Drosophila Contributions towards Understanding Neurofibromatosis 1
Abstract
:1. Introduction
1.1. Neurofibromin and Neurofibromatosis 1
1.2. Variability of Symptoms
1.3. The Necessity of Animal Models
2. Drosophila Models of Neurofibromatosis 1
2.1. The Drosophila Nf1 Gene and Protein (dNf1) Isoforms
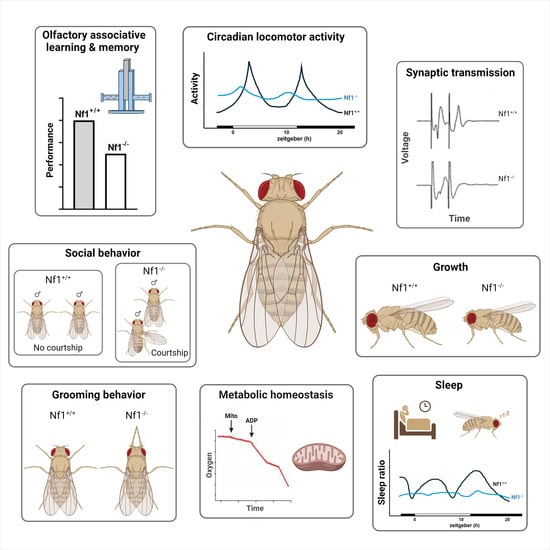
2.2. Modeling Νf1 Manifestations in Drosophila
2.2.1. Associative Learning and Memory Defects
2.2.2. Autism Spectrum Disorder Manifestations
2.2.3. Sleep Fragmentation and Activity Deficits
2.2.4. Stature and Metabolic Homeostasis Dysregulation
| Deficient Phenotype | Mutation | Implicated Pathway | Drug Treatment | Refs. |
|---|---|---|---|---|
| Olfactory associative learning and memory | null mutants; RNAi KD in OK72/GABA neurons | cAMP Alk/Ras1 | TAE684 | [51,52,72,73,101] |
| Circadian rhythms | null mutants; RNAi KD in MB neurons | Ras cAMP | [55,56,74] | |
| Locomotor activity | null mutants; pan-neuronal RNAi KD | MPH | [75,142] | |
| Sleep | null mutants; pan-neuronal RNAi KD; RNAi KD in GABAAR-expressing neurons (Rdl-Gal4); RNAi KD in MB neurons | Alk (Ras) cAMP | MPH | [56,76,140,141,142] |
| Grooming behavior | null mutants; RNAi KD pan-neuronal, in VNS and in ChAT- and oct-tyrR-expressing neurons | Ras | [54,75] | |
| Social behavior | null mutants; RNAi KD in Fru+ sensory neurons; RNAi KD in Ppk23+ sensory neurons | Ras | [77] | |
| Synaptic transmission | null mutants; pan-neuronal RNAi KD (pre-synaptic); RNAi KD in cholinergic neurons (ChaT-Gal4) | Ras BKCa channel cAMP | simvastatin BMS-04352 | [78,79,83,102] |
| Metabolic homeostasis | null mutants; pan-neuronal RNAi KD; RNAi KD in VNS, Oct-TyrR, and PCB neurons | cAMP Ras | [80,81,140] | |
| Growth | null mutants | Alk (Ras) cAMP | TAE684 | [6,52,70,71,102,105,153] |
3. Perspectives and Concluding Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Bergqvist, C.; Servy, A.; Valeyrie-Allanore, L.; Ferkal, S.; Combemale, P.; Wolkenstein, P.; Network, N.F.F. Neurofibromatosis 1 French national guidelines based on an extensive literature review since 1966. Orphanet J. Rare Dis. 2020, 15, 37. [Google Scholar] [CrossRef] [PubMed]
- Vandenbroucke, I.; Vandesompele, J.; De Paepe, A.; Messiaen, L. Quantification of NF1 transcripts reveals novel highly expressed splice variants. FEBS Lett. 2002, 522, 71–76. [Google Scholar] [CrossRef] [PubMed]
- Harrisingh, M.C.; Lloyd, A.C. Ras/Raf/ERK signalling and NF1. Cell Cycle 2004, 3, 1255–1258. [Google Scholar] [CrossRef] [PubMed]
- Johannessen, C.M.; Reczek, E.E.; James, M.F.; Brems, H.; Legius, E.; Cichowski, K. The NF1 tumor suppressor critically regulates TSC2 and mTOR. Proc. Natl. Acad. Sci. USA 2005, 102, 8573–8578. [Google Scholar] [CrossRef] [PubMed]
- Martin, G.A.; Viskochil, D.; Bollag, G.; McCabe, P.C.; Crosier, W.J.; Haubruck, H.; Conroy, L.; Clark, R.; O’Connell, P.; Cawthon, R.M.; et al. The GAP-related domain of the neurofibromatosis type 1 gene product interacts with ras p21. Cell 1990, 63, 843–849. [Google Scholar] [CrossRef]
- Tong, J.; Hannan, F.; Zhu, Y.; Bernards, A.; Zhong, Y. Neurofibromin regulates G protein-stimulated adenylyl cyclase activity. Nat. Neurosci. 2002, 5, 95–96. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.T.; Hinman, M.N.; Guo, X.; Sharma, A.; Arakawa, H.; Luo, G.; Lou, H. Neurofibromatosis type 1 alternative splicing is a key regulator of Ras/ERK signaling and learning behaviors in mice. Hum. Mol. Genet. 2017, 26, 3797–3807. [Google Scholar] [CrossRef]
- Gutmann, D.H.; Aylsworth, A.; Carey, J.C.; Korf, B.; Marks, J.; Pyeritz, R.E.; Rubenstein, A.; Viskochil, D. The diagnostic evaluation and multidisciplinary management of neurofibromatosis 1 and neurofibromatosis 2. JAMA 1997, 278, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Wallace, M.R.; Marchuk, D.A.; Andersen, L.B.; Letcher, R.; Odeh, H.M.; Saulino, A.M.; Fountain, J.W.; Brereton, A.; Nicholson, J.; Mitchell, A.L.e.a. Type 1 neurofibromatosis gene: Identification of a large transcript disrupted in three NF1 patients. Science 1990, 249, 181–186. [Google Scholar] [CrossRef]
- Yap, Y.S.; McPherson, J.R.; Ong, C.K.; Rozen, S.G.; Teh, B.T.; Lee, A.S.; Callen, D.F. The NF1 gene revisited—From bench to bedside. Oncotarget 2014, 5, 5873–5892. [Google Scholar] [CrossRef]
- Daston, M.M.; Scrable, H.; Nordlund, M.; Sturbaum, A.K.; Nissen, L.M.; Ratner, N. The protein product of the neurofibromatosis type 1 gene is expressed at highest abundance in neurons, Schwann cells, and oligodendrocytes. Neuron 1992, 8, 415–428. [Google Scholar] [CrossRef] [PubMed]
- Gutmann, D.H.; Wood, D.L.; Collins, F.S. Identification of the neurofibromatosis type 1 gene product. Proc. Natl. Acad. Sci. USA 1991, 88, 9658–9662. [Google Scholar] [CrossRef] [PubMed]
- Nishi, T.; Lee, P.S.; Oka, K.; Levin, V.A.; Tanase, S.; Morino, Y.; Saya, H. Differential expression of two types of the neurofibromatosis type 1 (NF1) gene transcripts related to neuronal differentiation. Oncogene 1991, 6, 1555–1559. [Google Scholar] [PubMed]
- Gutmann, D.H.; Ferner, R.E.; Listernick, R.H.; Korf, B.R.; Wolters, P.L.; Johnson, K.J. Neurofibromatosis type 1. Nat. Rev. Dis. Primers 2017, 3, 17004. [Google Scholar] [CrossRef]
- Koga, M.; Yoshida, Y.; Imafuku, S. Nutritional, muscular and metabolic characteristics in patients with neurofibromatosis type 1. J. Dermatol. 2016, 43, 799–803. [Google Scholar] [CrossRef] [PubMed]
- Peduto, C.; Zanobio, M.; Nigro, V.; Perrotta, S.; Piluso, G.; Santoro, C. Neurofibromatosis Type 1: Pediatric Aspects and Review of Genotype-Phenotype Correlations. Cancers 2023, 15, 1217. [Google Scholar] [CrossRef] [PubMed]
- Souza, M.; Jansen, A.; Martins, A.; Rodrigues, L.; Rezende, N. Body composition in adults with neurofibromatosis type 1. Rev. Assoc. Med. Bras. (1992) 2016, 62, 831–836. [Google Scholar] [CrossRef] [PubMed]
- Johnson, H.; Wiggs, L.; Stores, G.; Huson, S.M. Psychological disturbance and sleep disorders in children with neurofibromatosis type 1. Dev. Med. Child Neurol. 2005, 47, 237–242. [Google Scholar] [CrossRef]
- Leschziner, G.D.; Golding, J.F.; Ferner, R.E. Sleep disturbance as part of the neurofibromatosis type 1 phenotype in adults. Am. J. Med. Genet. A 2013, 161, 1319–1322. [Google Scholar] [CrossRef] [PubMed]
- Licis, A.K.; Vallorani, A.; Gao, F.; Chen, C.; Lenox, J.; Yamada, K.A.; Duntley, S.P.; Gutmann, D.H. Prevalence of Sleep Disturbances in Children with Neurofibromatosis Type 1. J. Child Neurol. 2013, 28, 1400–1405. [Google Scholar] [CrossRef]
- Morris, S.M.; Acosta, M.T.; Garg, S.; Green, J.; Huson, S.; Legius, E.; North, K.N.; Payne, J.M.; Plasschaert, E.; Frazier, T.W.; et al. Disease Burden and Symptom Structure of Autism in Neurofibromatosis Type 1: A Study of the International NF1-ASD Consortium Team (INFACT). JAMA Psychiatry 2016, 73, 1276–1284. [Google Scholar] [CrossRef] [PubMed]
- Shilyansky, C.; Lee, Y.S.; Silva, A.J. Molecular and Cellular Mechanisms of Learning Disabilities: A Focus on NF1. Annu. Rev. Neurosci. 2010, 33, 221–243. [Google Scholar] [CrossRef] [PubMed]
- Torres Nupan, M.M.; Velez Van Meerbeke, A.; Lopez Cabra, C.A.; Herrera Gomez, P.M. Cognitive and Behavioral Disorders in Children with Neurofibromatosis Type 1. Front. Pediatr. 2017, 5, 227. [Google Scholar] [CrossRef] [PubMed]
- Hyman, S.L.; Arthur Shores, E.; North, K.N. Learning disabilities in children with neurofibromatosis type 1: Subtypes, cognitive profile, and attention-deficit-hyperactivity disorder. Dev. Med. Child Neurol. 2006, 48, 973–977. [Google Scholar] [CrossRef]
- Hyman, S.; Gill, D.; Shores, E.; Steinberg, A.; Joy, P.; Gibikote, S.; North, K. Natural history of cognitive deficits and their relationship to MRI T2-hyperintensities in NF1. Neurology 2003, 60, 1139–1145. [Google Scholar] [CrossRef] [PubMed]
- North, K.; Hyman, S.; Barton, B. Cognitive deficits in neurofibromatosis 1. J. Child Neurol. 2002, 17, 605–612. [Google Scholar] [CrossRef] [PubMed]
- Gross, A.M.; Wolters, P.L.; Dombi, E.; Baldwin, A.; Whitcomb, P.; Fisher, M.J.; Weiss, B.; Kim, A.; Bornhorst, M.; Shah, A.C.; et al. Selumetinib in Children with Inoperable Plexiform Neurofibromas. N. Engl. J. Med. 2020, 382, 1430–1442. [Google Scholar] [CrossRef] [PubMed]
- Lidzba, K.; Granstroem, S.; Leark, R.A.; Kraegeloh-Mann, I.; Mautner, V.F. Pharmacotherapy of attention deficit in neurofibromatosis type 1: Effects on cognition. Neuropediatrics 2014, 45, 240–246. [Google Scholar] [CrossRef] [PubMed]
- Lion-Francois, L.; Gueyffier, F.; Mercier, C.; Gerard, D.; Herbillon, V.; Kemlin, I.; Rodriguez, D.; Ginhoux, T.; Peyric, E.; Coutinho, V.; et al. The effect of methylphenidate on neurofibromatosis type 1: A randomised, double-blind, placebo-controlled, crossover trial. Orphanet J. Rare Dis. 2014, 9, 142. [Google Scholar] [CrossRef]
- Mautner, V.F.; Kluwe, L.; Thakker, S.D.; Leark, R.A. Treatment of ADHD in neurofibromatosis type 1. Dev. Med. Child Neurol. 2002, 44, 164–170. [Google Scholar] [CrossRef]
- Bergoug, M.; Doudeau, M.; Godin, F.; Mosrin, C.; Vallée, B.; Bénédetti, H. Neurofibromin Structure, Functions and Regulation. Cells 2020, 9, 2365. [Google Scholar] [CrossRef] [PubMed]
- Kehrer-Sawatzki, H.; Mautner, V.F.; Cooper, D.N. Emerging genotype-phenotype relationships in patients with large NF1 deletions. Hum. Genet. 2017, 136, 349–376. [Google Scholar] [CrossRef] [PubMed]
- Koczkowska, M.; Callens, T.; Chen, Y.; Gomes, A.; Hicks, A.D.; Sharp, A.; Johns, E.; Uhas, K.A.; Armstrong, L.; Bosanko, K.A.; et al. Clinical spectrum of individuals with pathogenic NF1 missense variants affecting p.Met1149, p.Arg1276, and p.Lys1423: Genotype-phenotype study in neurofibromatosis type 1. Hum. Mutat. 2020, 41, 299–315. [Google Scholar] [CrossRef] [PubMed]
- Koczkowska, M.; Callens, T.; Gomes, A.; Sharp, A.; Chen, Y.; Hicks, A.D.; Aylsworth, A.S.; Azizi, A.A.; Basel, D.G.; Bellus, G.; et al. Expanding the clinical phenotype of individuals with a 3-bp in-frame deletion of the NF1 gene (c.2970_2972del): An update of genotype-phenotype correlation. Genet. Med. 2019, 21, 867–876. [Google Scholar] [CrossRef] [PubMed]
- Koczkowska, M.; Chen, Y.; Callens, T.; Gomes, A.; Sharp, A.; Johnson, S.; Hsiao, M.; Chen, Z.; Balasubramanian, M.; Barnett, C.; et al. Genotype-Phenotype Correlation in NF1: Evidence for a More Severe Phenotype Associated with Missense Mutations Affecting NF1 Codons 844-848. Am. J. Hum. Genet. 2018, 102, 69–87. [Google Scholar] [CrossRef] [PubMed]
- Anastasaki, C.; Orozco, P.; Gutmann, D. RAS and beyond: The many faces of the neurofibromatosis type 1 protein. Dis. Models Mech. 2022, 15, dmm049362. [Google Scholar] [CrossRef] [PubMed]
- Easton, D.F.; Ponder, M.A.; Huson, S.M.; Ponder, B.A. An analysis of variation in expression of neurofibromatosis (NF) type 1 (NF1): Evidence for modifying genes. Am. J. Hum. Genet. 1993, 53, 305–313. [Google Scholar] [PubMed]
- Rieley, M.B.; Stevenson, D.A.; Viskochil, D.H.; Tinkle, B.T.; Martin, L.J.; Schorry, E.K. Variable expression of neurofibromatosis 1 in monozygotic twins. Am. J. Med. Genet. A 2011, 155, 478–485. [Google Scholar] [CrossRef] [PubMed]
- Walker, J.A.; Upadhyaya, M. Emerging therapeutic targets for neurofibromatosis type 1. Expert Opin. Ther. Targets 2018, 22, 419–437. [Google Scholar] [CrossRef] [PubMed]
- Jiang, C.; McKay, R.M.; Le, L.Q. Tumorigenesis in neurofibromatosis type 1: Role of the microenvironment. Oncogene 2021, 40, 5781–5787. [Google Scholar] [CrossRef]
- Osum, S.H.; Watson, A.L.; Largaespada, D.A. Spontaneous and Engineered Large Animal Models of Neurofibromatosis Type 1. Int. J. Mol. Sci. 2021, 22, 1954. [Google Scholar] [CrossRef] [PubMed]
- Chaker-Margot, M.; Werten, S.; Dunzendorfer-Matt, T.; Lechner, S.; Ruepp, A.; Scheffzek, K.; Maier, T. Structural basis of activation of the tumor suppressor protein neurofibromin. Mol. Cell 2022, 82, 1288–1296.e1285. [Google Scholar] [CrossRef] [PubMed]
- Sherekar, M.; Han, S.W.; Ghirlando, R.; Messing, S.; Drew, M.; Rabara, D.; Waybright, T.; Juneja, P.; O’Neill, H.; Stanley, C.B.; et al. Biochemical and structural analyses reveal that the tumor suppressor neurofibromin (NF1) forms a high-affinity dimer. J. Biol. Chem. 2020, 295, 1105–1119. [Google Scholar] [CrossRef] [PubMed]
- Young, L.C.; Goldstein de Salazar, R.; Han, S.W.; Huang, Z.Y.S.; Merk, A.; Drew, M.; Darling, J.; Wall, V.; Grisshammer, R.; Cheng, A.; et al. Destabilizing NF1 variants act in a dominant negative manner through neurofibromin dimerization. Proc. Natl. Acad. Sci. USA 2023, 120, e2208960120. [Google Scholar] [CrossRef] [PubMed]
- Anastasaki, C.; Wegscheid, M.L.; Hartigan, K.; Papke, J.B.; Kopp, N.D.; Chen, J.; Cobb, O.; Dougherty, J.D.; Gutmann, D.H. Human iPSC-Derived Neurons and Cerebral Organoids Establish Differential Effects of Germline NF1 Gene Mutations. Stem Cell Rep. 2020, 14, 541–550. [Google Scholar] [CrossRef] [PubMed]
- Gutmann, D.H.; Giovannini, M. Mouse models of neurofibromatosis 1 and 2. Neoplasia 2002, 4, 279–290. [Google Scholar] [CrossRef]
- Isakson, S.H.; Rizzardi, A.E.; Coutts, A.W.; Carlson, D.F.; Kirstein, M.N.; Fisher, J.; Vitte, J.; Williams, K.B.; Pluhar, G.E.; Dahiya, S.; et al. Genetically engineered minipigs model the major clinical features of human neurofibromatosis type 1. Commun. Biol. 2018, 1, 158. [Google Scholar] [CrossRef]
- Miller, A.H.; Halloran, M.C. Mechanistic insights from animal models of neurofibromatosis type 1 cognitive impairment. Dis. Models Mech. 2022, 15, dmm049422. [Google Scholar] [CrossRef]
- Costa, R.M.; Silva, A.J. Molecular and cellular mechanisms underlying the cognitive deficits associated with neurofibromatosis 1. J. Child Neurol. 2002, 17, 622–626; discussion 627–629, 646–651. [Google Scholar] [CrossRef]
- Cui, Y.; Costa, R.M.; Murphy, G.G.; Elgersma, Y.; Zhu, Y.; Gutmann, D.H.; Parada, L.F.; Mody, I.; Silva, A.J. Neurofibromin regulation of ERK signaling modulates GABA release and learning. Cell 2008, 135, 549–560. [Google Scholar] [CrossRef]
- Georganta, E.; Moressis, A.; Skoulakis, E. Associative Learning Requires Neurofibromin to Modulate GABAergic Inputs to Drosophila Mushroom Bodies. J. Neurosci. 2021, 41, 5274–5286. [Google Scholar] [CrossRef]
- Gouzi, J.Y.; Moressis, A.; Walker, J.A.; Apostolopoulou, A.A.; Palmer, R.H.; Bernards, A.; Skoulakis, E.M.C. Drosophila Alk Tyrosine Kinase Controls Neurofibromin dependent Growth and Learning. PLoS Genet. 2011, 7, e1002281. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Cui, Y.; Kushner, S.A.; Brown, R.A.; Jentsch, J.D.; Frankland, P.W.; Cannon, T.D.; Silva, A.J. The HMG-CoA reductase inhibitor lovastatin reverses the learning and attention deficits in a mouse model of neurofibromatosis type 1. Curr. Biol. 2005, 15, 1961–1967. [Google Scholar] [CrossRef]
- King, L.B.; Boto, T.; Botero, V.; Aviles, A.M.; Jomsky, B.M.; Joseph, C.; Walker, J.A.; Tomchik, S.M. Developmental loss of neurofibromin across distributed neuronal circuits drives excessive grooming in Drosophila. PLoS Genet. 2020, 16, e1008920. [Google Scholar] [CrossRef]
- Williams, J.A.; Su, H.S.; Bernards, A.; Field, J.; Sehgal, A. A circadian output in Drosophila mediated by neurofibromatosis-1 and Ras/MAPK. Science 2001, 293, 2251–2256. [Google Scholar] [CrossRef]
- Bai, L.; Sehgal, A. Anaplastic Lymphoma Kinase Acts in the Drosophila Mushroom Body to Negatively Regulate Sleep. PLoS Genet. 2015, 11, e1005611. [Google Scholar] [CrossRef]
- Papanikolopoulou, K.; Mudher, A.; Skoulakis, E. An assessment of the translational relevance of Drosophila in drug discovery. Expert Opin. Drug Discov. 2019, 14, 303–313. [Google Scholar] [CrossRef]
- Reiter, L.T.; Potocki, L.; Chien, S.; Gribskov, M.; Bier, E. A systematic analysis of human disease-associated gene sequences in Drosophila melanogaster. Genome Res. 2001, 11, 1114–1125. [Google Scholar] [CrossRef] [PubMed]
- Kanca, O.; Zirin, J.; Garcia-Marques, J.; Knight, S.M.; Yang-Zhou, D.; Amador, G.; Chung, H.; Zuo, Z.; Ma, L.; He, Y.; et al. An efficient CRISPR-based strategy to insert small and large fragments of DNA using short homology arms. Elife 2019, 8, e51539. [Google Scholar] [CrossRef]
- Kanca, O.; Zirin, J.; Hu, Y.; Tepe, B.; Dutta, D.; Lin, W.W.; Ma, L.; Ge, M.; Zuo, Z.; Liu, L.P.; et al. An expanded toolkit for Drosophila gene tagging using synthesized homology donor constructs for CRISPR-mediated homologous recombination. Elife 2022, 11, e76077. [Google Scholar] [CrossRef]
- Zirin, J.; Bosch, J.; Viswanatha, R.; Mohr, S.E.; Perrimon, N. State-of-the-art CRISPR for in vivo and cell-based studies in Drosophila. Trends Genet. 2022, 38, 437–453. [Google Scholar] [CrossRef] [PubMed]
- Pitman, J.L.; DasGupta, S.; Krashes, M.J.; Leung, B.; Perrat, P.N.; Waddell, S. There are many ways to train a fly. Fly 2009, 3, 3–9. [Google Scholar] [CrossRef] [PubMed]
- Fenckova, M.; Blok, L.E.R.; Asztalos, L.; Goodman, D.P.; Cizek, P.; Singgih, E.L.; Glennon, J.C.; IntHout, J.; Zweier, C.; Eichler, E.E.; et al. Habituation Learning Is a Widely Affected Mechanism in Drosophila Models of Intellectual Disability and Autism Spectrum Disorders. Biol. Psychiatry 2019, 86, 294–305. [Google Scholar] [CrossRef]
- Foka, K.; Georganta, E.M.; Semelidou, O.; Skoulakis, E.M.C. Loss of the Schizophrenia-linked Furin protein from Drosophila mushroom body neurons results in antipsychotic-reversible habituation deficits. J. Neurosci. 2022, 42, 7496–7511. [Google Scholar] [CrossRef]
- Roussou, I.; Papanikolopoulou, K.; Savakis, C.; Skoulakis, E. Drosophila Bruton’s Tyrosine Kinase Regulates Habituation Latency and Facilitation in Distinct Mushroom Body Neurons. J. Neurosci. 2019, 39, 8730–8743. [Google Scholar] [CrossRef]
- Semelidou, O.; Acevedo, S.F.; Skoulakis, E.M. Temporally specific engagement of distinct neuronal circuits regulating olfactory habituation in Drosophila. Elife 2018, 7, e39569. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Sokolowski, M.B. How Social Experience and Environment Impacts Behavioural Plasticity in Drosophila. Fly 2022, 16, 68–84. [Google Scholar] [CrossRef]
- Coll-Tané, M.; Krebbers, A.; Castells-Nobau, A.; Zweier, C.; Schenck, A. Intellectual disability and autism spectrum disorders ‘on the fly’: Insights from Drosophila. Dis. Models Mech. 2019, 12, dmm039180. [Google Scholar] [CrossRef] [PubMed]
- Mariano, V.; Achsel, T.; Bagni, C.; Kanellopoulos, A.K. Modelling Learning and Memory in Drosophila to Understand Intellectual Disabilities. Neuroscience 2020, 445, 12–30. [Google Scholar] [CrossRef]
- The, I.; Hannigan, G.E.; Cowley, G.S.; Reginald, S.; Zhong, Y.; Gusella, J.F.; Hariharan, I.K.; Bernards, A. Rescue of a Drosophila NF1 mutant phenotype by protein kinase A. Science 1997, 276, 791–794. [Google Scholar] [CrossRef]
- Walker, J.A.; Tchoudakova, A.V.; McKenney, P.T.; Brill, S.; Wu, D.; Cowley, G.S.; Hariharan, I.K.; Bernards, A. Reduced growth of Drosophila neurofibromatosis 1 mutants reflects a non-cell-autonomous requirement for GTPase-Activating Protein activity in larval neurons. Genes Dev. 2006, 20, 3311–3323. [Google Scholar] [CrossRef] [PubMed]
- Buchanan, M.E.; Davis, R.L. A distinct set of Drosophila brain neurons required for neurofibromatosis type 1-dependent learning and memory. J. Neurosci. 2010, 30, 10135–10143. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.F.; Tong, J.; Hannan, F.; Luo, L.; Zhong, Y. A neurofibromatosis-1-regulated pathway is required for learning in Drosophila. Nature 2000, 403, 895–898. [Google Scholar] [CrossRef] [PubMed]
- Bai, L.; Lee, Y.; Hsu, C.; Williams, J.; Cavanaugh, D.; Zheng, X.; Stein, C.; Haynes, P.; Wang, H.; Gutmann, D.; et al. A Conserved Circadian Function for the Neurofibromatosis 1 Gene. Cell Rep. 2018, 22, 3416–3426. [Google Scholar] [CrossRef] [PubMed]
- King, L.B.; Koch, M.; Murphy, K.R.; Velazquez, Y.; Ja, W.W.; Tomchik, S.M. Neurofibromin Loss of Function Drives Excessive Grooming in Drosophila. G3 2016, 6, 1083–1093. [Google Scholar] [CrossRef] [PubMed]
- Durkin, J.; Poe, A.R.; Belfer, S.J.; Rodriguez, A.; Tang, S.H.; Walker, J.A.; Kayser, M.S. Neurofibromin 1 regulates early developmental sleep in Drosophila. Neurobiol. Sleep Circadian Rhythm. 2023, 15, 100101. [Google Scholar] [CrossRef]
- Moscato, E.H.; Dubowy, C.; Walker, J.A.; Kayser, M.S. Social Behavioral Deficits with Loss of Neurofibromin Emerge from Peripheral Chemosensory Neuron Dysfunction. Cell Rep. 2020, 32, 107856. [Google Scholar] [CrossRef]
- Dyson, A.; Ryan, M.; Garg, S.; Evans, D.G.; Baines, R.A. Loss of NF1 in Drosophila Larvae Causes Tactile Hypersensitivity and Impaired Synaptic Transmission at the Neuromuscular Junction. J. Neurosci. 2022, 42, 9450–9472. [Google Scholar] [CrossRef]
- Dyson, A.; Ryan, M.; Garg, S.; Evans, D.G.; Baines, R.A. A Targeted, Low-Throughput Compound Screen in a Drosophila Model of Neurofibromatosis Type 1 Identifies Simvastatin and BMS-204352 as Potential Therapies for Autism Spectrum Disorder (ASD). eNeuro 2023, 10, 1–15. [Google Scholar] [CrossRef]
- Botero, V.; Stanhope, B.A.; Brown, E.B.; Grenci, E.C.; Boto, T.; Park, S.J.; King, L.B.; Murphy, K.R.; Colodner, K.J.; Walker, J.A.; et al. Neurofibromin regulates metabolic rate via neuronal mechanisms in Drosophila. Nat. Commun. 2021, 12, 4285. [Google Scholar] [CrossRef]
- Tong, J.J.; Schriner, S.E.; McCleary, D.; Day, B.J.; Wallace, D.C. Life extension through neurofibromin mitochondrial regulation and antioxidant therapy for neurofibromatosis-1 in Drosophila melanogaster. Nat. Genet. 2007, 39, 476–485. [Google Scholar] [CrossRef] [PubMed]
- Shin, J.; Padmanabhan, A.; de Groh, E.D.; Lee, J.S.; Haidar, S.; Dahlberg, S.; Guo, F.; He, S.; Wolman, M.A.; Granato, M.; et al. Zebrafish neurofibromatosis type 1 genes have redundant functions in tumorigenesis and embryonic development. Dis. Models Mech. 2012, 5, 881–894. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.F.; The, I.; Hannan, F.; Bernards, A.; Zhong, Y. Requirement of Drosophila NF1 for activation of adenylyl cyclase by PACAP38-like neuropeptides. Science 1997, 276, 795–798. [Google Scholar] [CrossRef] [PubMed]
- Maixner, A.; Hecker, T.P.; Phan, Q.N.; Wassarman, D.A. A screen for mutations that prevent lethality caused by expression of activated sevenless and Ras1 in the Drosophila embryo. Dev. Genet. 1998, 23, 347–361. [Google Scholar] [CrossRef]
- Loucas, C.; Wolters, P.; Toledo-Tamula, M.A.; Rhodes, A.; Baldwin, A.; Goodwin, A.; Widemann, B.; Martin, S. Verbal learning and memory in youth with neurofibromatosis type 1 and plexiform neurofibromas: Relationships with disease severity. Eur. J. Paediatr. Neurol. 2022, 38, 7–12. [Google Scholar] [CrossRef] [PubMed]
- Johansson, E.; Kallionpää, R.A.; Böckerman, P.; Peltonen, J.; Peltonen, S. A rare disease and education: Neurofibromatosis type 1 decreases educational attainment. Clin. Genet. 2021, 99, 529–539. [Google Scholar] [CrossRef] [PubMed]
- Tully, T.; Quinn, W.G. Classical conditioning and retention in normal and mutant Drosophila melanogaster. J. Comp. Physiol. A 1985, 157, 263–277. [Google Scholar] [CrossRef] [PubMed]
- Davis, R.L. Learning and memory using Drosophila melanogaster: A focus on advances made in the fifth decade of research. Genetics 2023, 224, iyad085. [Google Scholar] [CrossRef] [PubMed]
- Crittenden, J.R.; Skoulakis, E.M.; Han, K.A.; Kalderon, D.; Davis, R.L. Tripartite mushroom body architecture revealed by antigenic markers. Learn. Mem. 1998, 5, 38–51. [Google Scholar] [CrossRef]
- Diggs-Andrews, K.A.; Gutmann, D.H. Modeling cognitive dysfunction in neurofibromatosis-1. Trends Neurosci. 2013, 36, 237–247. [Google Scholar] [CrossRef]
- Molosh, A.I.; Johnson, P.L.; Spence, J.P.; Arendt, D.; Federici, L.M.; Bernabe, C.; Janasik, S.P.; Segu, Z.M.; Khanna, R.; Goswami, C.; et al. Social learning and amygdala disruptions in Nf1 mice are rescued by blocking p21-activated kinase. Nat. Neurosci. 2014, 17, 1583–1590. [Google Scholar] [CrossRef] [PubMed]
- Omrani, A.; van der Vaart, T.; Mientjes, E.; van Woerden, G.M.; Hojjati, M.R.; Li, K.W.; Gutmann, D.H.; Levelt, C.N.; Smit, A.B.; Silva, A.J.; et al. HCN channels are a novel therapeutic target for cognitive dysfunction in Neurofibromatosis type 1. Mol. Psychiatry 2015, 20, 1311–1321. [Google Scholar] [CrossRef] [PubMed]
- Shilyansky, C.; Karlsgodt, K.H.; Cummings, D.M.; Sidiropoulou, K.; Hardt, M.; James, A.S.; Ehninger, D.; Bearden, C.E.; Poirazi, P.; Jentsch, J.D.; et al. Neurofibromin regulates corticostriatal inhibitory networks during working memory performance. Proc. Natl. Acad. Sci. USA 2010, 107, 13141–13146. [Google Scholar] [CrossRef] [PubMed]
- Ryu, H.-H.; Kang, M.; Park, J.; Park, S.-H.; Lee, Y.-S. Enriched expression of NF1 in inhibitory neurons in both mouse and human brain. Mol. Brain 2019, 12, 60. [Google Scholar] [CrossRef] [PubMed]
- Maffei, A.; Charrier, C.; Caiati, M.D.; Barberis, A.; Mahadevan, V.; Woodin, M.A.; Tyagarajan, S.K. Emerging Mechanisms Underlying Dynamics of GABAergic Synapses. J. Neurosci. 2017, 37, 10792–10799. [Google Scholar] [CrossRef] [PubMed]
- Ramamoorthi, K.; Lin, Y. The contribution of GABAergic dysfunction to neurodevelopmental disorders. Trends Mol. Med. 2011, 17, 452–462. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Jaenisch, R.; Sur, M. The role of GABAergic signalling in neurodevelopmental disorders. Nat. Rev. Neurosci. 2021, 22, 290–307. [Google Scholar] [CrossRef]
- Costa, R.M.; Federov, N.B.; Kogan, J.H.; Murphy, G.G.; Stern, J.; Ohno, M.; Kucherlapati, R.; Jacks, T.; Silva, A.J. Mechanism for the learning deficits in a mouse model of neurofibromatosis type 1. Nature 2002, 415, 526–530. [Google Scholar] [CrossRef] [PubMed]
- Palmer, R.H.; Vernersson, E.; Grabbe, C.; Hallberg, B. Anaplastic lymphoma kinase: Signalling in development and disease. Biochem. J. 2009, 420, 345–361. [Google Scholar] [CrossRef]
- Weiss, J.; Weber, S.; Marzulla, T.; Raber, J. Pharmacological inhibition of Anaplastic Lymphoma Kinase rescues spatial memory impairments in Neurofibromatosis 1 mutant mice. Behav. Brain Res. 2017, 332, 337–342. [Google Scholar] [CrossRef]
- Ho, I.S.; Hannan, F.; Guo, H.F.; Hakker, I.; Zhong, Y. Distinct functional domains of neurofibromatosis type 1 regulate immediate versus long-term memory formation. J. Neurosci. 2007, 27, 6852–6857. [Google Scholar] [CrossRef] [PubMed]
- Tsai, P.I.; Wang, M.; Kao, H.H.; Cheng, Y.J.; Walker, J.A.; Chen, R.H.; Chien, C.T. Neurofibromin mediates FAK signaling in confining synapse growth at Drosophila neuromuscular junctions. J. Neurosci. 2012, 32, 16971–16981. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Davis, R.L. The GABAergic anterior paired lateral neuron suppresses and is suppressed by olfactory learning. Nat. Neurosci. 2009, 12, 53–59. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Krause, W.C.; Davis, R.L. GABAA receptor RDL inhibits Drosophila olfactory associative learning. Neuron 2007, 56, 1090–1102. [Google Scholar] [CrossRef] [PubMed]
- Walker, J.A.; Gouzi, J.Y.; Long, J.B.; Huang, S.; Maher, R.C.; Xia, H.; Khalil, K.; Ray, A.; Van Vactor, D.; Bernards, R.; et al. Genetic and functional studies implicate synaptic overgrowth and ring gland cAMP/PKA signaling defects in the Drosophila melanogaster neurofibromatosis-1 growth deficiency. PLoS Genet. 2013, 9, e1003958. [Google Scholar] [CrossRef] [PubMed]
- Bearden, C.E.; Hellemann, G.S.; Rosser, T.; Montojo, C.; Jonas, R.; Enrique, N.; Pacheco, L.; Hussain, S.A.; Wu, J.Y.; Ho, J.S.; et al. A randomized placebo-controlled lovastatin trial for neurobehavioral function in neurofibromatosis I. Ann. Clin. Transl. Neurol. 2016, 3, 266–279. [Google Scholar] [CrossRef] [PubMed]
- Jones, T.A. Diffraction methods for biological macromolecules. Interactive computer graphics: FRODO. Methods Enzymol. 1985, 115, 157–171. [Google Scholar] [PubMed]
- Krab, L.C.; de Goede-Bolder, A.; Aarsen, F.K.; Pluijm, S.M.; Bouman, M.J.; van der Geest, J.N.; Lequin, M.; Catsman, C.E.; Arts, W.F.; Kushner, S.A.; et al. Effect of simvastatin on cognitive functioning in children with neurofibromatosis type 1: A randomized controlled trial. Jama 2008, 300, 287–294. [Google Scholar] [CrossRef] [PubMed]
- Mainberger, F.; Jung, N.H.; Zenker, M.; Wahllander, U.; Freudenberg, L.; Langer, S.; Berweck, S.; Winkler, T.; Straube, A.; Heinen, F.; et al. Lovastatin improves impaired synaptic plasticity and phasic alertness in patients with neurofibromatosis type 1. BMC Neurol. 2013, 13, 131. [Google Scholar] [CrossRef]
- Payne, J.M.; Barton, B.; Ullrich, N.J.; Cantor, A.; Hearps, S.J.; Cutter, G.; Rosser, T.; Walsh, K.S.; Gioia, G.A.; Wolters, P.L.; et al. Randomized placebo-controlled study of lovastatin in children with neurofibromatosis type 1. Neurology 2016, 87, 2575–2584. [Google Scholar] [CrossRef]
- Stivaros, S.; Garg, S.; Tziraki, M.; Cai, Y.; Thomas, O.; Mellor, J.; Morris, A.A.; Jim, C.; Szumanska-Ryt, K.; Parkes, L.M.; et al. Randomised controlled trial of simvastatin treatment for autism in young children with neurofibromatosis type 1 (SANTA). Mol. Autism 2018, 9, 12. [Google Scholar] [CrossRef] [PubMed]
- van der Vaart, T.; Plasschaert, E.; Rietman, A.B.; Renard, M.; Oostenbrink, R.; Vogels, A.; de Wit, M.C.; Descheemaeker, M.J.; Vergouwe, Y.; Catsman-Berrevoets, C.E.; et al. Simvastatin for cognitive deficits and behavioural problems in patients with neurofibromatosis type 1 (NF1-SIMCODA): A randomised, placebo-controlled trial. Lancet Neurol. 2013, 12, 1076–1083. [Google Scholar] [CrossRef] [PubMed]
- Plasschaert, E.; Descheemaeker, M.J.; Van Eylen, L.; Noens, I.; Steyaert, J.; Legius, E. Prevalence of Autism Spectrum Disorder symptoms in children with neurofibromatosis type 1. Am. J. Med. Genet. B Neuropsychiatr. Genet. 2015, 168, 72–80. [Google Scholar] [CrossRef] [PubMed]
- Qiao, B.; Li, C.; Allen, V.W.; Shirasu-Hiza, M.; Syed, S. Automated analysis of long-term grooming behavior in Drosophila using a k-nearest neighbors classifier. Elife 2018, 7, e34497. [Google Scholar] [CrossRef]
- Cichowski, K.; Jacks, T. NF1 tumor suppressor gene function: Narrowing the GAP. Cell 2001, 104, 593–604. [Google Scholar] [CrossRef] [PubMed]
- Hampel, S.; Franconville, R.; Simpson, J.H.; Seeds, A.M. A neural command circuit for grooming movement control. Elife 2015, 4, e08758. [Google Scholar] [CrossRef] [PubMed]
- Hampel, S.; McKellar, C.E.; Simpson, J.H.; Seeds, A.M. Simultaneous activation of parallel sensory pathways promotes a grooming sequence in Drosophila. Elife 2017, 6, e28804. [Google Scholar] [CrossRef] [PubMed]
- Seeds, A.M.; Ravbar, P.; Chung, P.; Hampel, S.; Midgley, F.M., Jr.; Mensh, B.D.; Simpson, J.H. A suppression hierarchy among competing motor programs drives sequential grooming in Drosophila. Elife 2014, 3, e02951. [Google Scholar] [CrossRef] [PubMed]
- Tomchek, S.D.; Dunn, W. Sensory processing in children with and without autism: A comparative study using the short sensory profile. Am. J. Occup. Ther. 2007, 61, 190–200. [Google Scholar] [CrossRef]
- Lnenicka, G.A. Crayfish and Drosophila NMJs. Neurosci. Lett. 2020, 732, 135110. [Google Scholar] [CrossRef]
- Mosca, T.J.; Carrillo, R.A.; White, B.H.; Keshishian, H. Dissection of synaptic excitability phenotypes by using a dominant-negative Shaker K+ channel subunit. Proc. Natl. Acad. Sci. USA 2005, 102, 3477–3482. [Google Scholar] [CrossRef] [PubMed]
- Takarae, Y.; Sweeney, J. Neural Hyperexcitability in Autism Spectrum Disorders. Brain Sci 2017, 7, 129. [Google Scholar] [CrossRef] [PubMed]
- Littleton, J.T.; Stern, M.; Perin, M.; Bellen, H.J. Calcium dependence of neurotransmitter release and rate of spontaneous vesicle fusions are altered in Drosophila synaptotagmin mutants. Proc. Natl. Acad. Sci. USA 1994, 91, 10888–10892. [Google Scholar] [CrossRef] [PubMed]
- Littleton, J.T.; Bellen, H.J.; Perin, M.S. Expression of synaptotagmin in Drosophila reveals transport and localization of synaptic vesicles to the synapse. Development 1993, 118, 1077–1088. [Google Scholar] [CrossRef]
- Chisholm, A.; Anderson, V.; Pride, N.; Malarbi, S.; North, K.; Payne, J. Social function and autism spectrumbdisorder in children and adults with neurofibromatosis type 1: A systematic review and meta-analysis. Neuropsychol. Rev. 2018, 28, 317–340. [Google Scholar] [CrossRef] [PubMed]
- Payne, J.M.; Walsh, K.S.; Pride, N.A.; Haebich, K.M.; Maier, A.; Chisholm, A.; Glad, D.M.; Casnar, C.L.; Rouel, M.; Lorenzo, J.; et al. Social skills and autism spectrum disorder symptoms in children with neurofibromatosis type 1: Evidence for clinical trial outcomes. Dev. Med. Child Neurol. 2020, 62, 813–819. [Google Scholar] [CrossRef] [PubMed]
- Cheyne, J.E.; Zabouri, N.; Baddeley, D.; Lohmann, C. Spontaneous Activity Patterns Are Altered in the Developing Visual Cortex of the Fmr1 Knockout Mouse. Front. Neural Circuits 2019, 13, 57. [Google Scholar] [CrossRef] [PubMed]
- Orefice, L.L.; Zimmerman, A.L.; Chirila, A.M.; Sleboda, S.J.; Head, J.P.; Ginty, D.D. Peripheral Mechanosensory Neuron Dysfunction Underlies Tactile and Behavioral Deficits in Mouse Models of ASDs. Cell 2016, 166, 299–313. [Google Scholar] [CrossRef] [PubMed]
- Orefice, L.L. Peripheral Somatosensory Neuron Dysfunction: Emerging Roles in Autism Spectrum Disorders. Neuroscience 2020, 445, 120–129. [Google Scholar] [CrossRef]
- Carotenuto, M.; Messina, G.; Esposito, M.; Santoro, C.; Iacono, D.; Spruyt, K. Polysomnographic study in pediatric neurofibromatosis type 1. Front. Neurol. 2023, 14, 1213430. [Google Scholar] [CrossRef]
- Maraña Pérez, A.I.; Duat Rodríguez, A.; Soto Insuga, V.; Domínguez Carral, J.; Puertas Martín, V.; González Gutiérrez Solana, L. Prevalence of sleep disorders in patients with neurofibromatosis type 1. Neurologia 2015, 30, 561–565. [Google Scholar] [CrossRef]
- Vassallo, G.; Mughal, Z.; Robinson, L.; Weisberg, D.; Roberts, S.; Hupton, E.; Eelloo, J.; Burkitt Wright, E.; Garg, S.; Lewis, L.; et al. Perceived fatigue in children and young adults with neurofibromatosis type 1. J. Paediatr. Child Health 2020, 56, 878–883. [Google Scholar] [CrossRef]
- Cirelli, C.; Tononi, G. Is sleep essential? PLoS Biol. 2008, 6, e216. [Google Scholar] [CrossRef]
- Jordan, K.W.; Carbone, M.A.; Yamamoto, A.; Morgan, T.J.; Mackay, T.F. Quantitative genomics of locomotor behavior in Drosophila melanogaster. Genome Biol. 2007, 8, R172. [Google Scholar] [CrossRef]
- Hendricks, J.C.; Finn, S.M.; Panckeri, K.A.; Chavkin, J.; Williams, J.A.; Sehgal, A.; Pack, A.I. Rest in Drosophila is a sleep-like state. Neuron 2000, 25, 129–138. [Google Scholar] [CrossRef]
- Huber, R.; Hill, S.L.; Holladay, C.; Biesiadecki, M.; Tononi, G.; Cirelli, C. Sleep homeostasis in Drosophila melanogaster. Sleep 2004, 27, 628–639. [Google Scholar] [CrossRef]
- Anastasaki, C.; Rensing, N.; Johnson, K.J.; Wong, M.; Gutmann, D.H. Neurofibromatosis type 1 (Nf1)-mutant mice exhibit increased sleep fragmentation. J. Sleep Res. 2019, 28, e12816. [Google Scholar] [CrossRef]
- Weiss, J.B.; Weber, S.J.; Torres, E.R.S.; Marzulla, T.; Raber, J. Genetic inhibition of Anaplastic Lymphoma Kinase rescues cognitive impairments in Neurofibromatosis 1 mutant mice. Behav. Brain Res. 2017, 321, 148–156. [Google Scholar] [CrossRef]
- Weiss, J.B.; Raber, J. Inhibition of Anaplastic Lymphoma Kinase (Alk) as Therapeutic Target to Improve Brain Function in Neurofibromatosis Type 1 (Nf1). Cancers 2023, 15, 4579. [Google Scholar] [CrossRef]
- Maurer, G.W.; Malita, A.; Nagy, S.; Koyama, T.; Werge, T.M.; Halberg, K.A.; Texada, M.J.; Rewitz, K. Analysis of genes within the schizophrenia-linked 22q11.2 deletion identifies interaction of night owl/LZTR1 and NF1 in GABAergic sleep control. PLoS Genet. 2020, 16, e1008727. [Google Scholar] [CrossRef]
- Brown, E.B.; Zhang, J.; Lloyd, E.; Lanzon, E.; Botero, V.; Tomchik, S.; Keene, A.C. Neurofibromin 1 mediates sleep depth in Drosophila. PLoS Genet. 2023, 19, e1011049. [Google Scholar] [CrossRef] [PubMed]
- van der Voet, M.; Harich, B.; Franke, B.; Schenck, A. ADHD-associated dopamine transporter, latrophilin and neurofibromin share a dopamine-related locomotor signature in Drosophila. Mol. Psychiatry 2016, 21, 565–573. [Google Scholar] [CrossRef] [PubMed]
- Shang, Y.; Haynes, P.; Pírez, N.; Harrington, K.I.; Guo, F.; Pollack, J.; Hong, P.; Griffith, L.C.; Rosbash, M. Imaging analysis of clock neurons reveals light buffers the wake-promoting effect of dopamine. Nat. Neurosci. 2011, 14, 889–895. [Google Scholar] [CrossRef] [PubMed]
- Anastasaki, C.; Woo, A.S.; Messiaen, L.M.; Gutmann, D.H. Elucidating the impact of neurofibromatosis-1 germline mutations on neurofibromin function and dopamine-based learning. Hum. Mol. Genet. 2015, 24, 3518–3528. [Google Scholar] [CrossRef] [PubMed]
- Brown, J.A.; Gianino, S.M.; Gutmann, D.H. Defective cAMP generation underlies the sensitivity of CNS neurons to neurofibromatosis-1 heterozygosity. J. Neurosci. 2010, 30, 5579–5589. [Google Scholar] [CrossRef] [PubMed]
- Diggs-Andrews, K.A.; Tokuda, K.; Izumi, Y.; Zorumski, C.F.; Wozniak, D.F.; Gutmann, D.H. Dopamine deficiency underlies learning deficits in neurofibromatosis-1 mice. Ann. Neurol. 2013, 73, 309–315. [Google Scholar] [CrossRef] [PubMed]
- Volkow, N.D.; Wang, G.; Fowler, J.S.; Logan, J.; Gerasimov, M.; Maynard, L.; Ding, Y.; Gatley, S.J.; Gifford, A.; Franceschi, D. Therapeutic doses of oral methylphenidate significantly increase extracellular dopamine in the human brain. J. Neurosci. 2001, 21, Rc121. [Google Scholar] [CrossRef] [PubMed]
- Volkow, N.D.; Wang, G.J.; Kollins, S.H.; Wigal, T.L.; Newcorn, J.H.; Telang, F.; Fowler, J.S.; Zhu, W.; Logan, J.; Ma, Y.; et al. Evaluating dopamine reward pathway in ADHD: Clinical implications. Jama 2009, 302, 1084–1091. [Google Scholar] [CrossRef] [PubMed]
- Tettamanti, G.; Mogensen, H.; Kampitsi, C.E.; Nordgren, A.; Feychting, M. Birth Characteristics Among Children Diagnosed with Neurofibromatosis Type 1 and Tuberous Sclerosis. J. Pediatr. 2021, 239, 200–205.e202. [Google Scholar] [CrossRef]
- Peltonen, S.; Poyhonen, M. Clinical Diagnosis and Atypical Forms of NF1. In Neurofibromatosis Type 1. Molecular and Cellular Biology; Upadhyaya, M., Cooper, D., Eds.; Springer: Berlin/Heidelberg, Germany, 2012; pp. 17–30. [Google Scholar]
- Masgras, I.; Rasola, A. Metabolic features of NF1-associated tumors. In Clinical and Basic Aspects of Neurofibromatosis Type 1; Nakayama, J., Yoshida, Y., Eds.; IntechOpen: London, UK, 2021. [Google Scholar]
- Apostolova, I.; Derlin, T.; Salamon, J.; Amthauer, H.; Granstrom, S.; Brenner, W.; Mautner, V.F.; Buchert, R. Cerebral glucose metabolism in adults with neurofibromatosis type 1. Brain Res. 2015, 1625, 97–101. [Google Scholar] [CrossRef]
- Walker, J.A.; Bernards, A. A Drosophila screen identifies neurofibromatosis-1 genetic modifiers involved in systemic and synaptic growth. Rare Dis. 2014, 2, e28341. [Google Scholar] [CrossRef] [PubMed]
- Souza, M.L.R.; Jansen, A.K.; Rodrigues, L.O.C.; Vilela, D.L.S.; Kakehasi, A.M.; Martins, A.S.; Souza, J.F.; Rezende, N.A. Reduced bone mineral content and density in neurofibromatosis type 1 and its association with nutrient intake. Rev. Assoc. Med. Bras. (1992) 2020, 66, 666–672. [Google Scholar] [CrossRef] [PubMed]
- Stevenson, D.A.; Viscogliosi, G.; Leoni, C. Bone health in RASopathies. Am. J. Med. Genet. C Semin. Med. Genet. 2022, 190, 459–470. [Google Scholar] [CrossRef] [PubMed]
- Visnapuu, V.; Peltonen, S.; Alivuotila, L.; Happonen, R.P.; Peltonen, J. Craniofacial and oral alterations in patients with Neurofibromatosis 1. Orphanet J. Rare Dis. 2018, 13, 131. [Google Scholar] [CrossRef] [PubMed]
- Souza, M.L.R.; Jansen, A.K.; Rodrigues, L.O.C.; Vilela, D.L.S.; Kakehasi, A.M.; Martins, A.S.; Souza, J.F.; Rezende, N.A. Increased resting metabolism in neurofibromatosis type 1. Clin. Nutr. ESPEN 2019, 32, 44–49. [Google Scholar] [CrossRef] [PubMed]
- Anastasaki, C.; Gutmann, D.H. Neuronal NF1/RAS regulation of cyclic AMP requires atypical PKC activation. Hum. Mol. Genet. 2014, 23, 6712–6721. [Google Scholar] [CrossRef]
- Hannan, F.; Ho, I.; Tong, J.J.; Zhu, Y.; Nurnberg, P.; Zhong, Y. Effect of neurofibromatosis type I mutations on a novel pathway for adenylyl cyclase activation requiring neurofibromin and Ras. Hum. Mol. Genet. 2006, 15, 1087–1098. [Google Scholar] [CrossRef]
- Anastasaki, C.; Mo, J.; Chen, J.K.; Chatterjee, J.; Pan, Y.; Scheaffer, S.M.; Cobb, O.; Monje, M.; Le, L.Q.; Gutmann, D.H. Neuronal hyperexcitability drives central and peripheral nervous system tumor progression in models of neurofibromatosis-1. Nat. Commun. 2022, 13, 2785. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Atsoniou, K.; Giannopoulou, E.; Georganta, E.-M.; Skoulakis, E.M.C. Drosophila Contributions towards Understanding Neurofibromatosis 1. Cells 2024, 13, 721. https://doi.org/10.3390/cells13080721
Atsoniou K, Giannopoulou E, Georganta E-M, Skoulakis EMC. Drosophila Contributions towards Understanding Neurofibromatosis 1. Cells. 2024; 13(8):721. https://doi.org/10.3390/cells13080721
Chicago/Turabian StyleAtsoniou, Kalliopi, Eleni Giannopoulou, Eirini-Maria Georganta, and Efthimios M. C. Skoulakis. 2024. "Drosophila Contributions towards Understanding Neurofibromatosis 1" Cells 13, no. 8: 721. https://doi.org/10.3390/cells13080721