Expression of PDLIM5 Spliceosomes and Regulatory Functions on Myogenesis in Pigs
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Materials
2.2. RNA Extraction and Expression Analysis
2.3. SNP Screening, Genotyping, and Correlation Analysis
2.4. Dual-Luciferase Reporter Assay
2.5. Primary Myoblasts Isolation and Culture
2.6. Plasmid Construction and Transfection
2.7. Cell Counting Kit-8 (CCK8) and EdU Assays
2.8. Immunofluorescence
2.9. Statistical Analysis
3. Results
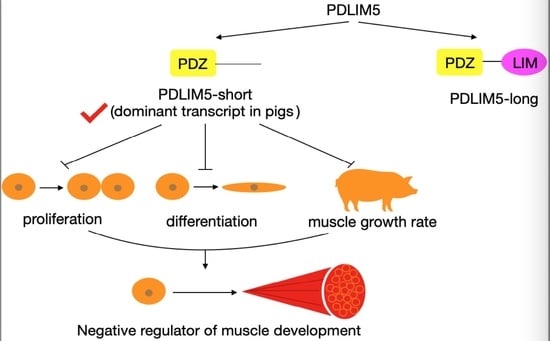
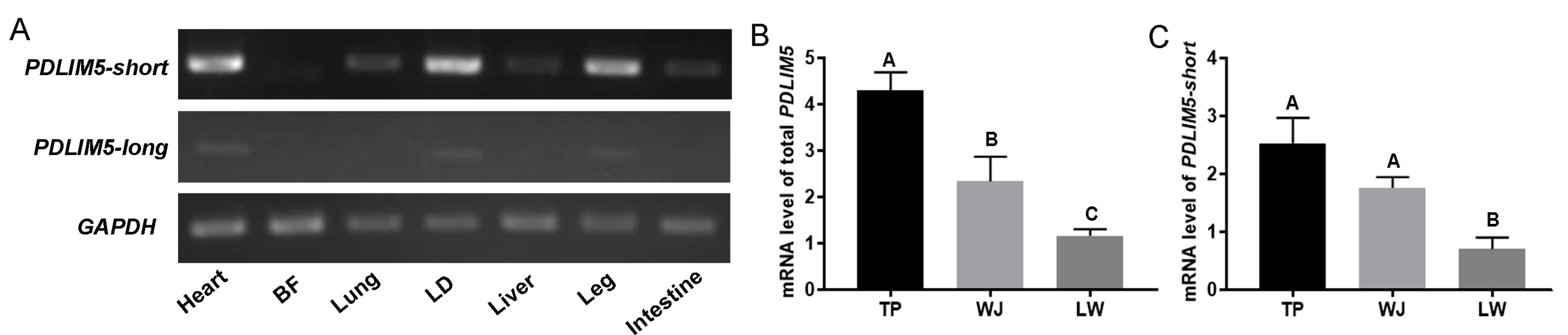
3.1. Expression and Identification of Main PDLIM5 Isoforms in Pigs
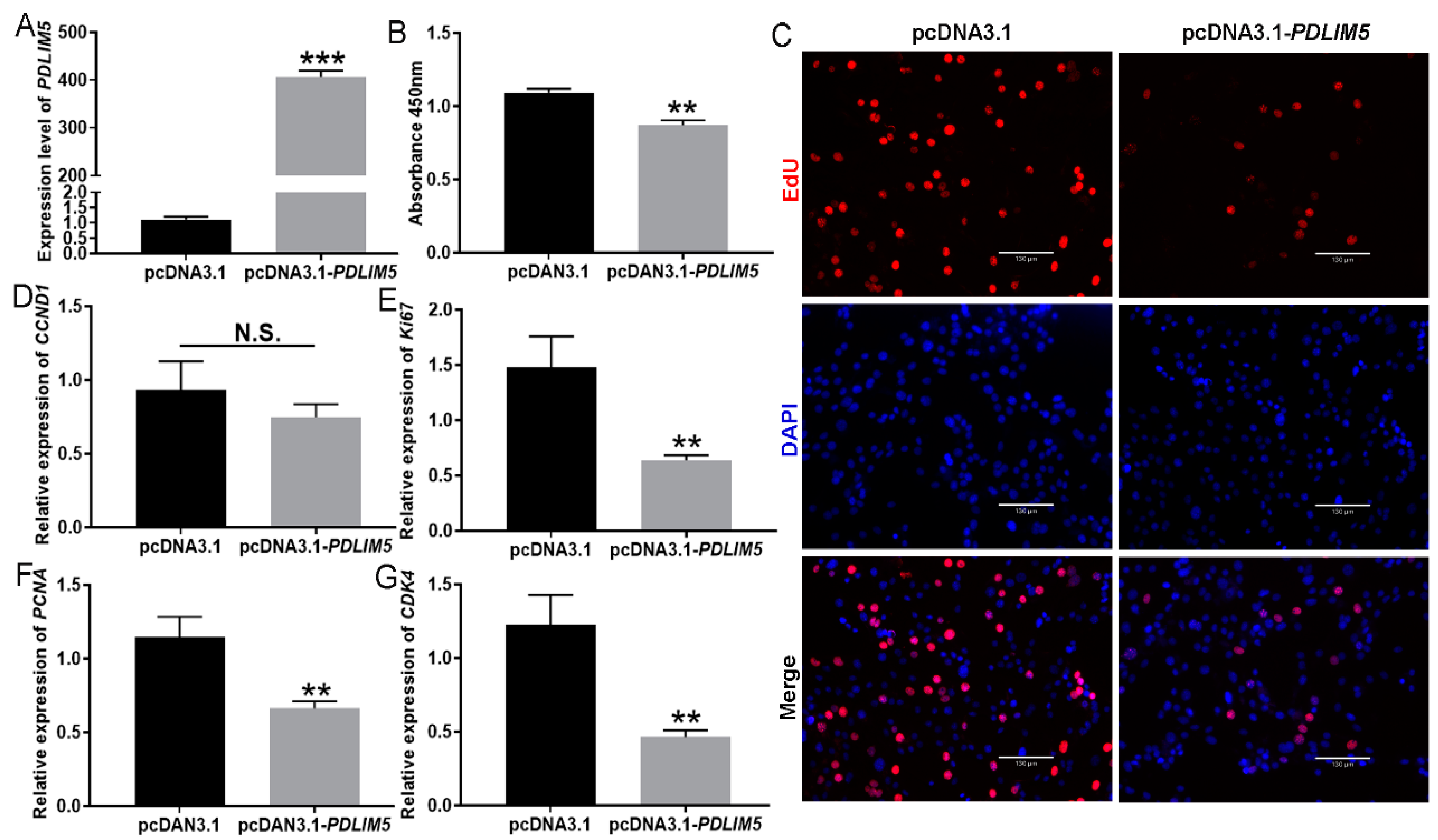
3.2. Inhibition of Inhibited Myoblast Proliferation by PDLIM5
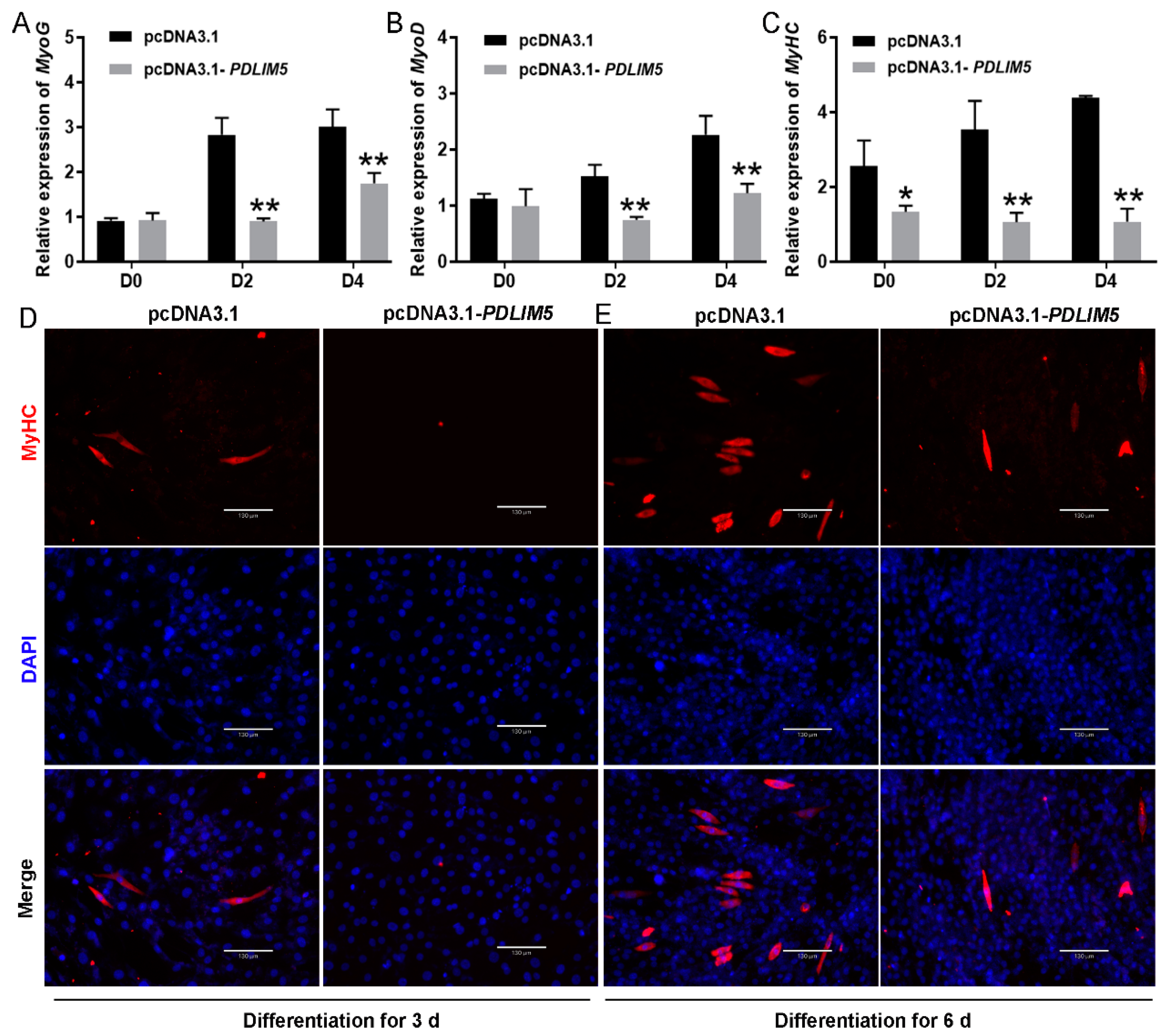
3.3. PDLIM5 Suppressed Myogenic Differentiation
3.4. Screening of SNP Sites in the PDLIM5 Gene
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Buckingham, M.; Vincent, S.D. Distinct and dynamic myogenic populations in the vertebrate embryo. Curr. Opin. Genet. Dev. 2009, 19, 444–453. [Google Scholar] [CrossRef] [PubMed]
- Wigmore, P.M.; Stickland, N.C. Muscle development in large and small pig fetuses. J. Anat. 1983, 137, 235–245. [Google Scholar] [PubMed]
- Reis, E.P.; Paixão, D.M.; Brustolini, O.J.; Silva, F.F.; Silva, W.; Araújo, F.M.; Salim, A.C.; Oliveira, G.; Guimarães, S.E. Expression of myogenes in longissimus dorsi muscle during prenatal development in commercial and local Piau pigs. Genet. Mol. Biol. 2016, 39, 589–599. [Google Scholar] [CrossRef] [PubMed]
- Knight, J.D.; Kothary, R. The myogenic kinome: Protein kinases critical to mammalian skeletal myogenesis. Skelet. Muscle. 2011, 1, 29–47. [Google Scholar] [CrossRef] [PubMed]
- Wei, D.; Zhang, J.; Raza, S.H.A.; Song, Y.; Jiang, C.; Song, X.; Wu, H.; Alotaibi, M.A.; Albiheyri, R.; Al-Zahrani, M.; et al. Interaction of MyoD and MyoG with Myoz2 gene in bovine myoblast differentiation. Res. Vet. Sci. 2022, 152, 569–578. [Google Scholar] [CrossRef] [PubMed]
- Skapek, S.X.; Rhee, J.; Kim, P.S.; Novitch, B.G.; Lassar, A.B. Cyclin-mediated inhibition of muscle gene expression via a mechanism that is independent of pRB hyperphosphorylation. Mol. Cell. Biol. 1996, 16, 7043–7053. [Google Scholar] [CrossRef]
- Saab, R.; Bills, J.L.; Miceli, A.P.; Anderson, C.M.; Khoury, J.D.; Fry, D.W.; Navid, F.; Houghton, P.J.; Skapek, S.X. Pharmacologic inhibition of cyclin-dependent kinase 4/6 activity arrests proliferation in myoblasts and rhabdomyosarcoma-derived cells. Mol. Cancer Ther. 2006, 5, 1299–1308. [Google Scholar] [CrossRef]
- Skapek, S.X.; Rhee, J.; Spicer, D.B.; Lassar, A.B. Inhibition of myogenic differentiation in proliferating myoblasts by cyclin D1-dependent kinase. Science 1995, 267, 1022–1024. [Google Scholar] [CrossRef]
- Karimi Majd, S.; Gholami, M.; Bazgir, B. PAX7 and MyoD proteins expression in response to eccentric and concentric resistance exercise in active young men. Cell J. 2023, 25, 135–142. [Google Scholar] [CrossRef]
- Mohammadabadi, M.; Bordbar, F.; Jensen, J.; Du, M.; Guo, W. Key genes regulating skeletal muscle development and growth in farm animals. Animals 2021, 11, 835. [Google Scholar] [CrossRef]
- Kuroda, S.; Tokunaga, C.; Kiyohara, Y.; Higuchi, O.; Konishi, H.; Mizuno, K.; Gill, G.N.; Kikkawa, U. Protein-protein interaction of zinc finger LIM domains with protein kinase C. J. Biol. Chem. 1996, 271, 31029–31032. [Google Scholar] [CrossRef] [PubMed]
- Te Velthuis, A.J.; Bagowski, C.P. PDZ and LIM domain-encoding genes: Molecular interactions and their role in development. Sci. World J. 2007, 7, 1470–1492. [Google Scholar] [CrossRef] [PubMed]
- Niederländer, N.; Fayein, N.A.; Auffray, C.; Pomiès, P. Characterization of a new human isoform of the enigma homolog family specifically expressed in skeletal muscle. Biochem. Biophys. Res. Commun. 2004, 325, 1304–1311. [Google Scholar] [CrossRef] [PubMed]
- Nakagawa, N.; Hoshijima, M.; Oyasu, M.; Saito, N.; Tanizawa, K.; Kuroda, S. ENH, containing PDZ and LIM domains, heart/skeletal muscle-specific protein, associates with cytoskeletal proteins through the PDZ domain. Biochem. Biophys. Res. Commun. 2000, 272, 505–512. [Google Scholar] [CrossRef] [PubMed]
- Cheng, H.; Kimura, K.; Peter, A.K.; Cui, L.; Ouyang, K.; Shen, T.; Liu, Y.; Gu, Y.; Dalton, N.D.; Evans, S.M.; et al. Loss of enigma homolog protein results in dilated cardiomyopathy. Circ. Res. 2010, 107, 348–356. [Google Scholar] [CrossRef]
- Ueki, N.; Seki, N.; Yano, K.; Masuho, Y.; Saito, T.; Muramatsu, M. Isolation, tissue expression, and chromosomal assignment of a human LIM protein gene, showing homology to rat enigma homologue (ENH). J. Hum. Genet. 1999, 44, 256–260. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, T.; Wälchli, S.; Fujita, T.; Ryser, S.; Hoshijima, M.; Schlegel, W.; Kuroda, S.; Maturana, A.D. Splice variants of enigma homolog, differentially expressed during heart development, promote or prevent hypertrophy. Cardiovasc. Res. 2010, 86, 374–382. [Google Scholar] [CrossRef] [PubMed]
- Ito, J.; Hashimoto, T.; Nakamura, S.; Aita, Y.; Yamazaki, T.; Schlegel, W.; Takimoto, K.; Maturana, A.D. Splicing transitions of the anchoring protein ENH during striated muscle development. Biochem. Biophys. Res. Commun. 2012, 421, 232–238. [Google Scholar] [CrossRef] [PubMed]
- Verdonschot, J.A.J.; Robinson, E.L.; James, K.N.; Mohamed, M.W.; Claes, G.R.F.; Casas, K.; Vanhoutte, E.K.; Hazebroek, M.R.; Kringlen, G.; Pasierb, M.M.; et al. Mutations in PDLIM5 are rare in dilated cardiomyopathy but are emerging as potential disease modifiers. Mol. Genet. Genomic. Med. 2020, 8, e1049–e1063. [Google Scholar] [CrossRef]
- Huang, X.; Qu, R.; Ouyang, J.; Zhong, S.; Dai, J. An Overview of the Cytoskeleton-Associated Role of PDLIM5. Front. Physiol. 2020, 11, 975–983. [Google Scholar] [CrossRef]
- Su, Y.; Hiemstra, T.F.; Yan, Y.; Li, J.; Karet, H.I.; Rosen, L.; Moreno, P.; Karet Frankl, F.E. PDLIM5 links kidney anion exchanger 1 (kAE1) to ILK and is required for membrane targeting of kAE1. Sci. Rep. 2017, 7, 39701–39712. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.L.; Gou, C.Y.; Wang, W.H.; Li, Y.; Cui, Y.; Duan, J.J.; Chen, Y. A novel effect of PDLIM5 in α7 nicotinic acetylcholine receptor upregulation and surface expression. Cell. Mol. Life Sci. 2022, 79, 64–75. [Google Scholar] [CrossRef] [PubMed]
- Srivastava, Y.; Donta, M.; Mireles, L.L.; Paulucci-Holthauzen, A.; Waxham, M.N.; McCrea, P.D. Role of a PDLIM5: PalmD complex in directing dendrite morphology. bioRxiv 2023, 1, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Wang, M.W.; Sun, Y.Y.; Hu, X.Y.; Geng, P.P.; Shu, H.; Wang, X.N.; Wang, H.; Zhang, J.F.; Cheng, H.Q.; et al. Nicotine pretreatment alleviates MK-801-induced behavioral and cognitive deficits in mice by regulating PDLIM5/CRTC1 in the PFC. Acta. Pharmacol. Sin. 2023, 44, 780–790. [Google Scholar] [CrossRef] [PubMed]
- Ito, J.; Iijima, M.; Yoshimoto, N.; Niimi, T.; Kuroda, S.; Maturana, A.D. Scaffold protein enigma homolog activates CREB whereas a short splice variant prevents CREB activation in cardiomyocytes. Cell. Signal. 2015, 27, 2425–2433. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Chen, L.; Huang, H.; Lv, J.M.; Chen, M.; Qu, F.J.; Pan, X.W.; Li, L.; Yin, L.; Cui, X.G.; et al. High expression of PDLIM5 facilitates cell tumorigenesis and migration by maintaining AMPK activation in prostate cancer. Oncotarget 2017, 8, 98117–98134. [Google Scholar] [CrossRef]
- Shi, Y.; Wang, X.; Xu, Z.; He, Y.; Guo, C.; He, L.; Huan, C.; Cai, C.; Huang, J.; Zhang, J.; et al. PDLIM5 inhibits STUB1-mediated degradation of SMAD3 and promotes the migration and invasion of lung cancer cells. J. Biol. Chem. 2020, 295, 13798–13811. [Google Scholar] [CrossRef] [PubMed]
- Wei, X.; Zhang, Y.; Yu, S.; Li, S.; Jiang, W.; Zhu, Y.; Xu, Y.; Yang, C.; Tian, G.; Mi, J.; et al. PDLIM5 identified by label-free quantitative proteomics as a potential novel biomarker of papillary thyroid carcinoma. Biochem. Biophys. Res. Commun. 2018, 499, 338–344. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Zhou, G.; Zhou, Q.; Tang, H.; Ibe, J.C.; Cheng, H.; Gou, D.; Chen, J.; Yuan, J.X.; Raj, J.U. Loss of microRNA-17-92 in smooth muscle cells attenuates experimental pulmonary hypertension via induction of PDZ and LIM domain 5. Am. J. Respir. Crit. Care. Med. 2015, 191, 678–692. [Google Scholar] [CrossRef]
- Liu, Z.; Liu, W.; Xiao, Z.; Wang, G.; Yin, S.; Zhu, F.; Wang, H.; Cheng, J.; Wang, X.; He, X.; et al. A major single nucleotide polymorphism of the PDLIM5 gene associated with recurrent major depressive disorder. J. Psychiatry. Neurosci. 2008, 33, 43–46. [Google Scholar]
- Owusu, D.; Pan, Y.; Xie, C.; Harirforoosh, S.; Wang, K.S. Polymorphisms in PDLIM5 gene are associated with alcohol dependence, type 2 diabetes, and hypertension. J. Psychiatr. Res. 2017, 84, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Nakatani, M.; Ito, J.; Koyama, R.; Iijima, M.; Yoshimoto, N.; Niimi, T.; Kuroda, S.; Maturana, A.D. Scaffold protein enigma homolog 1 overcomes the repression of myogenesis activation by inhibitor of DNA binding 2. Biochem. Biophys. Res. Commun. 2016, 474, 413–420. [Google Scholar] [CrossRef] [PubMed]
- Qiu, H.; Liu, N.; Luo, L.; Zhong, J.; Tang, Z.; Kang, K.; Qu, J.; Peng, W.; Liu, L.; Li, L.; et al. MicroRNA-17-92 regulates myoblast proliferation and differentiation by targeting the ENH1/Id1 signaling axis. Cell Death Differ. 2016, 23, 1658–1669. [Google Scholar] [CrossRef] [PubMed]
- He, H.; Yin, H.; Yu, X.; Zhang, Y.; Ma, M.; Li, D.; Zhu, Q. PDLIM5 affects chicken skeletal muscle satellite cell proliferation and differentiation via the p38-MAPK Pathway. Animals 2021, 11, 1016. [Google Scholar] [CrossRef] [PubMed]
- Shang, P.; Wang, Z.; Chamba, Y.; Zhang, B.; Zhang, H.; Wu, C. A comparison of prenatal muscle transcriptome and proteome profiles between pigs with divergent growth phenotypes. J. Cell Biochem. 2019, 120, 5277–5286. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Fu, Y.; Shang, P.; Zhang, B.; Tian, X.; Nie, R.; Zhang, R.; Zhang, H. Function of the porcine TRPC1 gene in myogenesis and muscle growth. Cells 2021, 10, 147. [Google Scholar] [CrossRef] [PubMed]
- Guy, P.M.; Kenny, D.A.; Gill, G.N. The PDZ domain of the LIM protein enigma binds to beta-tropomyosin. Mol. Biol. Cell 1999, 10, 1973–1984. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Ruiz-Lozano, P.; Martone, M.E.; Chen, J. Cypher, a striated muscle-restricted PDZ and LIM domain-containing protein, binds to alpha-actinin-2 and protein kinase C. J. Biol. Chem. 1999, 274, 19807–19813. [Google Scholar] [CrossRef]
- Lasorella, A.; Iavarone, A. The protein ENH is a cytoplasmic sequestration factor for Id2 in normal and tumor cells from the nervous system. Proc. Natl. Acad. Sci. USA 2006, 103, 4976–4981. [Google Scholar] [CrossRef]
- Gan, P.; Wang, Z.; Morales, M.G.; Zhang, Y.; Bassel-Duby, R.; Liu, N.; Olson, E.N. RBPMS is an RNA-binding protein that mediates cardiomyocyte binucleation and cardiovascular development. Dev. Cell 2022, 57, 959–973. [Google Scholar] [CrossRef] [PubMed]
- Ito, J.; Takita, M.; Takimoto, K.; Maturana, A.D. Enigma homolog 1 promotes myogenic gene expression and differentiation of C2C12 cells. Biochem. Biophys. Res. Commun. 2013, 435, 483–487. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Fang, J.; Lv, J.; Pan, Z.; Yin, X.; Cheng, H.; Guo, X. Novel polymorphisms in PDLIM3 and PDLIM5 gene encoding Z-line proteins increase risk of idiopathic dilated cardiomyopathy. J. Cell. Mol. Med. 2019, 23, 7054–7062. [Google Scholar] [CrossRef] [PubMed]

| Loci | Breed | Sample Size | Genotype Frequency | Allele Frequency | χ2 Value (p-Value) | |||
|---|---|---|---|---|---|---|---|---|
| −258 A > T | AA | AT | TT | A | T | |||
| DSE | 16 | 0.688 | 0.187 | 0.125 | 0.781 | 0.219 | 0.881 (0.664) | |
| TP | 34 | 0.471 | 0.353 | 0.176 | 0.647 | 0.353 | 1.105 (0.576) | |
| LW | 24 | 1 | 0 | 0 | 1 | 0 | / | |
| −191 T > G | TT | TG | GG | T | G | |||
| DSE | 16 | 0.750 | 0.188 | 0.062 | 0.844 | 0.156 | 1.155 (0.561) | |
| TP | 34 | 0.059 | 0.412 | 0.529 | 0.265 | 0.735 | 0.022 (0.989) | |
| LW | 24 | 0.916 | 0.042 | 0.042 | 0.938 | 0.063 | 2.023 (0.364) | |
| Loci | Genotype (Sample Size) | Days to 30 kg | Days to 90 kg |
|---|---|---|---|
| −258 A > T | AA (n = 78) | 92.15 ± 4.91 C | 183.53 ± 6.21 C |
| AT (n = 16) | 96.36 ± 4.84 B | 190.99 ± 5.93 B | |
| TT (n = 3) | 107.07 ± 7.32 A | 202.93 ± 6.32 A | |
| −191 T > G | TT (n = 74) | 91.85 ± 5.73 C | 183.57 ± 6.55 C |
| TG (n = 19) GG (n = 4) | 96.26 ± 5.13 B 105.11 ± 7.45 A | 191.16 ± 4.62 B 201.01 ± 6.41 A |
| Genotype (Sample Size) | Days to 30 kg | Days to 90 kg |
|---|---|---|
| TG/TG (n = 3) | 107.08 ± 7.73 A | 202.93 ± 6.27 A |
| AT/AG (n = 9) | 95.14 ± 5.49 B,C | 189.78 ± 6.56 A,B |
| AT/AT (n = 69) | 91.62 ± 5.89 C | 183.33 ± 5.53 B |
| AT/TT (n = 5) | 94.59 ± 4.61 B,C | 190.99 ± 7.13 A,B |
| AT/TG (n = 10) | 97.27 ± 4.25 A,B | 190.66 ± 4.82 A,B |
| AG/TG (n = 1) | 99.19 | 195.24 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fu, Y.; Li, S.; Nie, J.; Yan, D.; Zhang, B.; Hao, X.; Zhang, H. Expression of PDLIM5 Spliceosomes and Regulatory Functions on Myogenesis in Pigs. Cells 2024, 13, 720. https://doi.org/10.3390/cells13080720
Fu Y, Li S, Nie J, Yan D, Zhang B, Hao X, Zhang H. Expression of PDLIM5 Spliceosomes and Regulatory Functions on Myogenesis in Pigs. Cells. 2024; 13(8):720. https://doi.org/10.3390/cells13080720
Chicago/Turabian StyleFu, Yu, Shixin Li, Jingru Nie, Dawei Yan, Bo Zhang, Xin Hao, and Hao Zhang. 2024. "Expression of PDLIM5 Spliceosomes and Regulatory Functions on Myogenesis in Pigs" Cells 13, no. 8: 720. https://doi.org/10.3390/cells13080720