Diverse Roads Taken by 13C-Glucose-Derived Metabolites in Breast Cancer Cells Exposed to Limiting Glucose and Glutamine Conditions
Abstract
1. Introduction
2. Materials and Method
2.1. Cell Culture
2.2. Cell Cultures and Preparation of Cell Lysates for Biochemical Analyses
2.3. Cell Number Determination
2.4. Glucose Determination
2.5. Lactate Determinations of Cells and Culture Supernatants
2.6. Determination of Enzyme Activities
2.7. 13-C-Labeling Experiments
2.8. Extraction of Polar Metabolites
2.9. Data Analysis
3. Results
3.1. Changes in the Warburg Effect in Limiting Nutrient Conditions
3.1.1. Cell Growth
3.1.2. Glucose Consumption
3.1.3. Intracellular Lactate Concentrations and Lactate Released
3.1.4. Activities of Pyruvate Kinase and LDH
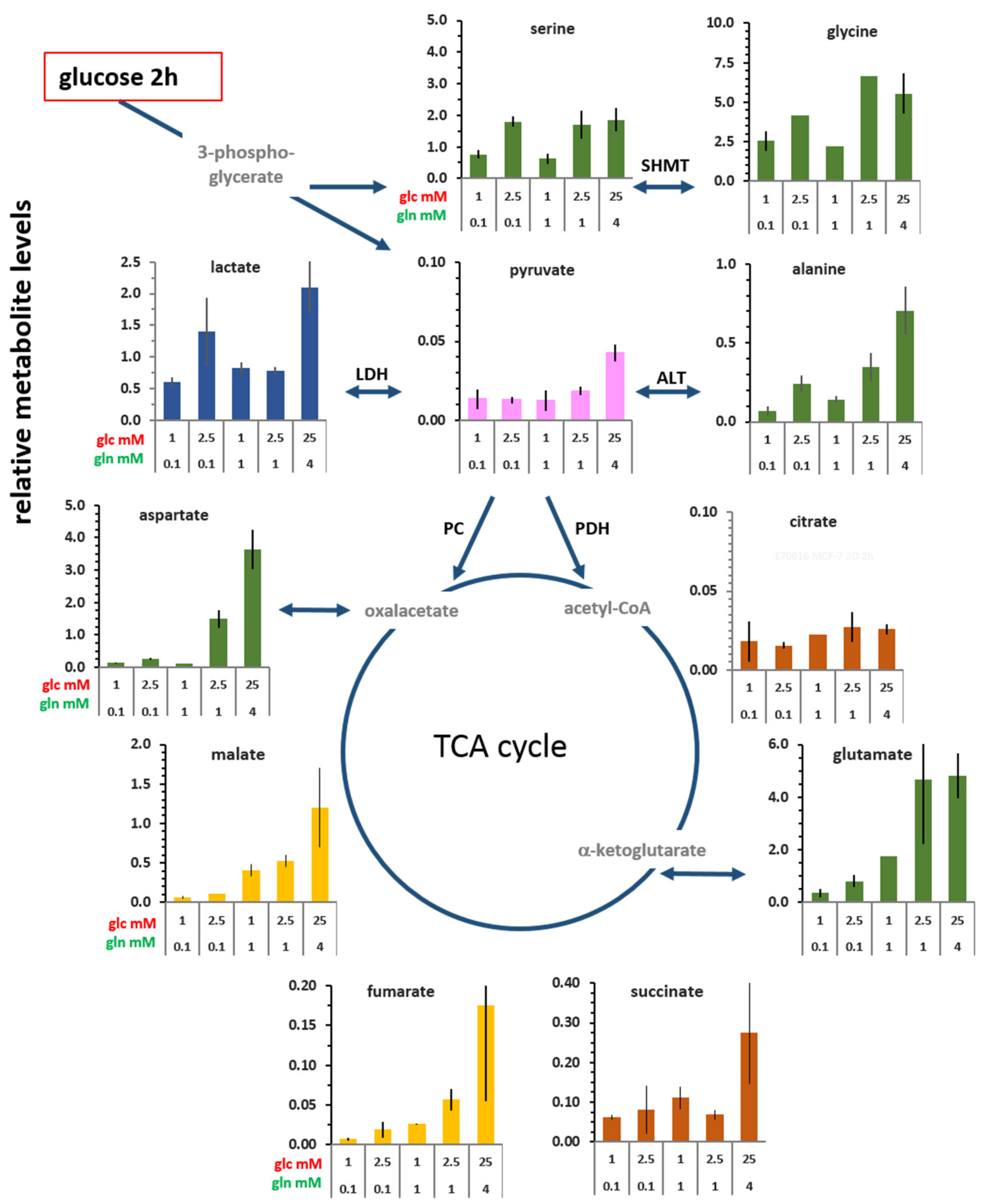
3.2. Changes in the Levels of Intracellular Metabolites
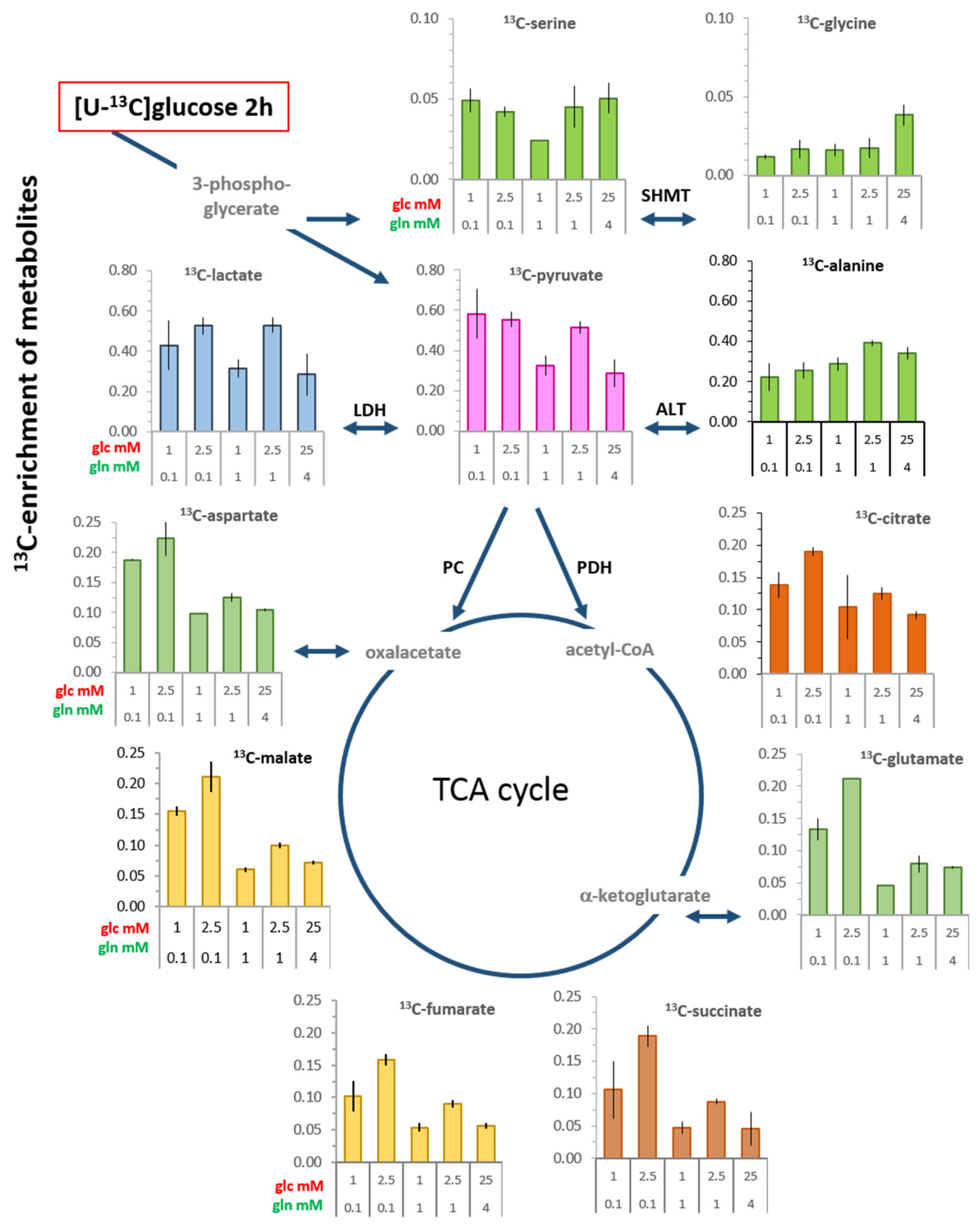
3.3. 13C-enrichment of Glucose-Derived Metabolites in Limiting Nutrient Conditions
3.3.1. Glycolysis-Associated Metabolites
3.3.2. TCA Cycle-Associated Metabolites
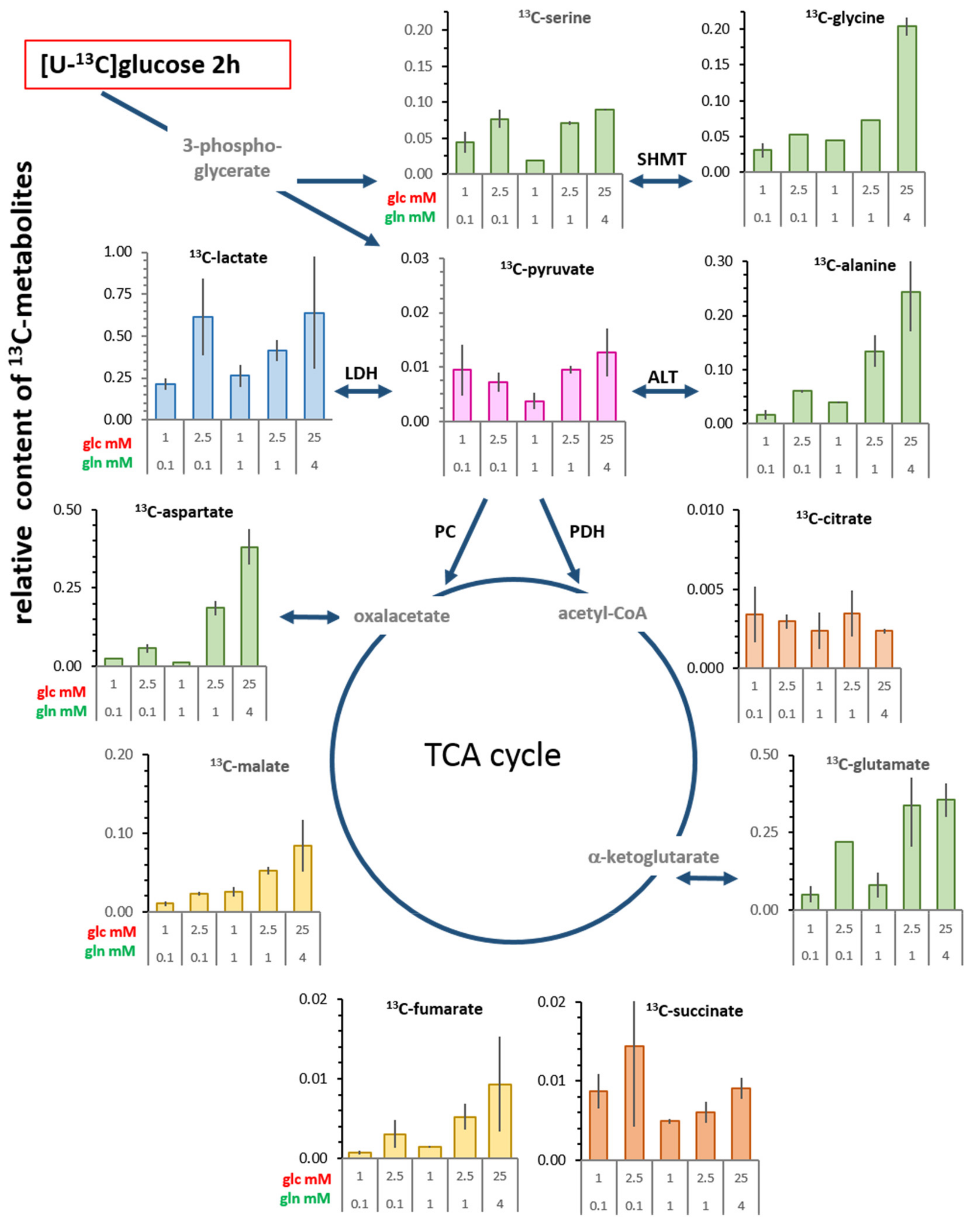
3.4. Changes in Levels of Intracellular 13C-metabolites
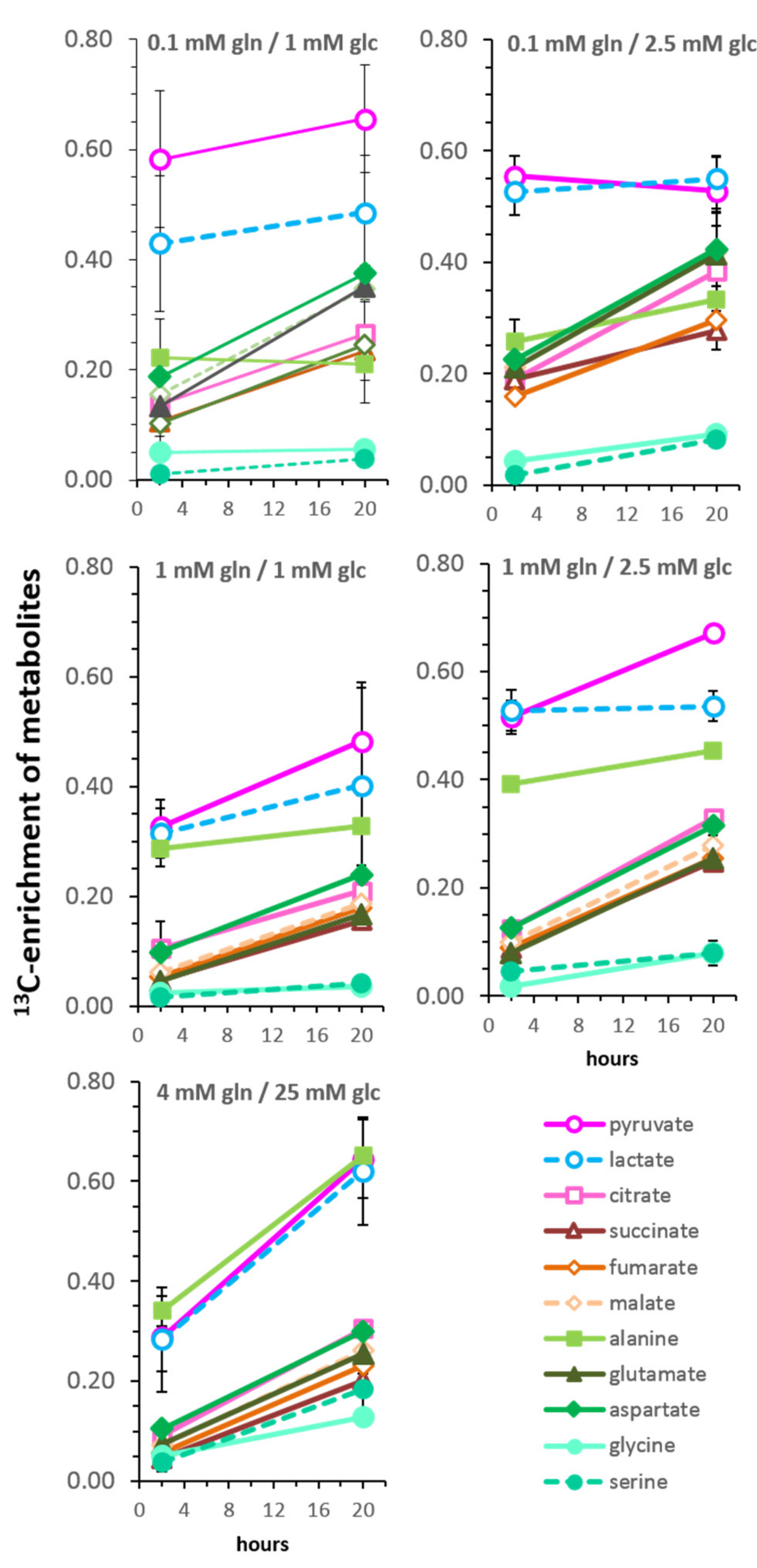
3.5. Kinetics of 13C-Enrichment and Evidence for Metabolic Reaction Compartments
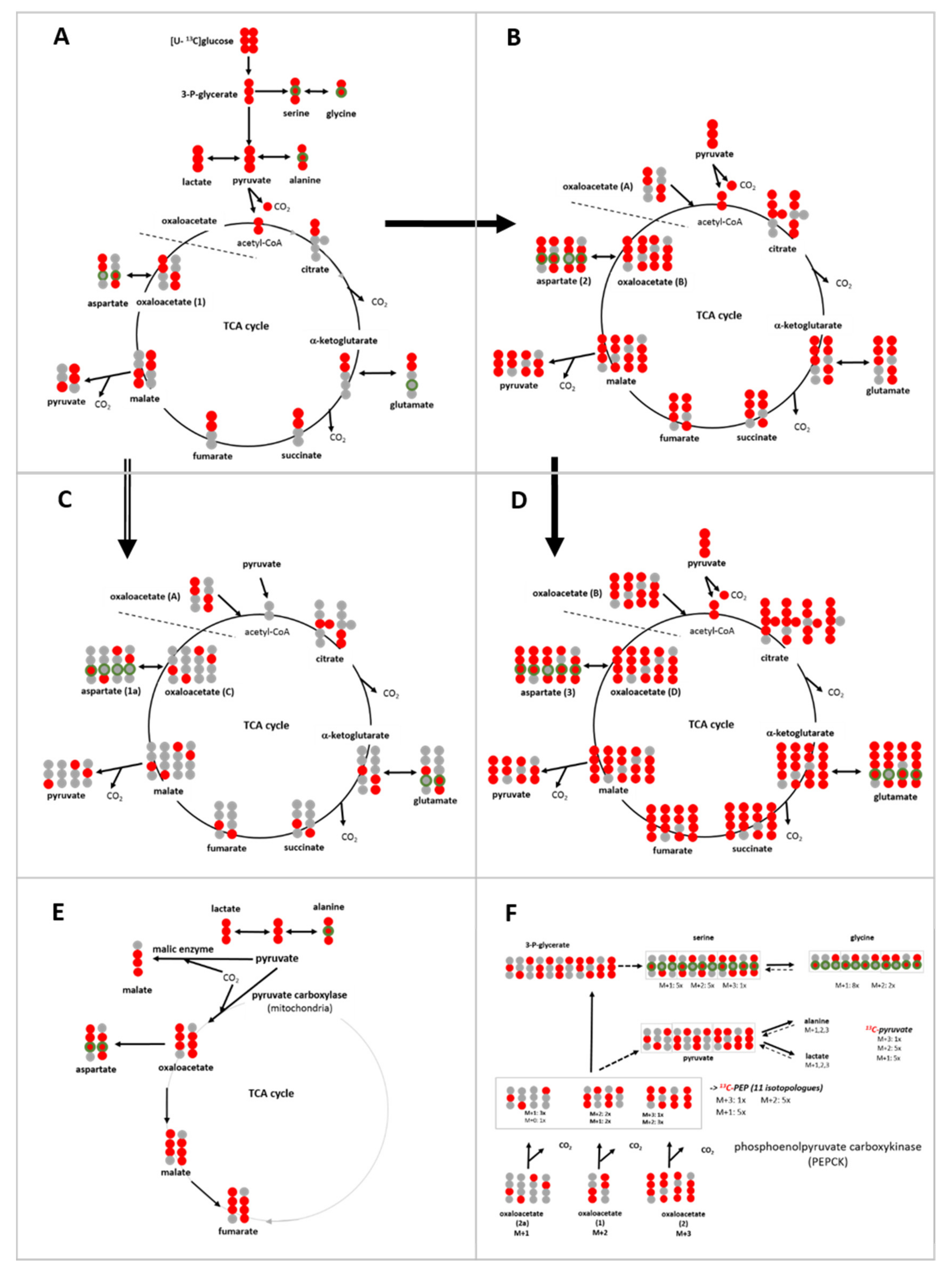
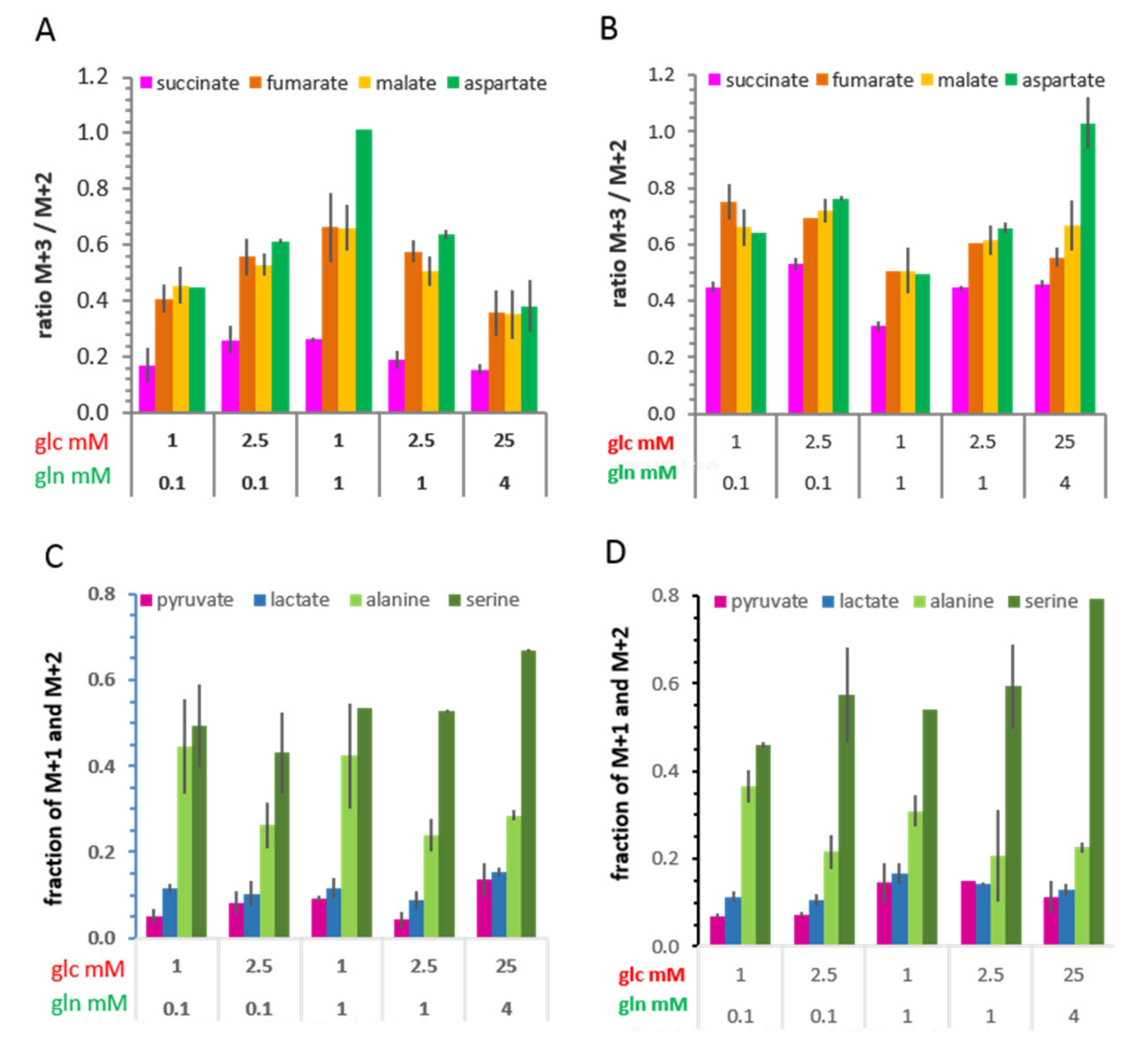
3.6. Isotopologue Profiles and Plasticity of Engaged Metabolic Pathways
3.6.1. 13C-Metabolites Related to Glycolysis
3.6.2. 13C-Metabolites Related to the TCA cycle and Pyruvate Anaplerosis
3.6.3. 13C-Metabolites Related to Gluconeogenesis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef] [PubMed]
- Vander Heiden, M.G.; DeBerardinis, R.J. Understanding the Intersections between Metabolism and Cancer Biology. Cell 2017, 168, 657–669. [Google Scholar] [CrossRef] [PubMed]
- Warburg, O.; Minami, S. Versuche an überlebendem Carcinomgewebe. Klin. Wochenschr. 1923, 2, 777–778. [Google Scholar] [CrossRef]
- Otto, A.M. Warburg effect(s)—A biographical sketch of Otto Warburg and his impacts on tumor metabolism. Cancer Metab. 2016, 4, 1–8. [Google Scholar] [CrossRef]
- Hosios, A.M.; Hecht, V.C.; Danai, L.V.; Johnson, M.O.; Rathmell, J.C.; Steinhauser, M.L.; Manalis, S.R.; Vander Heiden, M.G. Amino Acids Rather than Glucose Account for the Majority of Cell Mass in Proliferating Mammalian Cells. Dev. Cell 2016, 36, 540–549. [Google Scholar] [CrossRef] [PubMed]
- Daly, M.E.; Vale, C.; Walker, M.; Littlefield, A.; Alberti, K.G.; Mathers, J.C. Acute effects on insulin sensitivity and diurnal metabolic profiles of a high-sucrose compared with a high-starch diet. Am. J. Clin. Nutr. 1998, 67, 1186–1196. [Google Scholar] [CrossRef]
- Stein, W.H.; Moore, S. The Free Amino Acids of Human Blood Plasma. J. Biol. Chem. 1954, 211, 915–926. [Google Scholar]
- Felig, P.; Wahren, J.; Raf, L. Evidence of inter-organ amino-acid transport by blood cells in humans. Proc. Natl. Acad. Sci. USA 1973, 70, 1775–1779. [Google Scholar] [CrossRef]
- Gullino, P.M.; Clark, S.H.; Grantham, F.H. The interstitial fluid of solid tumors. Cancer Res. 1964, 24, 780–794. [Google Scholar]
- Schaefer, C.; Mayer, W.K.; Kruger, W.; Vaupel, P. Microregional distributions of glucose, lactate, ATP and tissue pH in experimental tumours upon local hyperthermia and/or hyperglycaemia. J. Cancer Res. Clin. Oncol. 1993, 119, 599–608. [Google Scholar] [CrossRef]
- Schroeder, T.; Yuan, H.; Viglianti, B.L.; Peltz, C.; Asopa, S.; Vujaskovic, Z.; Dewhirst, M.W. Spatial heterogeneity and oxygen dependence of glucose consumption in R3230Ac and fibrosarcomas of the Fischer 344 rat. Cancer Res. 2005, 65, 5163–5171. [Google Scholar] [CrossRef] [PubMed]
- Vaupel, P. Pathophysiology of solid tumors. In The Impact of Tumor biology on Cancer Treatment and Multidisciplinary Strategies; Molls, M., Vaupel, P., Nieder, C., Anscher, M.S., Eds.; Springer: Berlin/Heidelberg, Germany, 2009; pp. 52–85. [Google Scholar]
- Kallinowski, F.; Runkel, S.; Fortmeyer, H.P.; Forster, H.; Vaupel, P. L-glutamine: A major substrate for tumor cells in vivo? J. Cancer Res. Clin. Oncol. 1987, 113, 209–215. [Google Scholar] [CrossRef] [PubMed]
- Yuneva, M. Finding an “Achilles’ heel” of cancer: The role of glucose and glutamine metabolism in the survival of transformed cells. Cell Cycle 2008, 7, 2083–2089. [Google Scholar] [CrossRef] [PubMed]
- Chen, V.; Shtivelman, E. CC3/TIP30 regulates metabolic adaptation of tumor cells to glucose limitation. Cell Cycle 2010, 9, 4941–4953. [Google Scholar] [CrossRef] [PubMed]
- Gaglio, D.; Metallo, C.M.; Gameiro, P.A.; Hiller, K.; Danna, L.S.; Balestrieri, C.; Alberghina, L.; Stephanopoulos, G.; Chiaradonna, F. Oncogenic K-Ras decouples glucose and glutamine metabolism to support cancer cell growth. Mol. Syst. Biol. 2011, 7. [Google Scholar] [CrossRef] [PubMed]
- Otto, A.M.; Hintermair, J.; Janzon, C. NADH-Linked Metabolic Plasticity of MCF-7 Breast Cancer Cells Surviving in a Nutrient-Deprived Microenvironment. J. Cell Biochem. 2015, 116, 822–835. [Google Scholar] [CrossRef] [PubMed]
- Buescher, J.M.; Antoniewicz, M.R.; Boros, L.G.; Burgess, S.C.; Brunengraber, H.; Clish, C.B.; DeBerardinis, R.J.; Feron, O.; Frezza, C.; Ghesquiere, B.; et al. A roadmap for interpreting 13C metabolite labeling patterns from cells. Curr. Opin. Biotechnol. 2015, 34, 189–201. [Google Scholar] [CrossRef] [PubMed]
- Dong, W.; Keibler, M.A.; Stephanopoulos, G. Review of metabolic pathways activated in cancer cells as determined through isotopic labeling and network analysis. Metab. Eng. 2017, 43, 113–124. [Google Scholar] [CrossRef] [PubMed]
- Fan, T.W.; Lane, A.N.; Higashi, R.M.; Farag, M.A.; Gao, H.; Bousamra, M.; Miller, D.M. Altered regulation of metabolic pathways in human lung cancer discerned by (13)C stable isotope-resolved metabolomics (SIRM). Mol. Cancer 2009, 8, 41. [Google Scholar] [CrossRef]
- Davidson, S.M.; Papagiannakopoulos, T.; Olenchock, B.A.; Heyman, J.E.; Keibler, M.A.; Luengo, A.; Bauer, M.R.; Jha, A.K.; O’Brien, J.P.; Pierce, K.A.; et al. Environment Impacts the Metabolic Dependencies of Ras-Driven Non-Small Cell Lung Cancer. Cell Metab. 2016, 23, 517–528. [Google Scholar] [CrossRef] [PubMed]
- Hensley, C.T.; Faubert, B.; Yuan, Q.; Lev-Cohain, N.; Jin, E.; Kim, J.; Jiang, L.; Ko, B.; Skelton, R.; Loudat, L.; et al. Metabolic Heterogeneity in Human Lung Tumors. Cell 2016, 164, 681–694. [Google Scholar] [CrossRef] [PubMed]
- Phannasil, P.; Thuwajit, C.; Warnnissorn, M.; Wallace, J.C.; MacDonald, M.J.; Jitrapakdee, S. Pyruvate Carboxylase Is Up-Regulated in Breast Cancer and Essential to Support Growth and Invasion of MDA-MB-231 Cells. Plos ONE 2015, 10. [Google Scholar] [CrossRef] [PubMed]
- Christen, S.; Lorendeau, D.; Schmieder, R.; Broekaert, D.; Metzger, K.; Veys, K.; Elia, I.; Buescher, J.M.; Orth, M.F.; Davidson, S.M.; et al. Breast Cancer-Derived Lung Metastases Show Increased Pyruvate Carboxylase-Dependent Anaplerosis. Cell Rep. 2016, 17, 837–848. [Google Scholar] [CrossRef] [PubMed]
- Leithner, K.; Hrzenjak, A.; Trotzmuller, M.; Moustafa, T.; Kofeler, H.C.; Wohlkoenig, C.; Stacher, E.; Lindenmann, J.; Harris, A.L.; Olschewski, A.; et al. PCK2 activation mediates an adaptive response to glucose depletion in lung cancer. Oncogene 2014. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.V.; Oesterreich, S.; Davidson, N.E. MCF-7 Cells—Changing the Course of Breast Cancer Research and Care for 45 Years. J. Natl. Cancer Inst. 2015, 107. [Google Scholar] [CrossRef] [PubMed]
- Maughan, R.J. A simple, rapid method for the determination of glucose, lactate, pyruvate, alanine, 3-hydroxybutyrate and acetoacetate on a single 20-µL blood sample. Clin. Chim. Acta 1982, 122, 231–240. [Google Scholar] [CrossRef]
- Chen, F.; Rydzewski, K.; Kutzner, E.; Häuslein, I.; Schunder, E.; Wang, X.; Meighen-Berger, K.; Grunow, R.; Eisenreich, W.; Heuner, K. Differential substrate usage and metabolic fluxes in Francisella tularensis subspecies holoarctica and Francisella novocida. Front. Cell Infect. Microbiol. 2017, 7, 275. [Google Scholar] [CrossRef]
- Ahmed, Z.; Zeeshan, S.; Huber, C.; Hensel, M.; Schomburg, D.; Münch, R.; Eylert, E.; Eisenreich, W.; Dandekar, T. ‘Isotopo’ a database application for facile analysis and management of mass isotopomer data. Database: J. Biol. Databases Curation 2014, 2014, bau077. [Google Scholar] [CrossRef]
- Medina, R.A.; Meneses, A.M.; Vera, J.C.; Guzman, C.; Nualart, F.; Astuya, A.; de los Angeles Garcia, M.; Kato, S.; Carvajal, A.; Pinto, M.; et al. Estrogen and Progesterone Up-Regulate Glucose Transporter Expression in ZR-75-1 Human Breast Cancer Cells. Endocrinology 2003, 144, 4527–4535. [Google Scholar] [CrossRef]
- DeBerardinis, R.J.; Mancuso, A.; Daikhin, E.; Nissim, I.; Yudkoff, M.; Wehrli, S.; Thompson, C.B. Beyond aerobic glycolysis: Transformed cells can engage in glutamine metabolism that exceeds the requirement for protein and nucleotide synthesis. Proc. Natl. Acad. Sci. USA 2007, 104, 19345–19350. [Google Scholar] [CrossRef]
- Metallo, C.M.; Walther, J.L.; Stephanopoulos, G. Evaluation of 13C isotopic tracers for metabolic flux analysis in mammalian cells. J. Biotechnol. 2009, 144, 167–174. [Google Scholar] [CrossRef] [PubMed]
- Ovadi, J.; Saks, V. On the origin of intracellular compartmentation and organized metabolic systems. Mol. Cell Biochem. 2004, 256, 5–12. [Google Scholar] [CrossRef] [PubMed]
- Labuschagne, C.F.; van den Broek, N.J.F.; Mackay, G.M.; Vousden, K.H.; Maddocks, O.D.K. Serine, but Not Glycine, Supports One-Carbon Metabolism and Proliferation of Cancer Cells. Cell Rep. 2014, 7, 1248–1258. [Google Scholar] [CrossRef] [PubMed]
- Méndez-Lucas, A.; Hyroššová, P.; Novellasdemunt, L.; Viñals, F.; Perales, J.C. Mitochondrial Phosphoenolpyruvate Carboxykinase (PEPCK-M) Is a Pro-survival, Endoplasmic Reticulum (ER) Stress Response Gene Involved in Tumor Cell Adaptation to Nutrient Availability. J. Biol. Chem. 2014, 289, 22090–22102. [Google Scholar] [CrossRef] [PubMed]
- Montal, E.D.; Dewi, R.; Bhalla, K.; Ou, L.; Hwang, B.J.; Ropell, A.E.; Gordon, C.; Liu, W.-J.; DeBerardinis, R.J.; Sudderth, J.; et al. PEPCK Coordinates the Regulation of Central Carbon Metabolism to Promote Cancer Cell Growth. Mol. Cell 2015, 60, 571–583. [Google Scholar] [CrossRef] [PubMed]
- Ferguson, B.S.; Rogatzki, M.J.; Goodwin, M.L.; Kane, D.A.; Rightmire, Z.; Gladden, L.B. Lactate metabolism: Historical context, prior misinterpretations, and current understanding. Eur. J. Appl. Physiol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Dhup, S.; Dadhich, R.K.; Porporato, P.E.; Sonveaux, P. Multiple biological activities of lactic acid in cancer: Influences on tumor growth, angiogenesis and metastasis. Curr. Pharm. Design 2012, 18. [Google Scholar] [CrossRef] [PubMed]
- Fu, T.F.; Rife, J.P.; Schirch, V. The role of serine hydroxymethyltransferase isozymes in one-carbon metabolism in MCF-7 cells as determined by (13)C NMR. Arch. Biochem. Biophys. 2001, 393, 42–50. [Google Scholar] [CrossRef] [PubMed]
- Girgis, S.; Nasrallah, I.M.; Suh, J.R.; Oppenheim, E.; Zanetti, K.A.; Mastri, M.G.; Stover, P.J. Molecular cloning, characterization and alternative splicing of the human cytoplasmic serine hydroxymethyltransferase gene. Gene 1998, 210, 315–324. [Google Scholar] [CrossRef]
- Stover, P.J.; Chen, L.H.; Suh, J.R.; Stover, D.M.; Keyomarsi, K.; Shane, B. Molecular cloning, characterization, and regulation of the human mitochondrial serine hydroxymethyltransferase gene. J. Biol. Chem. 1997, 272, 1842–1848. [Google Scholar] [CrossRef]
- Kohnhorst, C.L.; Kyoung, M.; Jeon, M.; Schmitt, D.L.; Kennedy, E.L.; Ramirez, J.; Bracey, S.M.; Luu, B.T.; Russell, S.J.; An, S. Identification of a multienzyme complex for glucose metabolism in living cells. J. Biol. Chem. 2017, 292, 9191–9203. [Google Scholar] [CrossRef] [PubMed]
- Zwingmann, C.; Richter-Landsberg, C.; Leibfritz, D. 13C isotopomer analysis of glucose and alanine metabolism reveals cytosolic pyruvate compartmentation as part of energy metabolism in astrocytes. Glia 2001, 34, 200–212. [Google Scholar] [CrossRef] [PubMed]
- Glinghammar, B.; Rafter, I.; Lindstrom, A.K.; Hedberg, J.J.; Andersson, H.B.; Lindblom, P.; Berg, A.L.; Cotgreavei, I. Detection of the mitochondrial and catalytically active alanine aminotransferase in human tissues and plasma. Int. J. Mol. Med. 2009, 23, 621–631. [Google Scholar] [CrossRef] [PubMed]
- Choo, A.Y.; Kim, S.G.; Vander Heiden, M.G.; Mahoney, S.J.; Vu, H.; Yoon, S.O.; Cantley, L.C.; Blenis, J. Glucose addiction of TSC null cells is caused by failed mTORC1-dependent balancing of metabolic demand with supply. Mol. Cell 2010, 38, 487–499. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Li, J.J.; Guo, L.Y.; Li, P.; Zhao, Z.; Zhou, H.; Di, L.J. Molecular link between glucose and glutamine consumption in cancer cells mediated by CtBP and SIRT4. Oncogenesis 2018, 7, 26. [Google Scholar] [CrossRef] [PubMed]
- Fendt, S.-M.; Bell, E.L.; Keibler, M.A.; Olenchock, B.A.; Mayers, J.R.; Wasylenko, T.M.; Vokes, N.I.; Guarente, L.; Heiden, M.G.V.; Stephanopoulos, G. Reductive glutamine metabolism is a function of the α-ketoglutarate to citrate ratio in cells. Nature Commun. 2013, 4, 2236. [Google Scholar] [CrossRef] [PubMed]
- Brodsky, A.N.; Odenwelder, D.C.; Harcum, S. High extracellular lactate causes reductive carboxylation in breast tissue cell lines grown under normoxic conditions. PLOS ONE 2019, 14, e0213419. [Google Scholar] [CrossRef]
- Cheng, T.; Sudderth, J.; Yang, C.; Mullen, A.R.; Jin, E.S.; Mates, J.M.; DeBerardinis, R.J. Pyruvate carboxylase is required for glutamine-independent growth of tumor cells. Proc. Natl. Acad. Sci. USA 2011, 108, 8674–8679. [Google Scholar] [CrossRef]
- Scott, D.A.; Richardson, A.D.; Filipp, F.V.; Knutzen, C.A.; Chiang, G.G.; Ronai, Z.A.; Osterman, A.L.; Smith, J.W. Comparative Metabolic Flux Profiling of Melanoma Cell Lines—Beyond the Warburg Effect. J. Biol. Chem. 2011, 286, 42626–42634. [Google Scholar] [CrossRef]
- Willmann, L.; Schlimpert, M.; Halbach, S.; Erbes, T.; Stickeler, E.; Kammerer, B. Metabolic profiling of breast cancer: Differences in central metabolism between subtypes of breast cancer cell lines. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2015, 1000, 95–104. [Google Scholar] [CrossRef]
- Lane, A.N.; Tan, J.; Wang, Y.; Yan, J.; Higashi, R.M.; Fan, T.W. Probing the metabolic phenotype of breast cancer cells by multiple tracer stable isotope resolved metabolomics. Metab. Eng. 2017, 43, 125–136. [Google Scholar] [CrossRef] [PubMed]
- Kishimoto, S.; Brender, J.; Crooks, D.; Matsumoto, S.; Seki, T.; Oshima, N.; Merkle, H.; Lin, P.; Reed, G.; Chen, A.P.; et al. Imaging of Glucose Metabolism by 13C-MRI Distinguishes Pancreatic Cancer Subtypes in Mice. eLife Sci. 2019, 8, e46312. [Google Scholar] [CrossRef] [PubMed]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gkiouli, M.; Biechl, P.; Eisenreich, W.; Otto, A.M. Diverse Roads Taken by 13C-Glucose-Derived Metabolites in Breast Cancer Cells Exposed to Limiting Glucose and Glutamine Conditions. Cells 2019, 8, 1113. https://doi.org/10.3390/cells8101113
Gkiouli M, Biechl P, Eisenreich W, Otto AM. Diverse Roads Taken by 13C-Glucose-Derived Metabolites in Breast Cancer Cells Exposed to Limiting Glucose and Glutamine Conditions. Cells. 2019; 8(10):1113. https://doi.org/10.3390/cells8101113
Chicago/Turabian StyleGkiouli, Maria, Philipp Biechl, Wolfgang Eisenreich, and Angela M. Otto. 2019. "Diverse Roads Taken by 13C-Glucose-Derived Metabolites in Breast Cancer Cells Exposed to Limiting Glucose and Glutamine Conditions" Cells 8, no. 10: 1113. https://doi.org/10.3390/cells8101113
APA StyleGkiouli, M., Biechl, P., Eisenreich, W., & Otto, A. M. (2019). Diverse Roads Taken by 13C-Glucose-Derived Metabolites in Breast Cancer Cells Exposed to Limiting Glucose and Glutamine Conditions. Cells, 8(10), 1113. https://doi.org/10.3390/cells8101113





