Expression Profiling of Differentiating Emerin-Null Myogenic Progenitor Identifies Molecular Pathways Implicated in Their Impaired Differentiation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. RNA-seq
2.3. Pathway Analysis
2.4. EdU Incorporation and Immunofluorescence Microscopy
2.5. qPCR
2.6. Data Sharing Statement
3. Results
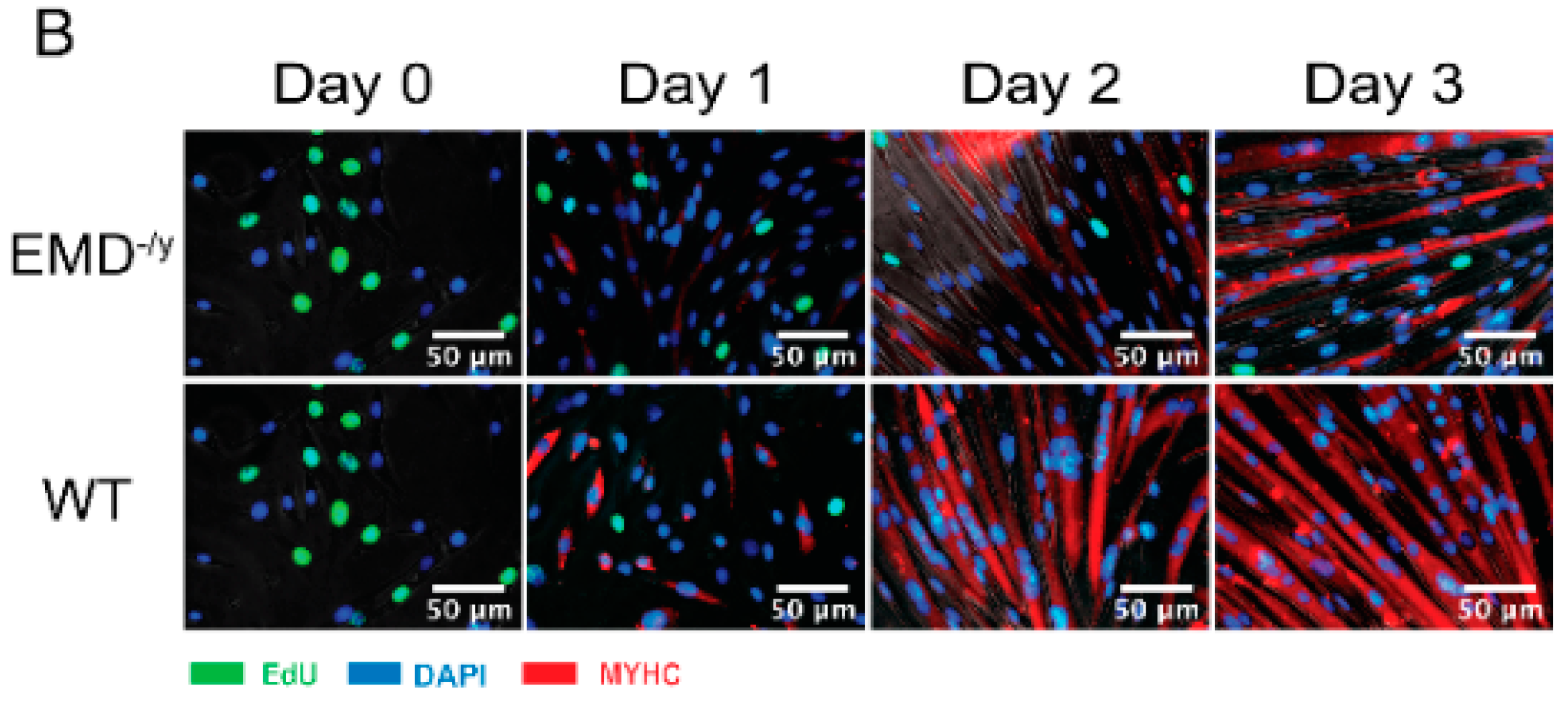
3.1. Emerin-Null Myogenic Progenitor Cells Display Impaired Myogenic Differentiation
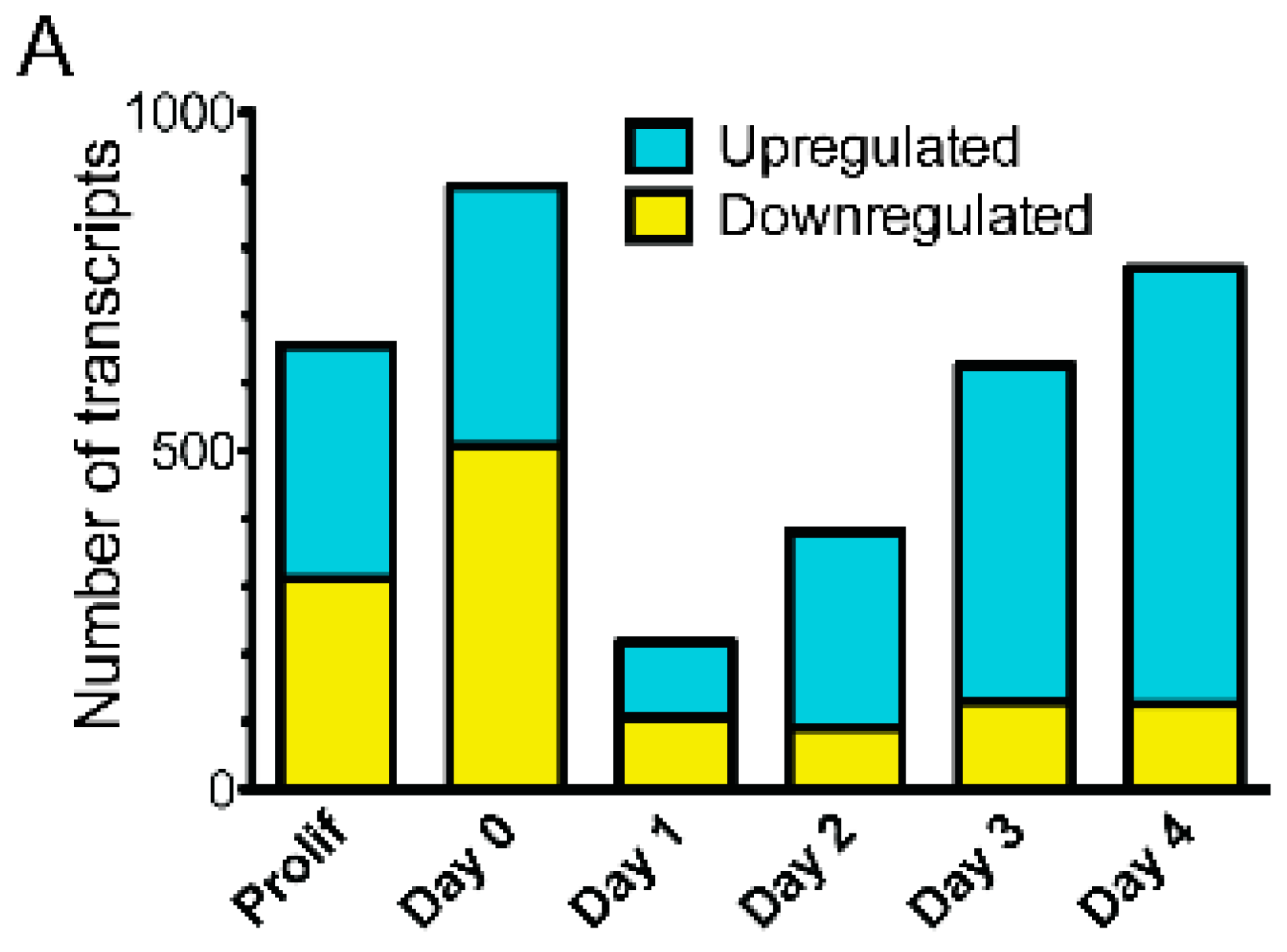
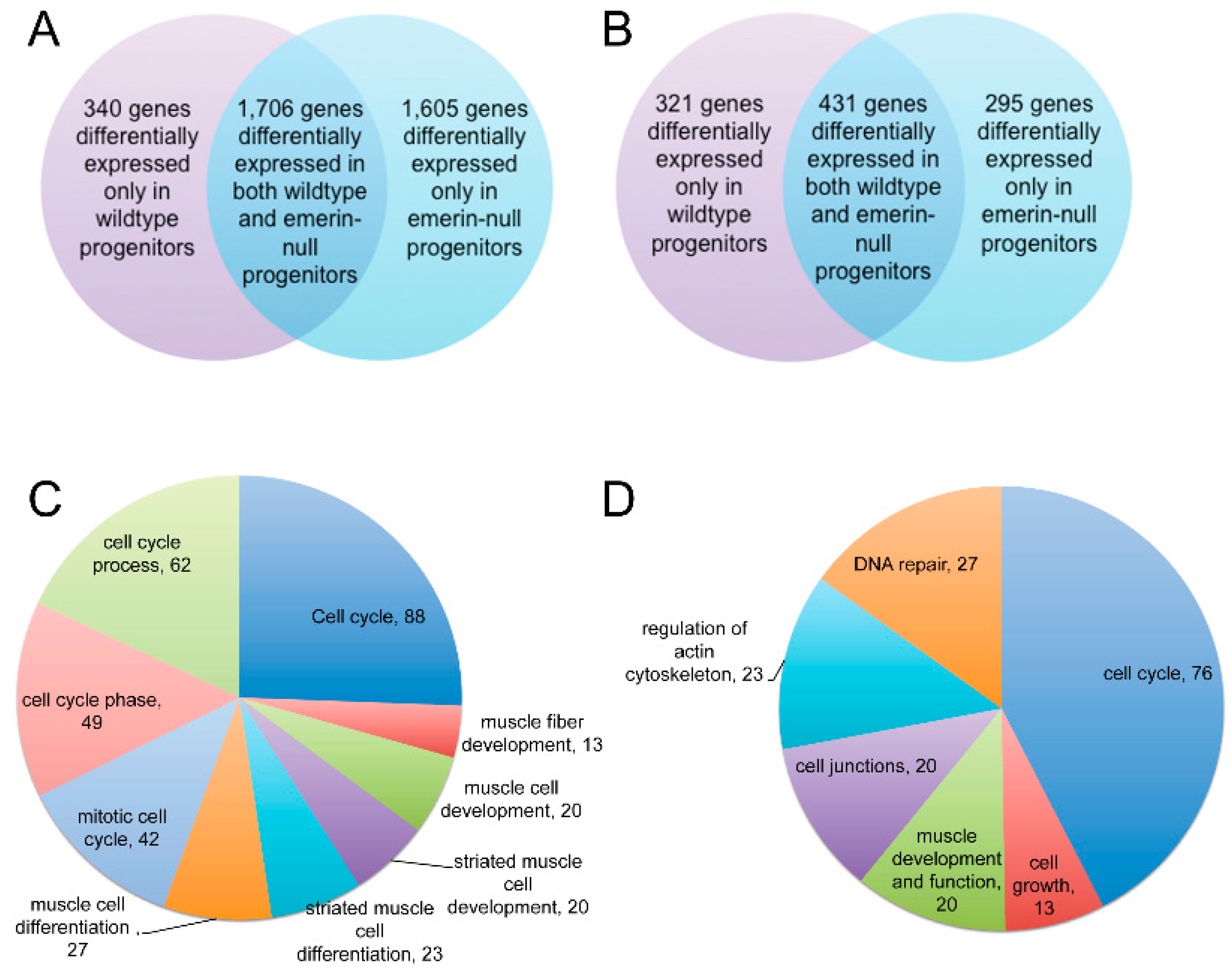
3.2. Emerin-Null Myogenic Progenitor Cells Show Extensive Transcriptional Changes Compared to Wildtype Cells at Each Day of Myogenic Differentiation
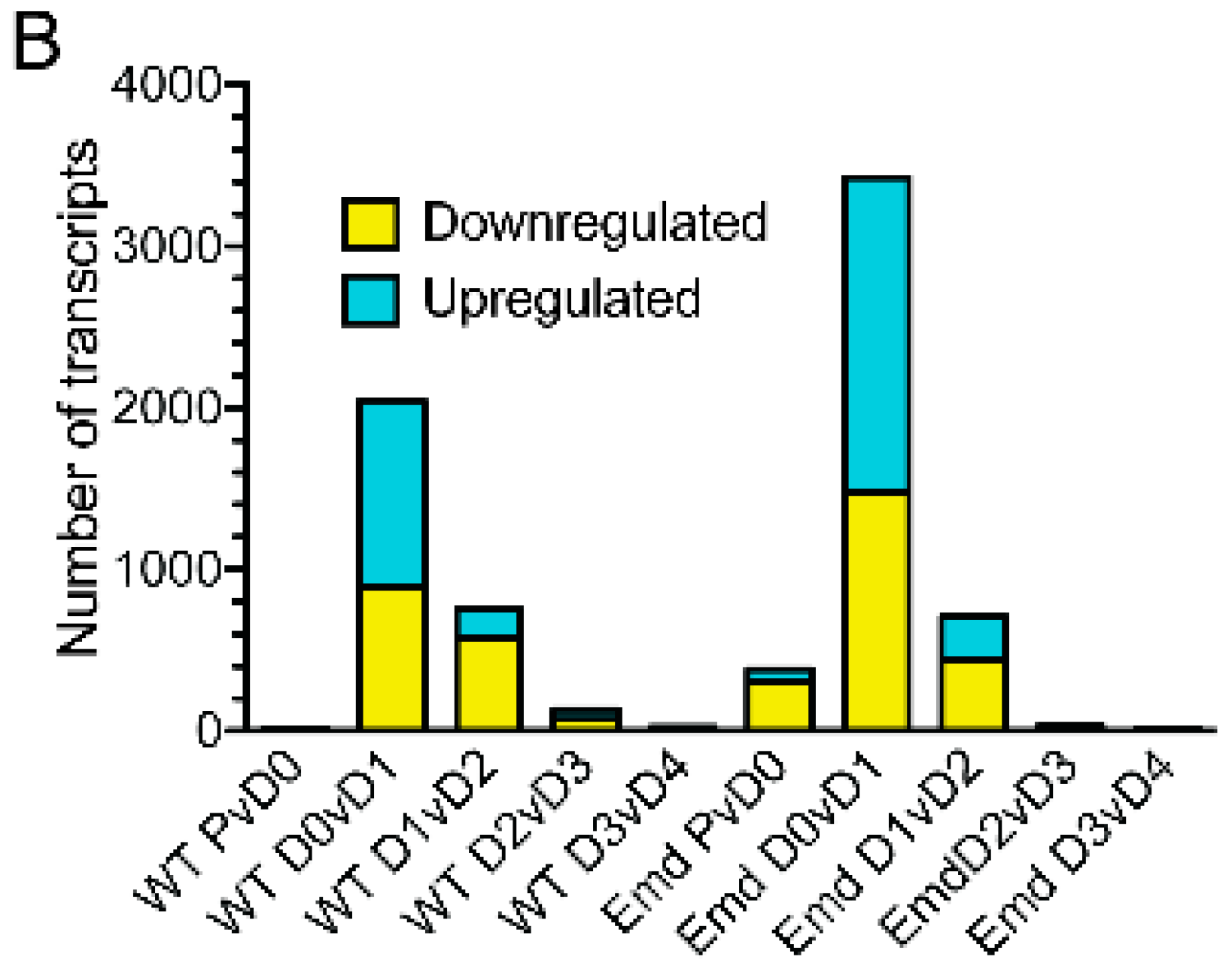
3.3. Emerin-Null and Wildtype Myogenic Progenitors Diverge Extensively in Transcript Expression During the Transitions from Days 0–2 of Myogenic Differentiation
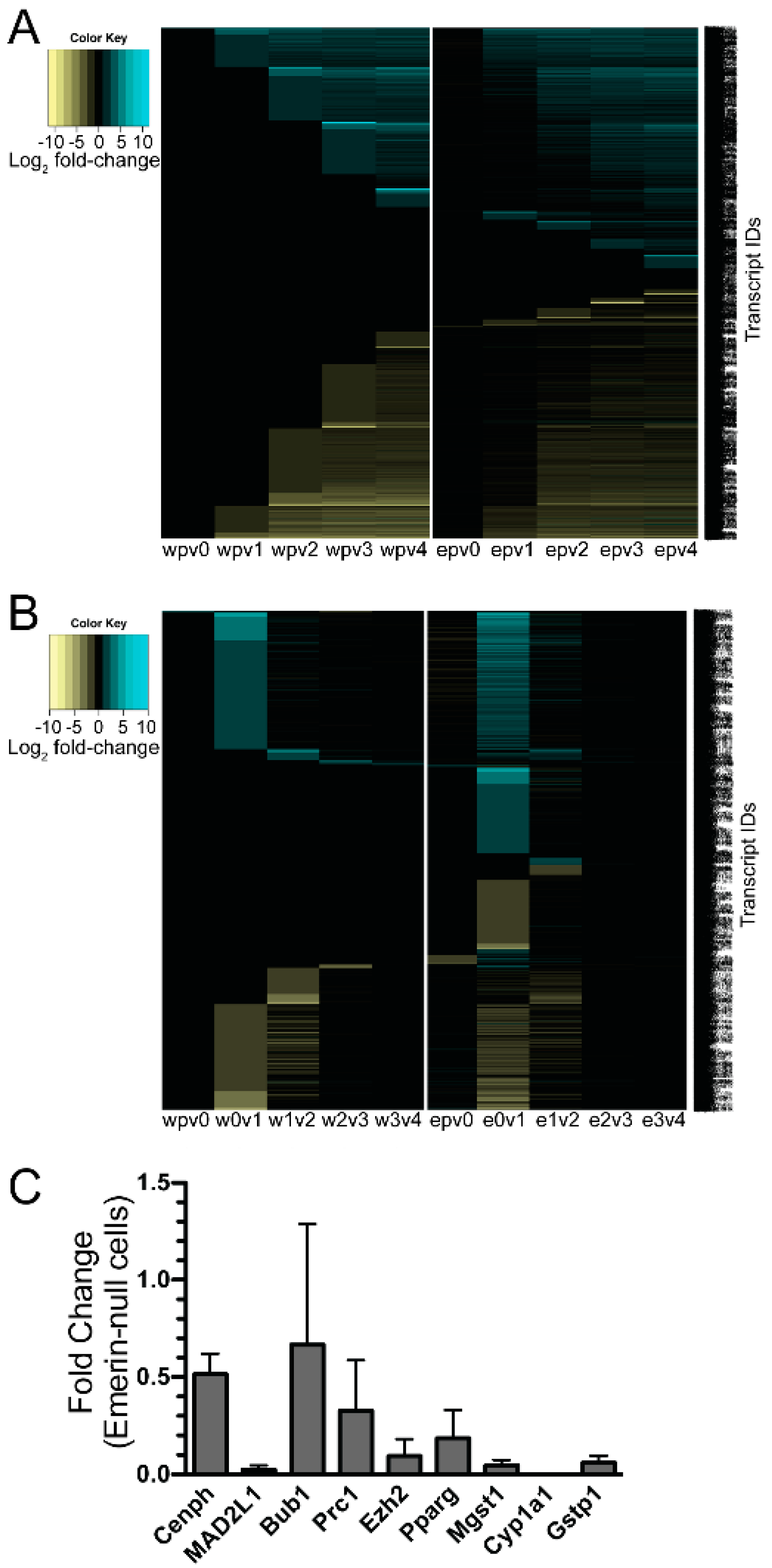
3.4. qPCR Was Done to Validate the Results from the RNAseq Analysis
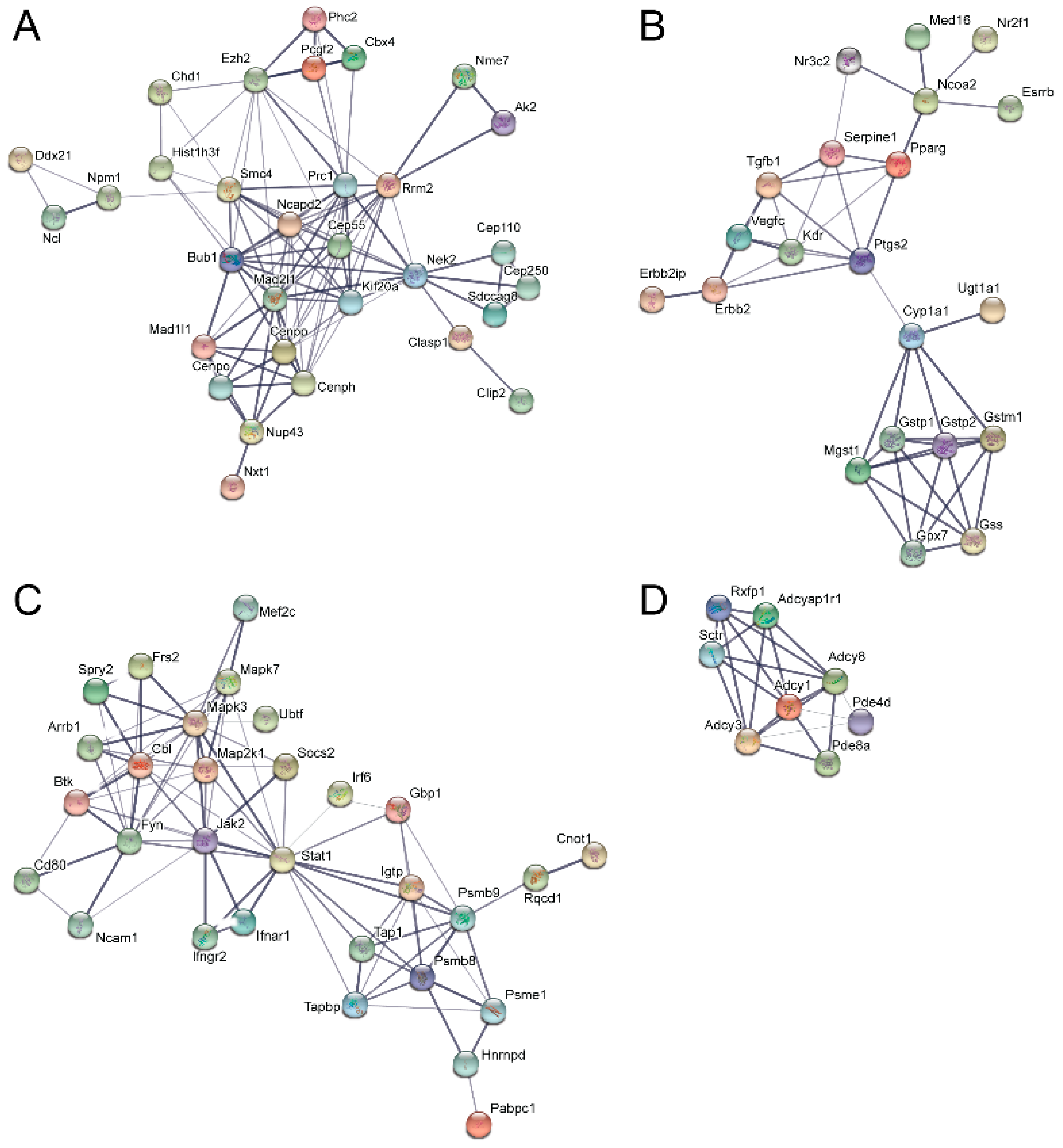
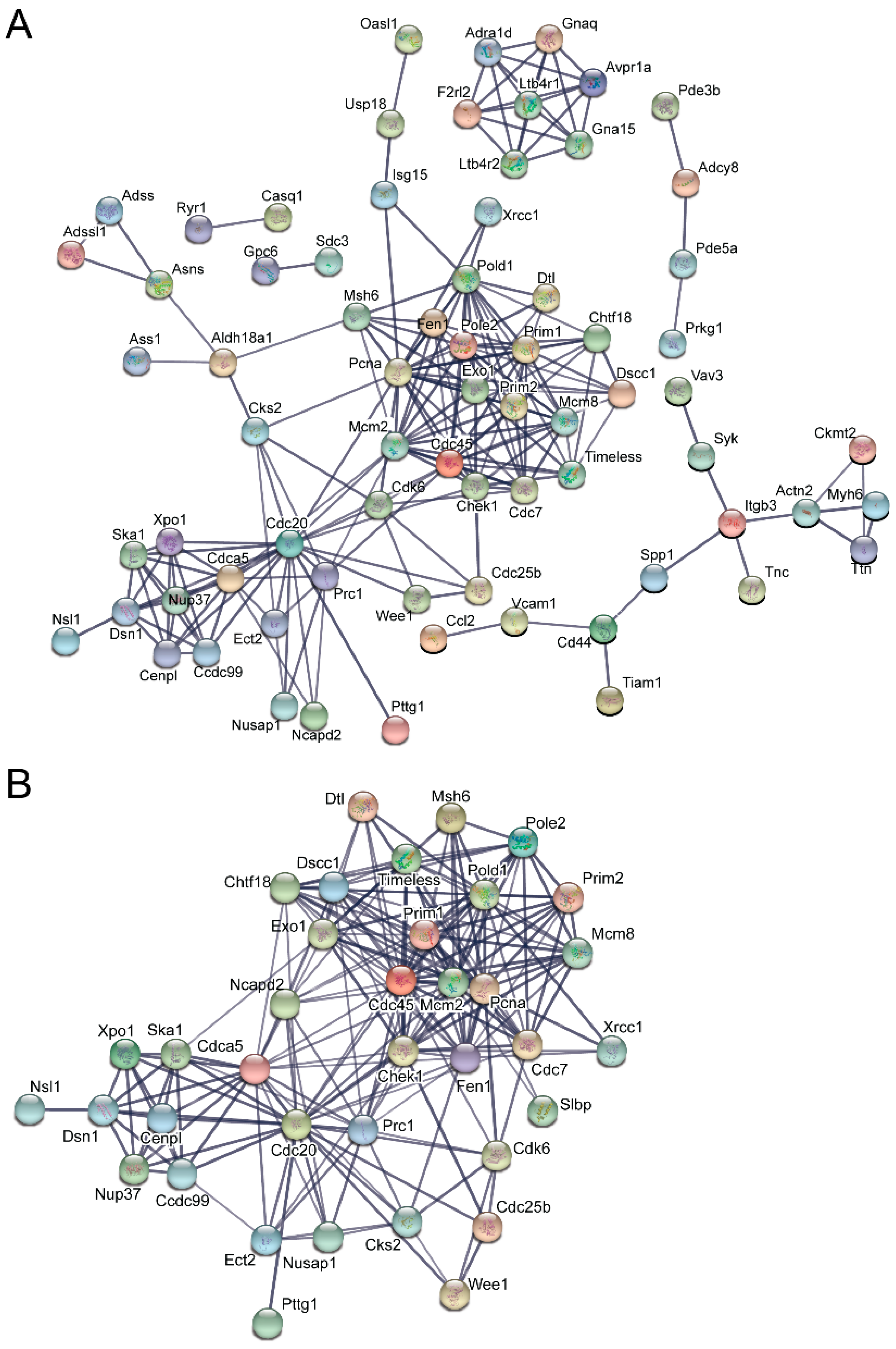
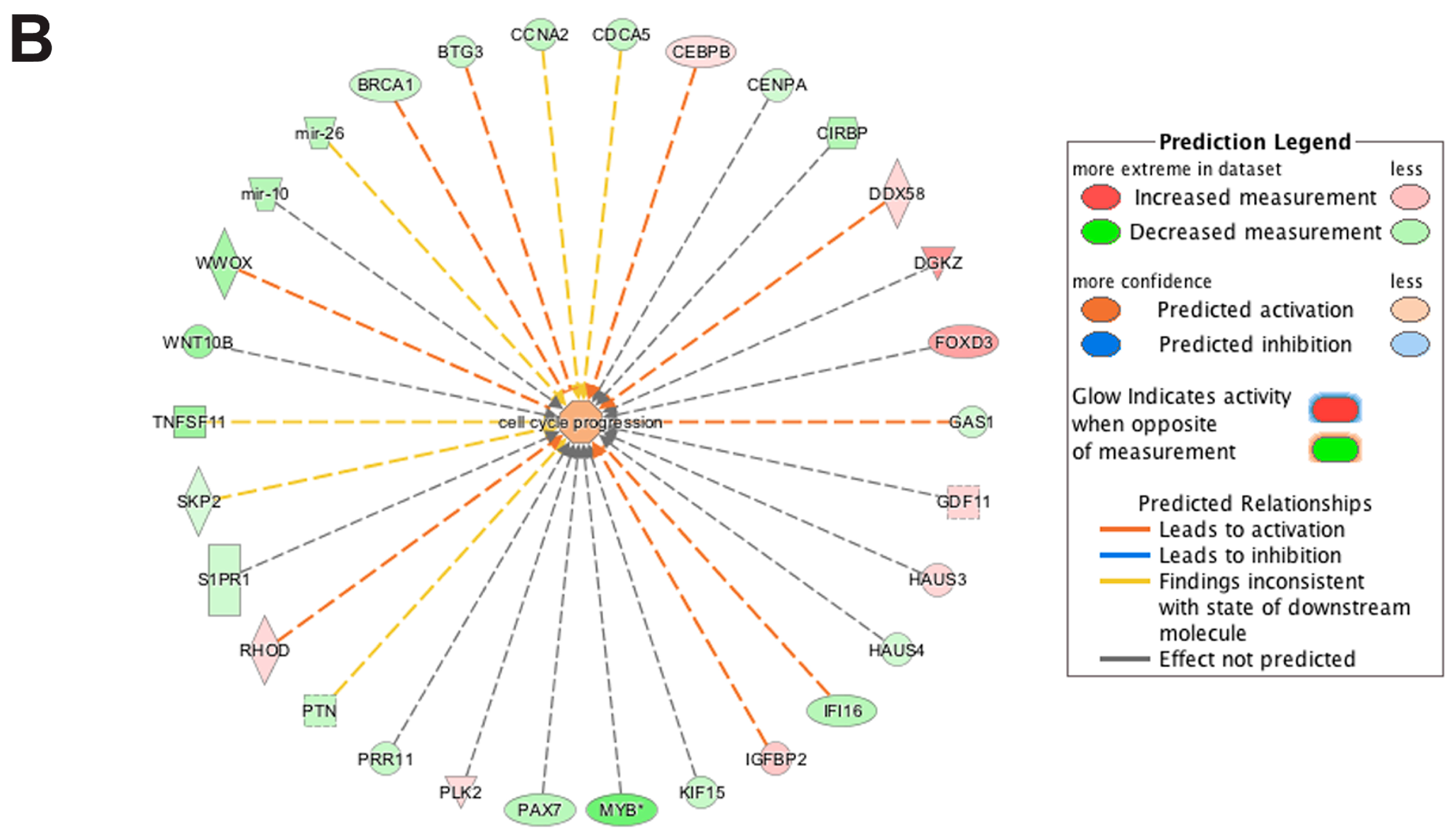
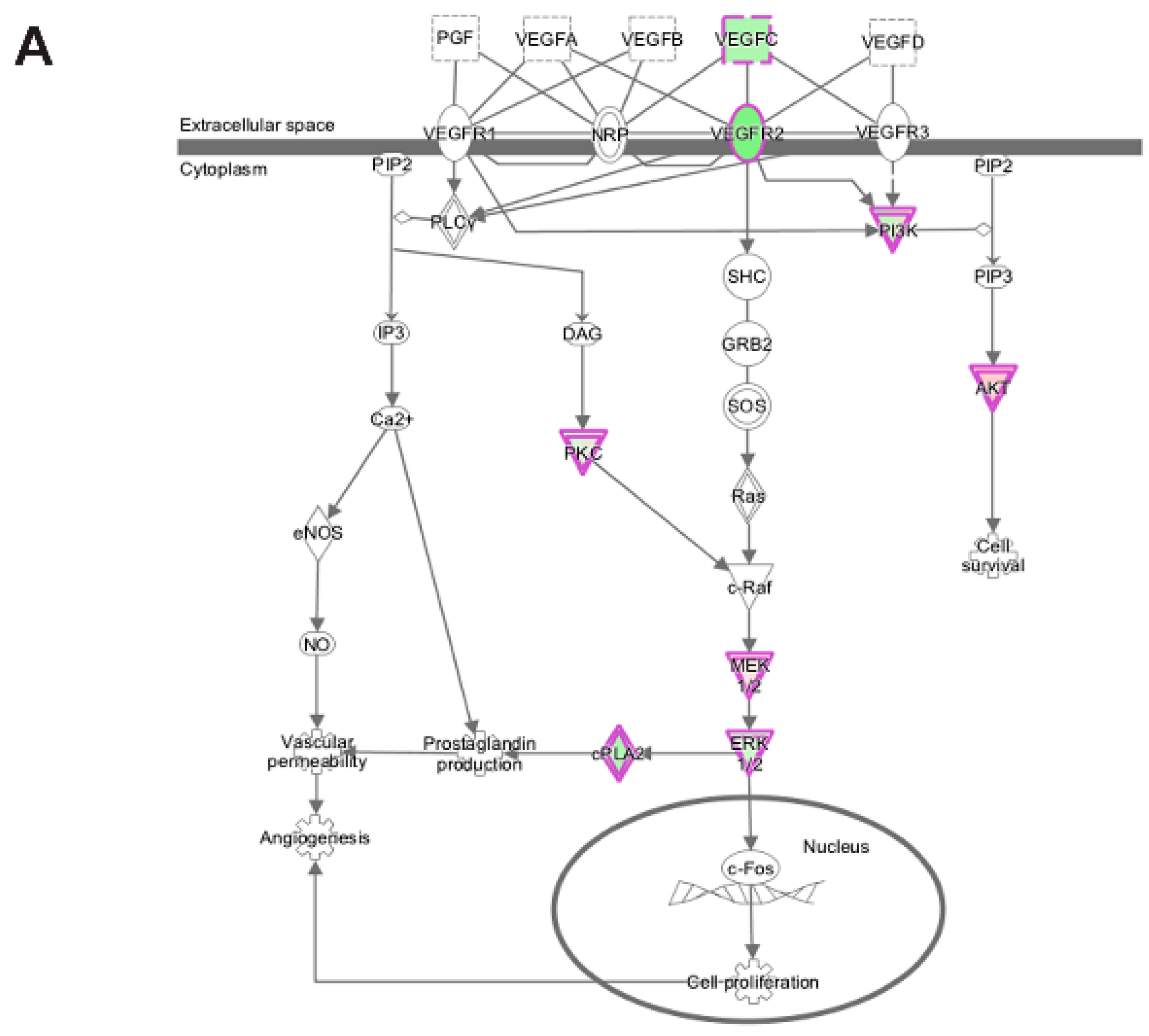
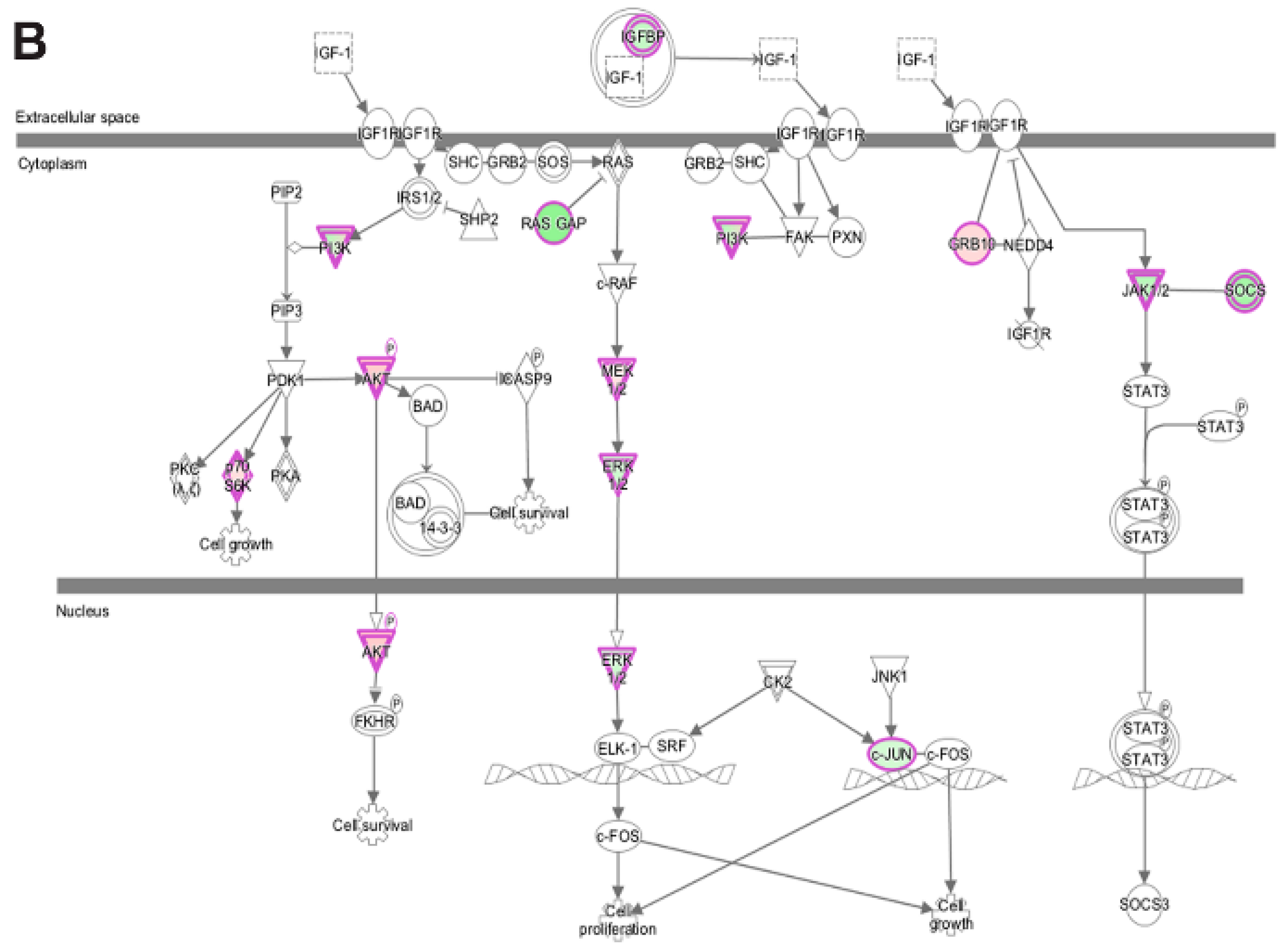
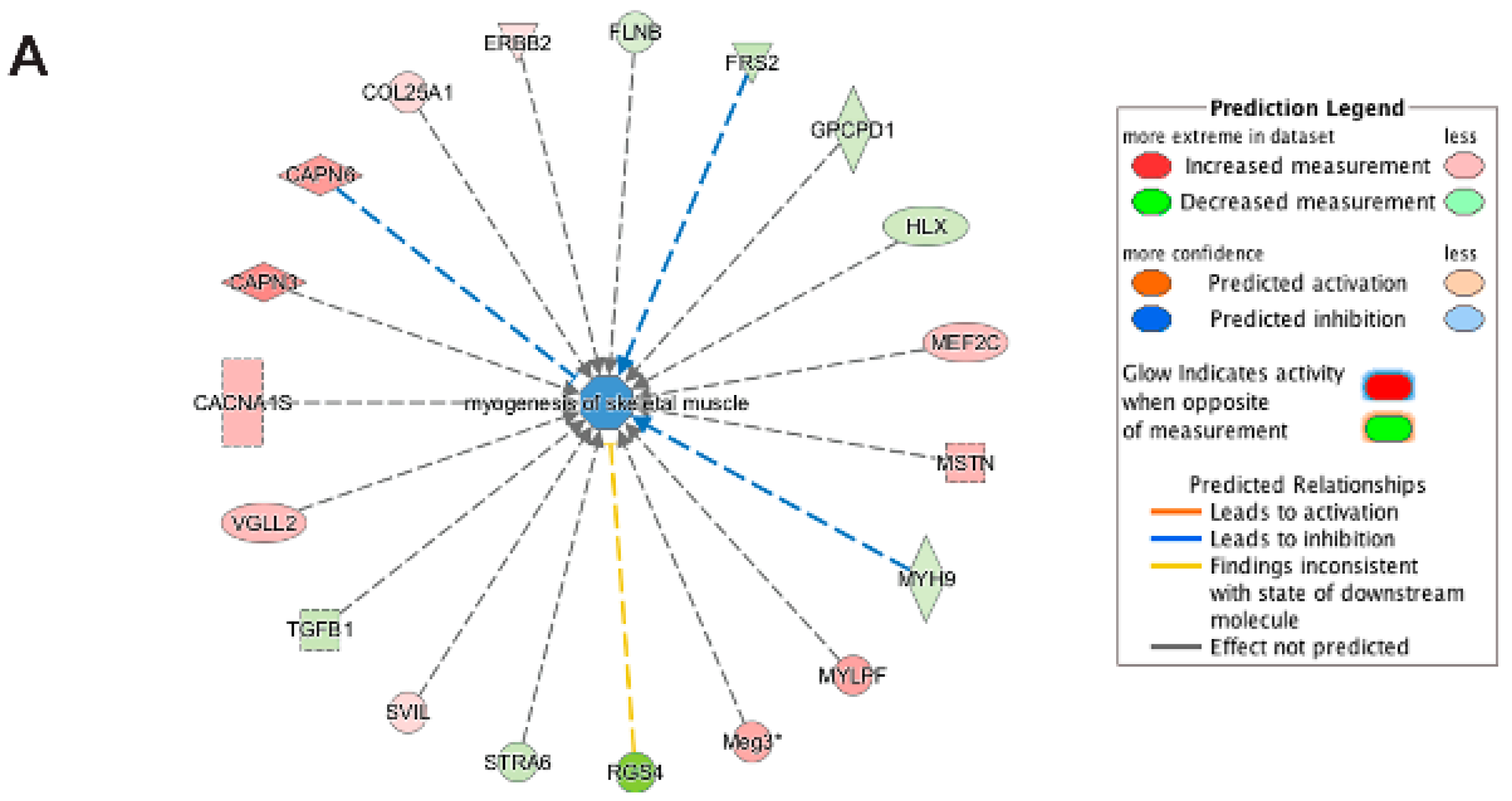
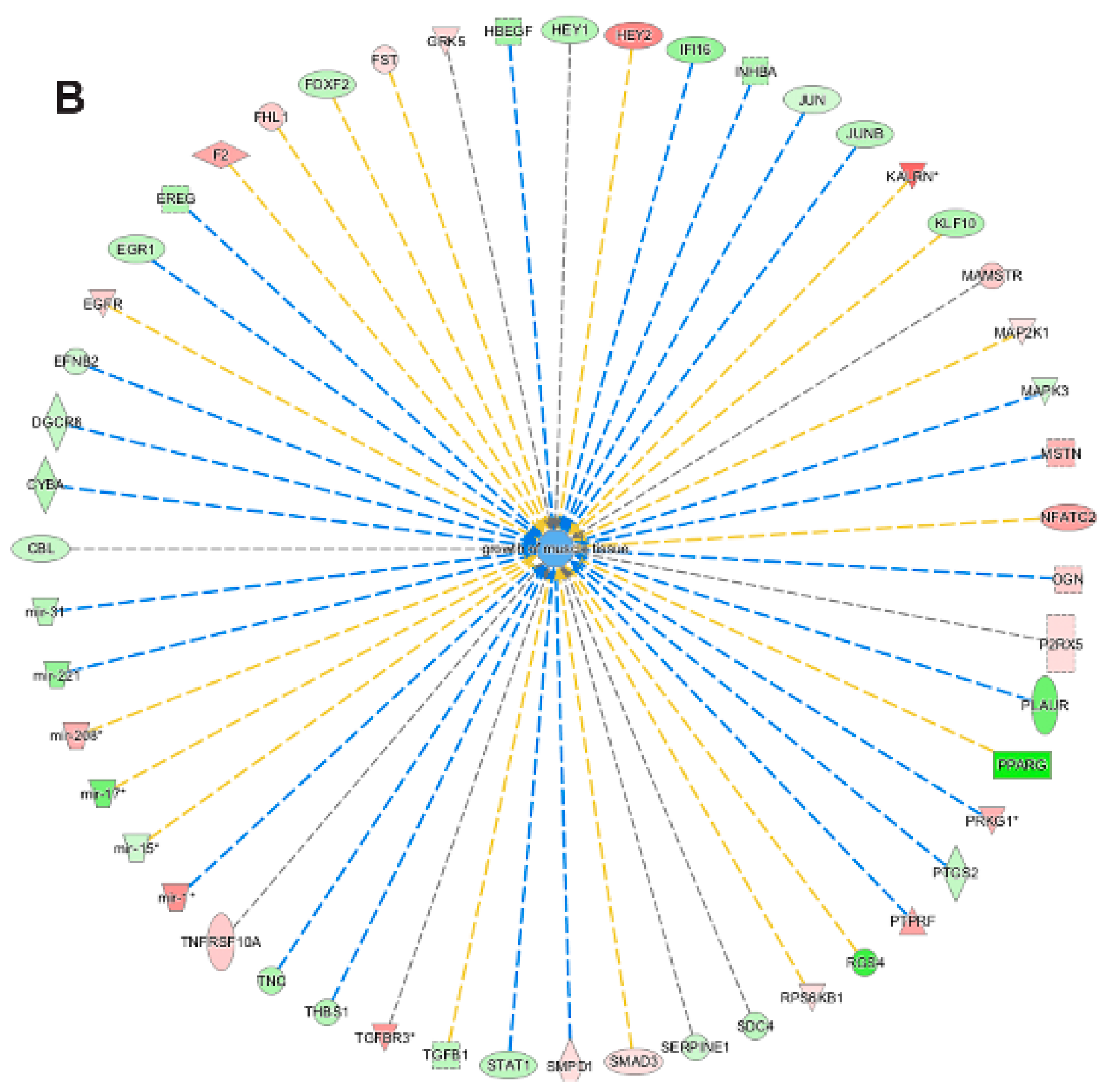
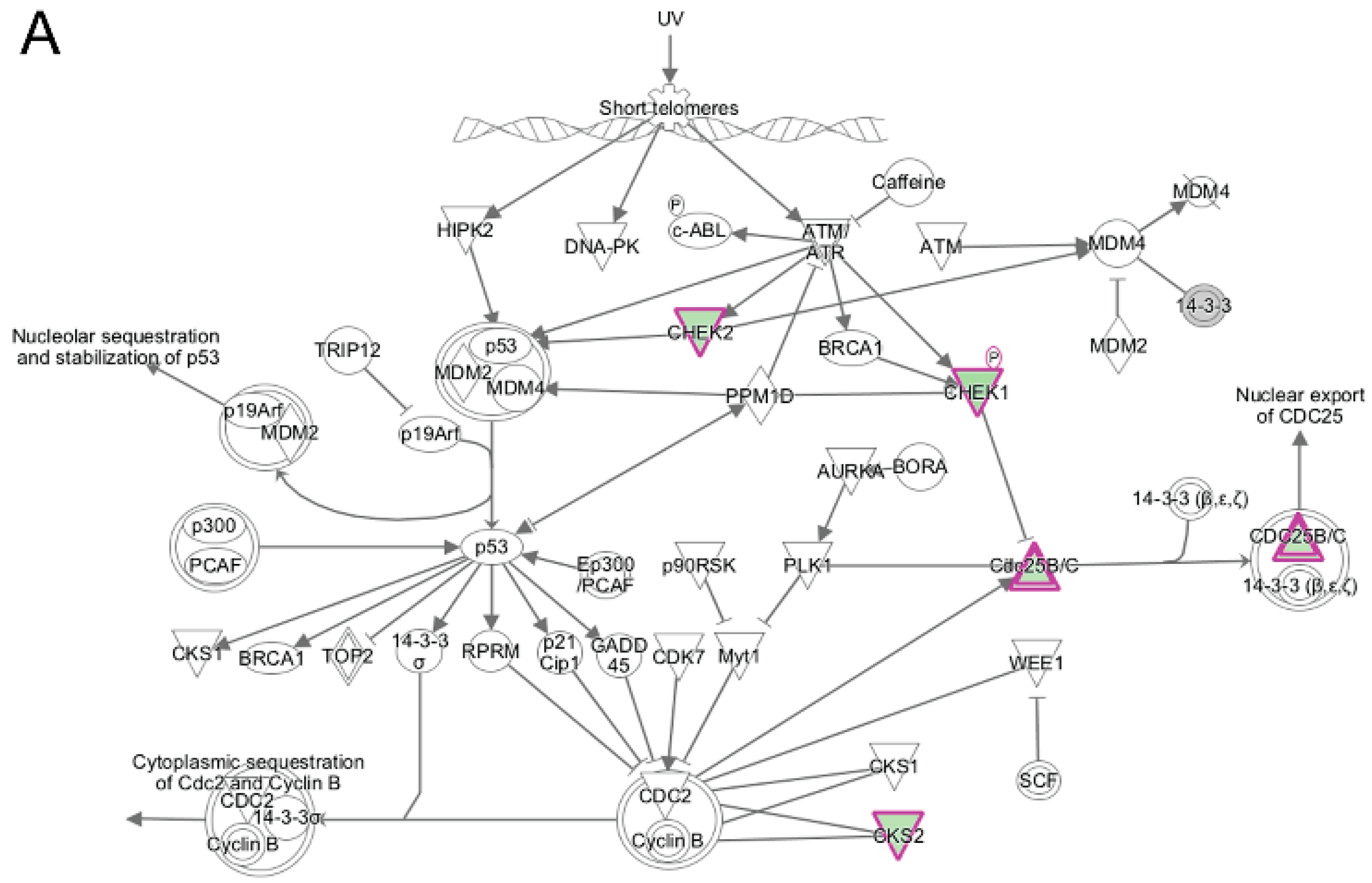
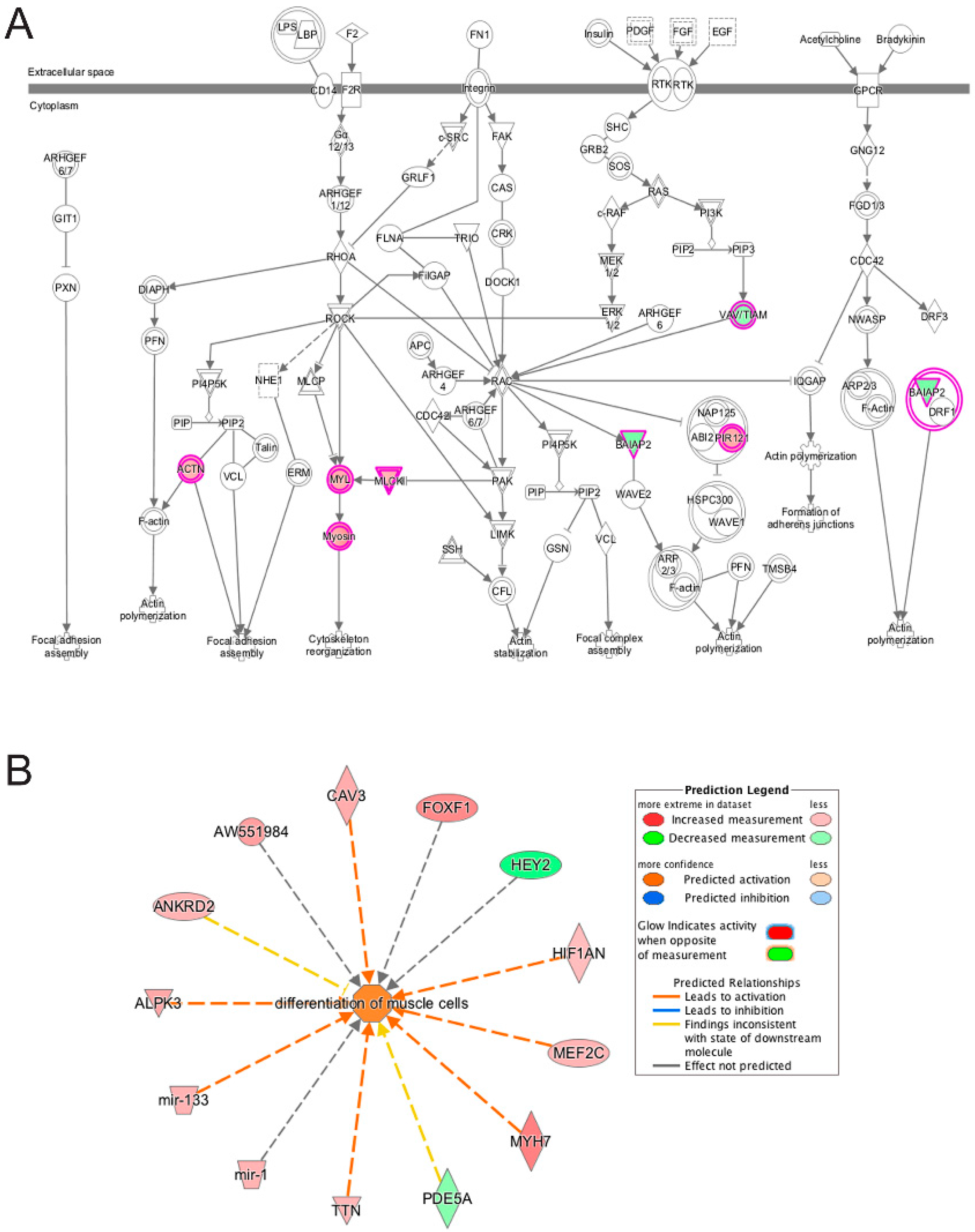
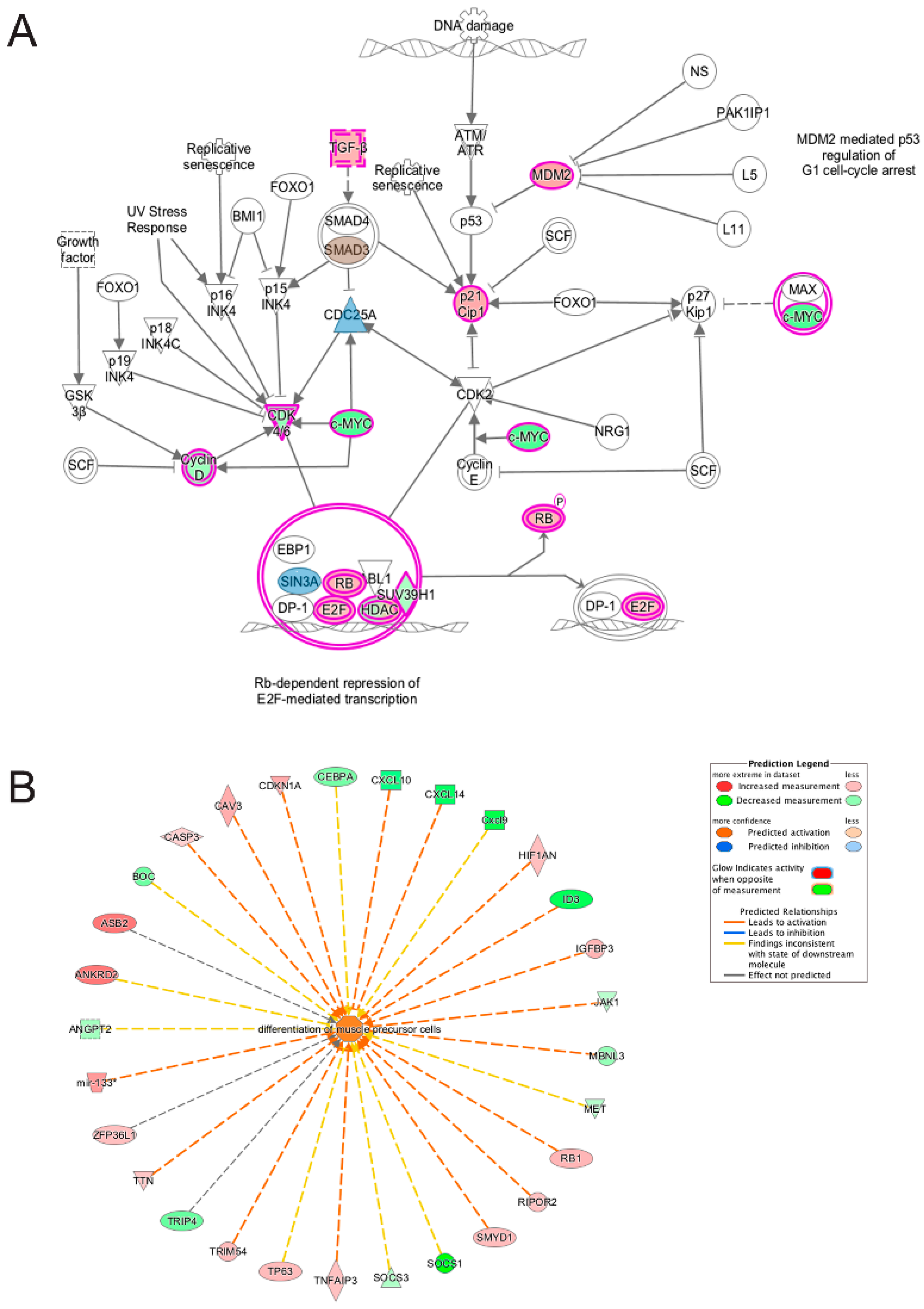
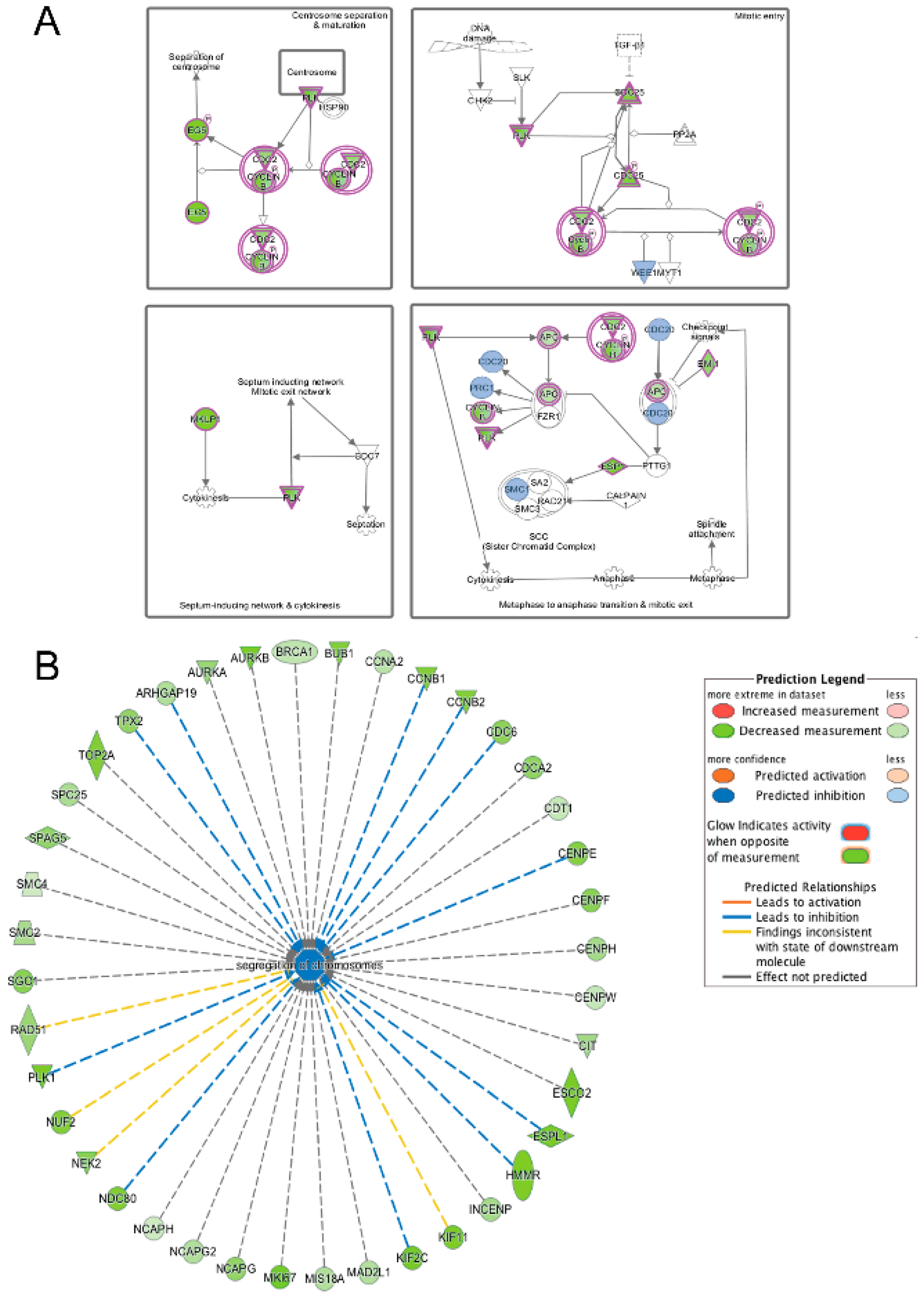
3.5. Pathway and Network Analysis of Differentially Expressed Transcripts during Myogenic Differentiation of Wildtype and Emerin-Null Progenitors
4. Discussion
How Does Loss of Emerin Cause Such Massive Changes in Gene Expression?
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Bione, S.; Maestrini, E.; Rivella, S.; Mancini, M.; Regis, S.; Romeo, G.; Toniolo, D. Identification of a novel X-linked gene responsible for Emery-Dreifuss muscular dystrophy. Nat. Genet. 1994, 8, 323–327. [Google Scholar] [CrossRef] [PubMed]
- Mendez-Lopez, I.; Worman, H.J. Inner nuclear membrane proteins: Impact on human disease. Chromosoma 2012, 121, 153–167. [Google Scholar] [CrossRef] [PubMed]
- Vlcek, S.; Foisner, R. Lamins and lamin-associated proteins in aging and disease. Curr. Opin. Cell Biol. 2007, 19, 298–304. [Google Scholar] [CrossRef] [PubMed]
- Worman, H.J. Nuclear lamins and laminopathies. J. Pathol. 2012, 226, 316–325. [Google Scholar] [CrossRef] [PubMed]
- Bonne, G.; Leturcq, F.; Ben Yaou, R. Emery-Dreifuss Muscular Dystrophy. In GeneReviews(R); Pagon, R.A., Adam, M.P., Ardinger, H.H., Wallace, S.E., Amemiya, A., Bean, L.J.H., Bird, T.D., Ledbetter, N., Mefford, H.C., Smith, R.J.H., et al., Eds.; University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- Melcon, G.; Kozlov, S.; Cutler, D.A.; Sullivan, T.; Hernandez, L.; Zhao, P.; Mitchell, S.; Nader, G.; Bakay, M.; Rottman, J.N.; et al. Loss of emerin at the nuclear envelope disrupts the Rb1/E2F and MyoD pathways during muscle regeneration. Hum. Mol. Genet. 2006, 15, 637–651. [Google Scholar] [CrossRef] [PubMed]
- Ozawa, R.; Hayashi, Y.K.; Ogawa, M.; Kurokawa, R.; Matsumoto, H.; Noguchi, S.; Nonaka, I.; Nishino, I. Emerin-lacking mice show minimal motor and cardiac dysfunctions with nuclear-associated vacuoles. Am. J. Pathol. 2006, 168, 907–917. [Google Scholar] [CrossRef] [PubMed]
- Bakay, M.; Wang, Z.; Melcon, G.; Schiltz, L.; Xuan, J.; Zhao, P.; Sartorelli, V.; Seo, J.; Pegoraro, E.; Angelini, C.; et al. Nuclear envelope dystrophies show a transcriptional fingerprint suggesting disruption of Rb-MyoD pathways in muscle regeneration. Brain 2006, 129, 996–1013. [Google Scholar] [CrossRef] [PubMed]
- Frock, R.L.; Kudlow, B.A.; Evans, A.M.; Jameson, S.A.; Hauschka, S.D.; Kennedy, B.K. Lamin A/C and emerin are critical for skeletal muscle satellite cell differentiation. Genes Dev. 2006, 20, 486–500. [Google Scholar] [CrossRef] [PubMed]
- Huber, M.D.; Guan, T.; Gerace, L. Overlapping functions of nuclear envelope proteins NET25 (Lem2) and emerin in regulation of extracellular signal-regulated kinase signaling in myoblast differentiation. Mol. Cell. Biol. 2009, 29, 5718–5728. [Google Scholar] [CrossRef] [PubMed]
- Collins, C.M.; Ellis, J.A.; Holaska, J.M. MAPK signaling pathways and HDAC3 activity are disrupted during differentiation of emerin-null myogenic progenitor cells. Dis. Models Mech. 2017, 10, 385–397. [Google Scholar] [CrossRef] [PubMed]
- Demmerle, J.; Koch, A.J.; Holaska, J.M. Emerin and histone deacetylase 3 (HDAC3) cooperatively regulate expression and nuclear positions of MyoD, Myf5, and Pax7 genes during myogenesis. Chromosome Res. 2013, 21, 765–779. [Google Scholar] [CrossRef] [PubMed]
- Koch, A.J.; Holaska, J.M. Loss of Emerin Alters Myogenic Signaling and miRNA Expression in Mouse Myogenic Progenitors. PLoS ONE 2012, 7, e37262. [Google Scholar] [CrossRef] [PubMed]
- Manilal, S.; Nguyen, T.M.; Sewry, C.A.; Morris, G.E. The Emery-Dreifuss muscular dystrophy protein, emerin, is a nuclear membrane protein. Hum. Mol. Genet. 1996, 5, 801–808. [Google Scholar] [CrossRef] [PubMed]
- Nagano, A.; Koga, R.; Ogawa, M.; Kurano, Y.; Kawada, J.; Okada, R.; Hayashi, Y.K.; Tsukahara, T.; Arahata, K. Emerin deficiency at the nuclear membrane in patients with Emery- Dreifuss muscular dystrophy. Nat. Genet. 1996, 12, 254–259. [Google Scholar] [CrossRef] [PubMed]
- Tunnah, D.; Sewry, C.A.; Vaux, D.; Schirmer, E.C.; Morris, G.E. The apparent absence of lamin B1 and emerin in many tissue nuclei is due to epitope masking. J. Mol. Histol. 2005, 36, 337–344. [Google Scholar] [CrossRef] [PubMed]
- Holaska, J.M.; Kowalski, A.M.; Wilson, K.L. Emerin caps the pointed end of actin filaments: Evidence for an actin cortical network at the nuclear inner membrane. PLoS Biol. 2004, 2, 1354–1362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holaska, J.M.; Rais-Bahrami, S.; Wilson, K.L. Lmo7 is an emerin-binding protein that regulates the transcription of emerin and many other muscle-relevant genes. Hum. Mol. Genet. 2006, 15, 3459–3472. [Google Scholar] [CrossRef] [PubMed]
- Holaska, J.M.; Wilson, K.L. Multiple roles for emerin: Implications for Emery-Dreifuss muscular dystrophy. Anat. Rec. A Discov. Mol. Cell. Evol. Biol. 2006, 288, 676–680. [Google Scholar] [CrossRef] [PubMed]
- Holaska, J.M.; Wilson, K.L. An emerin “proteome”: Purification of distinct emerin-containing complexes from HeLa cells suggests molecular basis for diverse roles including gene regulation, mRNA splicing, signaling, mechanosensing, and nuclear architecture. Biochemistry 2007, 46, 8897–8908. [Google Scholar] [CrossRef] [PubMed]
- Demmerle, J.; Koch, A.J.; Holaska, J.M. The Nuclear Envelope Protein Emerin Binds Directly to Histone Deacetylase 3 (HDAC3) and Activates HDAC3 Activity. J. Biol. Chem. 2012, 287, 22080–22088. [Google Scholar] [CrossRef] [PubMed]
- Markiewicz, E.; Tilgner, K.; Barker, N.; van de Wetering, M.; Clevers, H.; Dorobek, M.; Hausmanowa-Petrusewicz, I.; Ramaekers, F.C.; Broers, J.L.; Blankesteijn, W.M.; et al. The inner nuclear membrane protein emerin regulates beta-catenin activity by restricting its accumulation in the nucleus. EMBO J. 2006, 25, 3275–3285. [Google Scholar] [CrossRef] [PubMed]
- Haraguchi, T.; Holaska, J.M.; Yamane, M.; Wilson, K.L.; Hiraoka, Y. Emerin binding to Btf, a death-promoting transcriptional repressor, is disrupted by a missense mutation that causes Emery-Dreifuss muscular dystrophy. Eur. J. Biochem. 2004, 271, 1035–1045. [Google Scholar] [CrossRef] [PubMed]
- Dedeic, Z.; Cetera, M.; Cohen, T.V.; Holaska, J.M. Emerin inhibits Lmo7 binding to the Pax3 and MyoD promoters and expression of myoblast proliferation genes. J. Cell. Sci. 2011, 124, 1691–1702. [Google Scholar] [CrossRef] [PubMed]
- Massague, J.; Cheifetz, S.; Endo, T.; Nadal-Ginard, B. Type beta transforming growth factor is an inhibitor of myogenic differentiation. Proc. Natl. Acad. Sci. USA 1986, 83, 8206–8210. [Google Scholar] [CrossRef] [PubMed]
- Polesskaya, A.; Seale, P.; Rudnicki, M.A. Wnt signaling induces the myogenic specification of resident CD45+ adult stem cells during muscle regeneration. Cell 2003, 113, 841–852. [Google Scholar] [CrossRef]
- Ridgeway, A.G.; Petropoulos, H.; Wilton, S.; Skerjanc, I.S. Wnt signaling regulates the function of MyoD and myogenin. J. Biol. Chem. 2000, 275, 32398–32405. [Google Scholar] [CrossRef] [PubMed]
- Conboy, I.M.; Rando, T.A. The regulation of Notch signaling controls satellite cell activation and cell fate determination in postnatal myogenesis. Dev. Cell 2002, 3, 397–409. [Google Scholar] [CrossRef]
- Edwall, D.; Schalling, M.; Jennische, E.; Norstedt, G. Induction of insulin-like growth factor I messenger ribonucleic acid during regeneration of rat skeletal muscle. Endocrinology 1989, 124, 820–825. [Google Scholar] [CrossRef] [PubMed]
- Jennische, E.; Hansson, H.A. Regenerating skeletal muscle cells express insulin-like growth factor I. Acta Physiol. Scand. 1987, 130, 327–332. [Google Scholar] [CrossRef] [PubMed]
- Jennische, E.; Skottner, A.; Hansson, H.A. Satellite cells express the trophic factor IGF-I in regenerating skeletal muscle. Acta Physiol. Scand. 1987, 129, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Chang, N.C.; Rudnicki, M.A. Satellite cells: The architects of skeletal muscle. Curr. Top. Dev. Biol. 2014, 107, 161–181. [Google Scholar] [PubMed]
- Brack, A.S.; Conboy, I.M.; Conboy, M.J.; Shen, J.; Rando, T.A. A temporal switch from notch to Wnt signaling in muscle stem cells is necessary for normal adult myogenesis. Cell Stem Cell 2008, 2, 50–59. [Google Scholar] [CrossRef] [PubMed]
- Segales, J.; Perdiguero, E.; Munoz-Canoves, P. Regulation of Muscle Stem Cell Functions: A Focus on the p38 MAPK Signaling Pathway. Front. Cell Dev. Biol. 2016, 4, 91. [Google Scholar] [CrossRef] [PubMed]
- Rosenthal, S.M.; Cheng, Z.Q. Opposing early and late effects of insulin-like growth factor I on differentiation and the cell cycle regulatory retinoblastoma protein in skeletal myoblasts. Proc. Natl. Acad. Sci. USA 1995, 92, 10307–10311. [Google Scholar] [CrossRef] [PubMed]
- Muchir, A.; Pavlidis, P.; Bonne, G.; Hayashi, Y.K.; Worman, H.J. Activation of MAPK in hearts of EMD null mice: Similarities between mouse models of X-linked and autosomal dominant Emery Dreifuss muscular dystrophy. Hum. Mol. Genet. 2007, 16, 1884–1895. [Google Scholar] [CrossRef] [PubMed]
- Muchir, A.; Wu, W.; Worman, H.J. Reduced expression of A-type lamins and emerin activates extracellular signal-regulated kinase in cultured cells. Biochim. Biophys. Acta 2009, 1792, 75–81. [Google Scholar] [CrossRef] [PubMed]
- Mozzetta, C.; Consalvi, S.; Saccone, V.; Forcales, S.V.; Puri, P.L.; Palacios, D. Selective control of Pax7 expression by TNF-activated p38alpha/polycomb repressive complex 2 (PRC2) signaling during muscle satellite cell differentiation. Cell Cycle 2011, 10, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Palacios, D.; Mozzetta, C.; Consalvi, S.; Caretti, G.; Saccone, V.; Proserpio, V.; Marquez, V.E.; Valente, S.; Mai, A.; Forcales, S.V.; et al. TNF/p38alpha/polycomb signaling to Pax7 locus in satellite cells links inflammation to the epigenetic control of muscle regeneration. Cell Stem Cell 2010, 7, 455–469. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Woodring, P.J.; Bhakta, K.S.; Tamura, K.; Wen, F.; Feramisco, J.R.; Karin, M.; Wang, J.Y.; Puri, P.L. p38 and extracellular signal-regulated kinases regulate the myogenic program at multiple steps. Mol. Cell. Biol. 2000, 20, 3951–3964. [Google Scholar] [CrossRef] [PubMed]
- Favreau, C.; Delbarre, E.; Courvalin, J.C.; Buendia, B. Differentiation of C2C12 myoblasts expressing lamin A mutated at a site responsible for Emery-Dreifuss muscular dystrophy is improved by inhibition of the MEK-ERK pathway and stimulation of the PI3-kinase pathway. Exp. Cell Res. 2008, 314, 1392–1405. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Roberts, A.; Goff, L.; Pertea, G.; Kim, D.; Kelley, D.R.; Pimentel, H.; Salzberg, S.L.; Rinn, J.L.; Pachter, L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 2012, 7, 562–578. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Hendrickson, D.G.; Sauvageau, M.; Goff, L.; Rinn, J.L.; Pachter, L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 2013, 31, 46–53. [Google Scholar] [CrossRef] [PubMed]
- Moseley, J.B.; Nurse, P. Cdk1 and cell morphology: Connections and directions. Curr. Opin. Cell Biol. 2009, 21, 82–88. [Google Scholar] [CrossRef] [PubMed]
- Chiou, J.G.; Balasubramanian, M.K.; Lew, D.J. Cell Polarity in Yeast. Annu. Rev. Cell Dev. Biol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Rizzino, A.; Wuebben, E.L. Sox2/Oct4: A delicately balanced partnership in pluripotent stem cells and embryogenesis. Biochim. Biophys. Acta 2016, 1859, 780–791. [Google Scholar] [CrossRef] [PubMed]
- Onichtchouk, D.; Driever, W. Zygotic Genome Activators, Developmental Timing, and Pluripotency. Curr. Top. Dev. Biol. 2016, 116, 273–297. [Google Scholar] [PubMed]
- Bryan, B.A.; Walshe, T.E.; Mitchell, D.C.; Havumaki, J.S.; Saint-Geniez, M.; Maharaj, A.S.; Maldonado, A.E.; D’Amore, P.A. Coordinated vascular endothelial growth factor expression and signaling during skeletal myogenic differentiation. Mol. Biol. Cell 2008, 19, 994–1006. [Google Scholar] [CrossRef] [PubMed]
- Deasy, B.M.; Feduska, J.M.; Payne, T.R.; Li, Y.; Ambrosio, F.; Huard, J. Effect of VEGF on the regenerative capacity of muscle stem cells in dystrophic skeletal muscle. Mol. Ther. 2009, 17, 1788–1798. [Google Scholar] [CrossRef] [PubMed]
- Barre, B.; Avril, S.; Coqueret, O. Opposite regulation of myc and p21waf1 transcription by STAT3 proteins. J. Biol. Chem. 2003, 278, 2990–2996. [Google Scholar] [CrossRef] [PubMed]
- Kostyo, J.L. Rapid effects of growth hormone on amino acid transport and protein synthesis. Ann. N. Y. Acad. Sci. 1968, 148, 389–407. [Google Scholar] [CrossRef] [PubMed]
- Allen, R.E.; Boxhorn, L.K. Inhibition of skeletal muscle satellite cell differentiation by transforming growth factor-beta. J. Cell. Physiol. 1987, 133, 567–572. [Google Scholar] [CrossRef] [PubMed]
- Gardner, S.; Alzhanov, D.; Knollman, P.; Kuninger, D.; Rotwein, P. TGF-beta inhibits muscle differentiation by blocking autocrine signaling pathways initiated by IGF-II. Mol. Endocrinol. 2011, 25, 128–137. [Google Scholar] [CrossRef] [PubMed]
- Schabort, E.J.; van der Merwe, M.; Loos, B.; Moore, F.P.; Niesler, C.U. TGF-beta’s delay skeletal muscle progenitor cell differentiation in an isoform-independent manner. Exp. Cell Res. 2009, 315, 373–384. [Google Scholar] [CrossRef] [PubMed]
- Archambault, V.; Lepine, G.; Kachaner, D. Understanding the Polo Kinase machine. Oncogene 2015, 34, 4799–4807. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Jiang, Q.; Zhang, C. The role of mitotic kinases in coupling the centrosome cycle with the assembly of the mitotic spindle. J. Cell. Sci. 2014, 127, 4111–4122. [Google Scholar] [CrossRef] [PubMed]
- Cole, F.; Zhang, W.; Geyra, A.; Kang, J.S.; Krauss, R.S. Positive regulation of myogenic bHLH factors and skeletal muscle development by the cell surface receptor CDO. Dev. Cell 2004, 7, 843–854. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.S.; Yi, M.J.; Zhang, W.; Feinleib, J.L.; Cole, F.; Krauss, R.S. Netrins and neogenin promote myotube formation. J. Cell Biol. 2004, 167, 493–504. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Bae, G.U.; Leem, Y.E.; Choi, H.K.; Kang, T.M.; Cho, H.; Kim, S.T.; Kang, J.S. Phosphorylation of Stim1 at serine 575 via netrin-2/Cdo-activated ERK1/2 is critical for the promyogenic function of Stim1. Mol. Biol. Cell 2012, 23, 1376–1387. [Google Scholar] [CrossRef] [PubMed]
- Burkin, D.J.; Wallace, G.Q.; Nicol, K.J.; Kaufman, D.J.; Kaufman, S.J. Enhanced expression of the alpha 7 beta 1 integrin reduces muscular dystrophy and restores viability in dystrophic mice. J. Cell Biol. 2001, 152, 1207–1218. [Google Scholar] [CrossRef] [PubMed]
- Marshall, J.L.; Chou, E.; Oh, J.; Kwok, A.; Burkin, D.J.; Crosbie-Watson, R.H. Dystrophin and utrophin expression require sarcospan: Loss of alpha7 integrin exacerbates a newly discovered muscle phenotype in sarcospan-null mice. Hum. Mol. Genet. 2012, 21, 4378–4393. [Google Scholar] [CrossRef] [PubMed]
- Xiao, J.; Jethanandani, P.; Ziober, B.L.; Kramer, R.H. Regulation of alpha7 integrin expression during muscle differentiation. J. Biol. Chem. 2003, 278, 49780–49788. [Google Scholar] [CrossRef] [PubMed]
- Benavides Damm, T.; Egli, M. Calcium’s role in mechanotransduction during muscle development. Cell. Physiol. Biochem. 2014, 33, 249–272. [Google Scholar] [CrossRef] [PubMed]
- Muchir, A.; Shan, J.; Bonne, G.; Lehnart, S.E.; Worman, H.J. Inhibition of extracellular signal-regulated kinase signaling to prevent cardiomyopathy caused by mutation in the gene encoding A-type lamins. Hum. Mol. Genet. 2009, 18, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Dialynas, G.; Shrestha, O.K.; Ponce, J.M.; Zwerger, M.; Thiemann, D.A.; Young, G.H.; Moore, S.A.; Yu, L.; Lammerding, J.; Wallrath, L.L. Myopathic lamin mutations cause reductive stress and activate the nrf2/keap-1 pathway. PLoS Genet. 2015, 11, e1005231. [Google Scholar] [CrossRef] [PubMed]
- Webster, M.T.; Manor, U.; Lippincott-Schwartz, J.; Fan, C.M. Intravital Imaging Reveals Ghost Fibers as Architectural Units Guiding Myogenic Progenitors during Regeneration. Cell Stem Cell 2016, 18, 243–252. [Google Scholar] [CrossRef] [PubMed]
- Baghdadi, M.B.; Tajbakhsh, S. Regulation and phylogeny of skeletal muscle regeneration. Dev. Biol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Schiaffino, S.; Dyar, K.A.; Ciciliot, S.; Blaauw, B.; Sandri, M. Mechanisms regulating skeletal muscle growth and atrophy. FEBS J. 2013, 280, 4294–4314. [Google Scholar] [CrossRef] [PubMed]
- Paris, N.D.; Soroka, A.; Klose, A.; Liu, W.; Chakkalakal, J.V. Smad4 restricts differentiation to promote expansion of satellite cell derived progenitors during skeletal muscle regeneration. Elife 2016, 5. [Google Scholar] [CrossRef] [PubMed]
- Rossi, G.; Antonini, S.; Bonfanti, C.; Monteverde, S.; Vezzali, C.; Tajbakhsh, S.; Cossu, G.; Messina, G. Nfix Regulates Temporal Progression of Muscle Regeneration through Modulation of Myostatin Expression. Cell Rep. 2016, 14, 2238–2249. [Google Scholar] [CrossRef] [PubMed]
- Chazaud, B.; Sonnet, C.; Lafuste, P.; Bassez, G.; Rimaniol, A.C.; Poron, F.; Authier, F.J.; Dreyfus, P.A.; Gherardi, R.K. Satellite cells attract monocytes and use macrophages as a support to escape apoptosis and enhance muscle growth. J. Cell Biol. 2003, 163, 1133–1143. [Google Scholar] [CrossRef] [PubMed]
- Saclier, M.; Cuvellier, S.; Magnan, M.; Mounier, R.; Chazaud, B. Monocyte/macrophage interactions with myogenic precursor cells during skeletal muscle regeneration. FEBS J. 2013, 280, 4118–4130. [Google Scholar] [CrossRef] [PubMed]
- Saclier, M.; Yacoub-Youssef, H.; Mackey, A.L.; Arnold, L.; Ardjoune, H.; Magnan, M.; Sailhan, F.; Chelly, J.; Pavlath, G.K.; Mounier, R.; et al. Differentially activated macrophages orchestrate myogenic precursor cell fate during human skeletal muscle regeneration. Stem Cells 2013, 31, 384–396. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christov, C.; Chretien, F.; Abou-Khalil, R.; Bassez, G.; Vallet, G.; Authier, F.J.; Bassaglia, Y.; Shinin, V.; Tajbakhsh, S.; Chazaud, B.; et al. Muscle satellite cells and endothelial cells: Close neighbors and privileged partners. Mol. Biol. Cell 2007, 18, 1397–1409. [Google Scholar] [CrossRef] [PubMed]
- Rhoads, R.P.; Johnson, R.M.; Rathbone, C.R.; Liu, X.; Temm-Grove, C.; Sheehan, S.M.; Hoying, J.B.; Allen, R.E. Satellite cell-mediated angiogenesis in vitro coincides with a functional hypoxia-inducible factor pathway. Am. J. Physiol. Cell Physiol. 2009, 296, C1321–C1328. [Google Scholar] [CrossRef] [PubMed]
- Price, F.D.; von Maltzahn, J.; Bentzinger, C.F.; Dumont, N.A.; Yin, H.; Chang, N.C.; Wilson, D.H.; Frenette, J.; Rudnicki, M.A. Inhibition of JAK-STAT signaling stimulates adult satellite cell function. Nat. Med. 2014, 20, 1174–1181. [Google Scholar] [CrossRef] [PubMed]
- Tierney, M.T.; Aydogdu, T.; Sala, D.; Malecova, B.; Gatto, S.; Puri, P.L.; Latella, L.; Sacco, A. STAT3 signaling controls satellite cell expansion and skeletal muscle repair. Nat. Med. 2014, 20, 1182–1186. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Ma, K.; Wang, H.; Xiao, F.; Gao, Y.; Zhang, W.; Wang, K.; Gao, X.; Ip, N.; Wu, Z. JAK1-STAT1-STAT3, a key pathway promoting proliferation and preventing premature differentiation of myoblasts. J. Cell Biol. 2007, 179, 129–138. [Google Scholar] [CrossRef] [PubMed]
- Ooshio, T.; Irie, K.; Morimoto, K.; Fukuhara, A.; Imai, T.; Takai, Y. Involvement of LMO7 in the association of two cell-cell adhesion molecules, nectin and E-cadherin, through afadin and alpha-actinin in epithelial cells. J. Biol. Chem. 2004, 279, 31365–31373. [Google Scholar] [CrossRef] [PubMed]
- Holaska, J.; Lee, K.; Kowalski, A.; Wilson, K. Transcriptional repressor germ cell-less (GCL) and barrier-to-autointegration factor (BAF) compete for binding to emerin in vitro. J. Biol. Chem. 2003, 278, 6969–6975. [Google Scholar] [CrossRef] [PubMed]
- Chen, I.H.; Huber, M.; Guan, T.; Bubeck, A.; Gerace, L. Nuclear envelope transmembrane proteins (NETs) that are up-regulated during myogenesis. BMC Cell Biol. 2006, 7, 38. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.H.; Guan, T.; Datta, K.; Coppinger, J.; Yates, J., 3rd; Gerace, L. Regulation of myoblast differentiation by the nuclear envelope protein NET39. Mol. Cell. Biol. 2009, 29, 5800–5812. [Google Scholar] [CrossRef] [PubMed]
- Robson, M.I.; de Las Heras, J.I.; Czapiewski, R.; Le Thanh, P.; Booth, D.G.; Kelly, D.A.; Webb, S.; Kerr, A.R.; Schirmer, E.C. Tissue-Specific Gene Repositioning by Muscle Nuclear Membrane Proteins Enhances Repression of Critical Developmental Genes during Myogenesis. Mol. Cell. 2016, 62, 834–847. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Not available |



| Gene | Forward (5’-3’) | Reverse (5’-3’) |
|---|---|---|
| Cenph | ACATACATTCCAGGGCCTTATT | CTGCAGAGGATGCCACTTTA |
| Mad2l1 | AAGTCCGTCTACGCTCATTTAC | CTCAGACAAGTCCAGGAAGAAC |
| Bub1 | TGGGTTCTTTGCTGGTCTATG | CCCTACTAATATGCTGCCATTCT |
| Prc1 | CCTCTTCTGGTGTGCAGAAATA | CAAGAAACCCTCACTGGGATAG |
| Ezh2 | CAGCTCAAGAGGTTCAGAAGAG | GGGCGACCAAGAGTACATTATAG |
| Pparg | CTGGCCTCCCTGATGAATAAAG | AGGCTCCATAAAGTCACCAAAG |
| Mgst1 | ACCGCATTCCAGAGGATAAC | CGTCAGTGCGAACAAACTTC |
| Cyp1a1 | GTGAGCAAGGAGGCTAACTATC | GGCTACTGACACGACCAAATA |
| Gstp1 | GAGACCTCACCCTTTACCAATC | CCCATCATTCACCATATCCATCT |
| Gapdh | AACATTGGCATTGTGGAAGGGCTC | TGGAAGAGTGGGAGTTGCTGTTGA |
| Oaz1 | GAGCTGAATGCTGTGTTTGTC | AGGTCACCTGACCATCTTAAAC |
| Canonical Pathways Identified Unique to Wildtype Progenitors during Transition from Day 0 to Day 1 of Differentiation | Canonical Pathways Identified Unique to Emerin-Null Progenitors during Transition from Day 0 to Day 1 of Differentiation | Canonical Pathways Identified Unique to Wildtype Progenitors during Transition from Day 1 to Day 2 of Differentiation | Canonical Pathways Identified Unique to Emerin-Null Progenitors during Transition from Day 1 to Day 2 of Differentiation |
|---|---|---|---|
| OX40 Signaling Pathway | Growth Hormone Signaling | Cell cycle control of chromosomal replication | Netrin signaling |
| (p-value: <0.001) | (p-value: <0.001) | (p-value: <0.05) | (p-value: <0.001) |
| Cdc42 Signaling | STAT3 pathway | Cell cycle: G2/M DNA Damage Checkpoint Regulation | Calcium Signaling |
| (p-value: <0.001) | (p-value: <0.01) | (p-value: <0.05) | (p-value: <0.001) |
| Epoxysqualene Biosynthesis | TGF β Signaling | Actin Cytoskeleton Signaling | Actin cytoskeleton signaling |
| (p-value: <0.001) | (p-value: <0.01) | (p-value: <0.05) | (p-value: <0.001) |
| Glycine Biosynthesis I | EGF Signaling | Mitotic Roles of Polo-Like Kinase | Mitotic Roles of Polo-Like Kinase |
| (p-value: <0.05) | (p-value: <0.01) | (p-value: <0.05) | (p-value: <0.001) |
| Human Embryonic Stem Cell Pluripotency | HIPPO Pathway | ILK signaling | Axonal guidance signaling |
| (p-value: <0.05) | (p-value: <0.05) | (p-value: <0.05) | (p-value: <0.01) |
| Transcriptional Regulatory in Embryonic Stem cells | Glutamate Signaling | TGF β Signaling | Integrin signaling |
| (p-value: <0.05) | (p-value: <0.05) | (p-value: <0.05) | (p-value: <0.05) |
| IGF-1 Signaling | |||
| (p-value: <0.05) | |||
| VEGF Signaling | |||
| (p-value: <0.05) |
| Canonical Pathway Identified for Genes Common between Wildtype and Emerin-Null Cells during Transition from Day 0 to Day 1 of Differentiation | Canonical Pathway Identified for Genes Common between Wildtype and Emerin-Null Cells during Transition from Day 1 to Day 2 of Differentiation |
|---|---|
| cell cycle control of chromosomal replication | cell cycle control of chromosomal replication |
| (p-value: <0.001) | (p-value: <0.001) |
| ILK signaling | ATM signaling |
| (p-value: <0.001) | (p-value: <0.001) |
| Integrin signaling | Mitotic roles of polo-like kinase |
| (p-value: <0.01) | (p-value: <0.001) |
| Cell Cycle G1/S checkpoint regulation | Cell Cycle G2/M DNA damage checkpoint regulation |
| (p-value: <0.01) | (p-value: <0.001) |
| P38 MAPK signaling | Estrogen mediated S phase entry |
| (p-value: <0.01) | (p-value: <0.001) |
| Cdc42 Signaling | Role of CHK Proteins in Cell Cycle checkpoint control |
| (p-value: <0.01) | (p-value: <0.001) |
| Mitotic roles of polo-like kinase | Cyclins and cell cycle Regulation |
| (p-value: <0.01) | (p-value: <0.001) |
| p53 signaling | p53 signaling |
| (p-value: <0.05) | (p-value: <0.001) |
| PAK signaling | Aryl Hydrocarbon Receptor Signaling |
| (p-value: <0.05) | (p-value: <0.001) |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Iyer, A.; Koch, A.J.; Holaska, J.M. Expression Profiling of Differentiating Emerin-Null Myogenic Progenitor Identifies Molecular Pathways Implicated in Their Impaired Differentiation. Cells 2017, 6, 38. https://doi.org/10.3390/cells6040038
Iyer A, Koch AJ, Holaska JM. Expression Profiling of Differentiating Emerin-Null Myogenic Progenitor Identifies Molecular Pathways Implicated in Their Impaired Differentiation. Cells. 2017; 6(4):38. https://doi.org/10.3390/cells6040038
Chicago/Turabian StyleIyer, Ashvin, Adam J. Koch, and James M. Holaska. 2017. "Expression Profiling of Differentiating Emerin-Null Myogenic Progenitor Identifies Molecular Pathways Implicated in Their Impaired Differentiation" Cells 6, no. 4: 38. https://doi.org/10.3390/cells6040038




