 40. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Trajkovski, M. MiR-27 orchestrates the transcriptional regulation of brown adipogenesis. Metab.-Clin. Exp. 2014, 63, 272–282. [Google Scholar] [CrossRef]
- Blondin, D.P.; Nielsen, S.; Kuipers, E.N.; Severinsen, M.C.; Jensen, V.H.; Miard, S.; Jespersen, N.Z.; Kooijman, S.; Boon, M.R.; Fortin, M.; et al. Human Brown Adipocyte Thermogenesis Is Driven by β2-AR Stimulation. Cell Metab. 2020, 32, 287–300.e287. [Google Scholar] [CrossRef] [PubMed]
- Cero, C.; Lea, H.J.; Zhu, K.Y.; Shamsi, F.; Tseng, Y.H.; Cypess, A.M. β3-Adrenergic receptors regulate human brown/beige adipocyte lipolysis and thermogenesis. JCI Insight 2021, 6. [Google Scholar] [CrossRef] [PubMed]
- Riis-Vestergaard, M.J.; Richelsen, B.; Bruun, J.M.; Li, W.; Hansen, J.B.; Pedersen, S.B. Beta-1 and Not Beta-3 Adrenergic Receptors May Be the Primary Regulator of Human Brown Adipocyte Metabolism. J. Clin. Endocrinol. Metab. 2020, 105, e994–e1005. [Google Scholar] [CrossRef] [PubMed]





Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, S.; Ma, H.; Wang, L.; Wang, F.; Xia, J.; Liu, D.; Mu, L.; Yang, X.; Liu, D. The Role of β3-Adrenergic Receptors in Cold-Induced Beige Adipocyte Production in Pigs. Cells 2024, 13, 709. https://doi.org/10.3390/cells13080709
Yang S, Ma H, Wang L, Wang F, Xia J, Liu D, Mu L, Yang X, Liu D. The Role of β3-Adrenergic Receptors in Cold-Induced Beige Adipocyte Production in Pigs. Cells. 2024; 13(8):709. https://doi.org/10.3390/cells13080709
Chicago/Turabian StyleYang, Shuo, Hong Ma, Liang Wang, Fang Wang, Jiqiao Xia, Dongyu Liu, Linlin Mu, Xiuqin Yang, and Di Liu. 2024. "The Role of β3-Adrenergic Receptors in Cold-Induced Beige Adipocyte Production in Pigs" Cells 13, no. 8: 709. https://doi.org/10.3390/cells13080709