Nuclear Receptors as Regulators of Pituitary Corticotroph Pro-Opiomelanocortin Transcription
Abstract
:1. Introduction
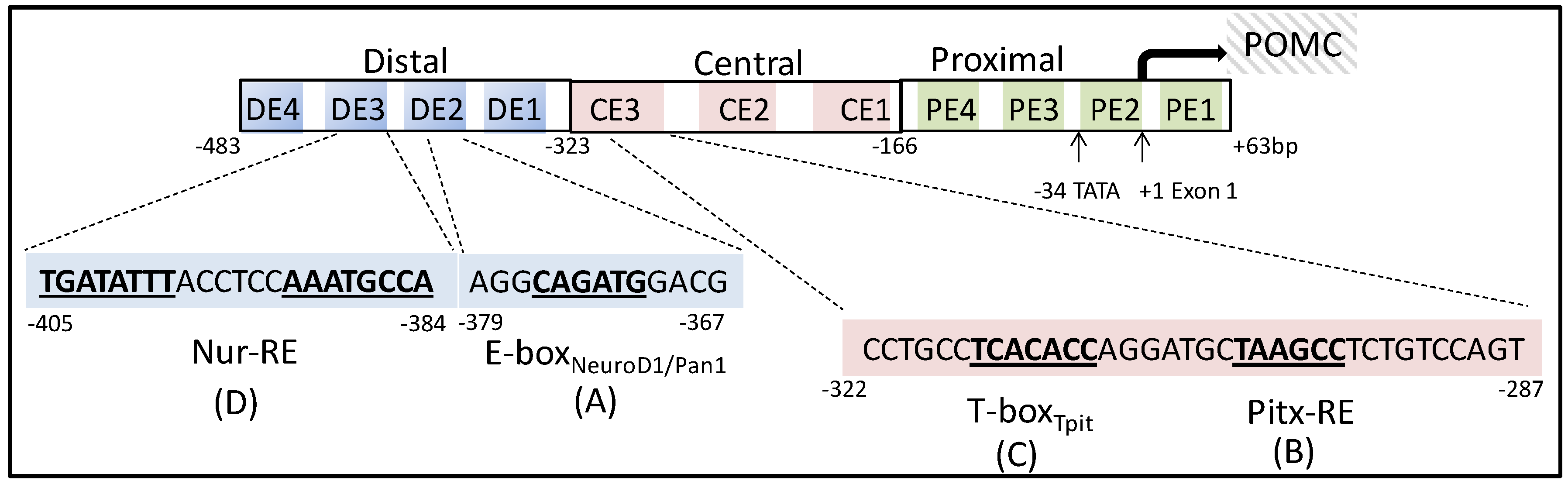
2. Pituitary Specific Regulators of Pro-Opiomelanocortin Transcription
3. Nuclear Receptors as Pro-Opiomelanocortin Transcription Regulators in Pituitary Corticotropes
3.1. Subfamily 1
3.1.1. Thyroid Hormone Receptors (TRα and TRβ)
3.1.2. Retinoic Acid Receptors (RARα, RARβ, RARγ)
3.1.3. Peroxisome Proliferator Activated Receptors (PPARα, PPARβ and PPARγ)
3.1.4. Reverse Erb Receptors (REV-ERBα and REV-ERβ)
3.1.5. Retinoic Acid Receptor-Related Orphan Receptors (RORα, RORβ and RORγ)
3.1.6. Liver X Receptors (LXRα and LXRβ)
3.1.7. Pregnane X Receptor (PXR) and Constitutive Androstane Receptor (CAR)
3.2. Subfamily 2
3.2.1. Retinoid X Receptors (RXRα, RXRβ, and RXRγ)
3.2.2. Testicular Orphan Nuclear Receptor-4 (TR4)
3.2.3. Chicken Ovalbumin Upstream Promoter Transcription Factors (COUP-TFα, COUP-TFβ and COUP-TFγ)
3.3. Subfamily 3
3.3.1. Estrogen Receptors (ERα and ERβ)
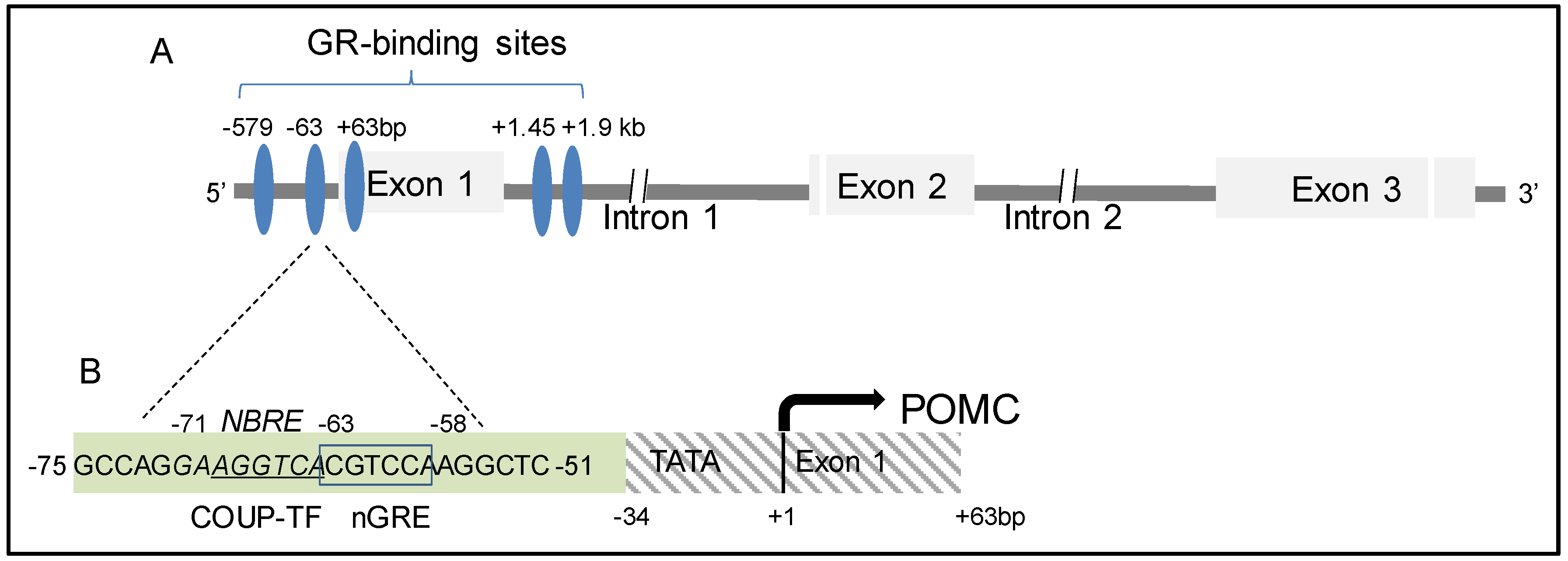
3.3.2. Glucocorticoid Receptor (GR)
3.4. Subfamily 4
3.4.1. Nerve Growth Factor Induced Clone B (NGFI-B, Nur77)
3.4.2. Nur-Related Factor 1 (Nurr1)
3.4.3. Neuron derived Orphan Receptor 1 (NOR-1)
3.5. Subfamily 5 and 6
4. Summary
Author Contributions
Funding
Conflicts of Interest
References
- Nussey, S.; Whitehead, S. The pituitary gland. In Endocrinology: An Integrated Approach; BIOS Scientific Publisher: Oxford, UK, 2001. [Google Scholar]
- Smith, S.M.; Vale, W.W. The role of the hypothalamic-pituitary-adrenal axis in neuroendocrine responses to stress. Dialogues Clin. Neurosci. 2006, 8, 383–395. [Google Scholar] [PubMed]
- Harno, E.; Ramamoorthy, T.G.; Coll, A.P.; White, A. POMC: The Physiological Power of Hormone Processing. Physiol. Rev. 2018, 98, 2381–2430. [Google Scholar] [CrossRef] [PubMed]
- Dores, R.M.; Baron, A.J. Evolution of POMC: Origin, phylogeny, posttranslational processing, and the melanocortins. Ann. N. Y. Acad. Sci. 2011, 1220, 34–48. [Google Scholar] [CrossRef] [PubMed]
- Sundström, G.; Dreborg, S.; Larhammar, D. Concomitant Duplications of Opioid Peptide and Receptor Genes before the Origin of Jawed Vertebrates. PLoS ONE 2010, 5, e10512. [Google Scholar] [CrossRef]
- Donohoue, P.A.; Ali, O. Disorders of the Body Mass. In Emery and Rimoin’s Principles and Practice of Medical Genetics, 6th ed.; Academic Press: Oxford, UK, 2013; pp. 1–37. [Google Scholar]
- Clark, A.J.L. 60 YEARS OF POMC: The proopiomelanocortin gene: Discovery, deletion and disease. J. Mol. Endocrinol. 2016, 56, T27–T37. [Google Scholar] [CrossRef] [Green Version]
- Deen, P.M.T.; Terwel, D.; Bussemakers, M.J.M.; Roubos, E.W.; Martens, G.J.M. Structural analysis of the entire proopiomelanocortin gene of Xenopus laevis. Eur. J. Biochem. 1991, 201, 129–137. [Google Scholar] [CrossRef]
- Jeannotte, L.; Trifiro, M.A.; Plante, R.K.; Chamberland, M.; Drouin, J. Tissue-specific activity of the pro-opiomelanocortin gene promoter. Mol. Cell. Biol. 1987, 7, 4058–4064. [Google Scholar] [CrossRef] [Green Version]
- Tremblay, Y.; Tretjakoff, I.; Peterson, A.; Antakly, T.; Zhang, C.X.; Drouin, J. Pituitary-specific expression and glucocorticoid regulation of a proopiomelanocortin fusion gene in transgenic mice. Proc. Natl Acad. Sci. USA 1988, 85, 8890–8894. [Google Scholar] [CrossRef] [Green Version]
- Therrien, M.; Drouin, J. Pituitary pro-opiomelanocortin gene expression requires synergistic interactions of several regulatory elements. Mol. Cell Biol. 1991, 11, 3492–3503. [Google Scholar] [CrossRef] [Green Version]
- Therrien, M.; Drouin, J. Cell-specific helix-loop-helix factor required for pituitary expression of the pro-opiomelanocortin gene. Mol. Cell Biol. 1993, 13, 2342–2353. [Google Scholar] [CrossRef]
- Poulin, G.; Turgeon, B.; Drouin, J. NeuroD1/beta2 contributes to cell-specific transcription of the proopiomelanocortin gene. Mol. Cell Biol. 1997, 17, 6673–6682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lamonerie, T.; Tremblay, J.J.; Lanctôt, C.; Therrien, M.; Gauthier, Y.; Drouin, J. Ptx1, a bicoid-related homeo box transcription factor involved in transcription of the pro-opiomelanocortin gene. Genes Dev. 1996, 10, 1284–1295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lamolet, B.; Pulichino, A.-M.; Lamonerie, T.; Gauthier, Y.; Brue, T.; Enjalbert, A.; Drouin, J. A Pituitary Cell-Restricted T Box Factor, Tpit, Activates POMC Transcription in Cooperation with Pitx Homeoproteins. Cell 2001, 104, 849–859. [Google Scholar] [CrossRef]
- Willson, T.M.; Moore, J.T. Minireview: Genomics Versus Orphan Nuclear Receptors-A Half-Time Report. Mol. Endocrinol. 2002, 16, 1135–1144. [Google Scholar] [CrossRef]
- Owen, G.I.; Zelent, A. Origins and evolutionary diversification of the nuclear receptor superfamily. Cell. Mol. Life Sci. 2000, 57, 809–827. [Google Scholar] [CrossRef]
- Germain, P.; Staels, B.; Dacquet, C.; Spedding, M.; Laudet, V. Overview of Nomenclature of Nuclear Receptors. Pharmacol. Rev. 2006, 58, 685–704. [Google Scholar] [CrossRef]
- Enmark, E.; Gustafsson, J.Å. Comparing nuclear receptors in worms, flies and humans. Trends Pharmacol. Sci. 2001, 22, 611–615. [Google Scholar] [CrossRef]
- Treviño, L.S.; Weigel, N.L. Phosphorylation: A Fundamental Regulator of Steroid Receptor Action. Trends Endocrinol. Metab. TEM 2013, 24, 515–524. [Google Scholar]
- Robinson-Rechavi, M.; Garcia, H.E.; Laudet, V. The nuclear receptor superfamily. J. Cell Sci. 2003, 116, 585–586. [Google Scholar] [CrossRef] [Green Version]
- Lindholm, J.; Laurberg, P. Hypothyroidism and Thyroid Substitution: Historical Aspects. J. Thyroid Res. 2011, 2011, 10. [Google Scholar] [CrossRef] [Green Version]
- Li, M.; Boyages, S.C. Detection of extended distribution of beta2-thyroid hormone receptor messenger ribonucleic acid (RNA) in adult rat brain using complementary RNA in situ hybridization histochemistry. Endocrinology 1996, 137, 1272–1275. [Google Scholar] [CrossRef]
- Bradley, D.J.; Towle, H.C.; Young, W.S., 3rd. Alpha and beta thyroid hormone receptor (TR) gene expression during auditory neurogenesis: Evidence for TR isoform-specific transcriptional regulation in vivo. Proc. Natl. Acad. Sci. USA 1994, 91, 439–443. [Google Scholar] [CrossRef] [Green Version]
- Lazar, M.A. Thyroid Hormone Receptors: Multiple Forms, Multiple Possibilities. Endocr. Rev. 1993, 14, 184–193. [Google Scholar]
- Furumoto, H.; Ying, H.; Chandramouli, G.V.R.; Zhao, L.; Walker, R.L.; Meltzer, P.S.; Willingham, M.C.; Cheng, S.Y. An unliganded thyroid hormone beta receptor activates the cyclin D1/cyclin-dependent kinase/retinoblastoma/E2F pathway and induces pituitary tumorigenesis. Mol. Cell. Biol. 2005, 25, 124–135. [Google Scholar] [CrossRef] [Green Version]
- Saltó, C.; Kindblom, J.M.; Johansson, C.; Wang, Z.; Gullberg, H.; Nordström, K.; Mansén, A.; Ohlsson, C.; Thorén, P.; Forrest, D.; et al. Ablation of TRα2 and a Concomitant Overexpression of α1 Yields a Mixed Hypo- and Hyperthyroid Phenotype in Mice. Molec. Endocrinol. 2001, 15, 2115–2128. [Google Scholar]
- Ng, L.; Rüsch, A.; Amma, L.L.; Nordström, K.; Erway, L.C.; Vennström, B.; Forrest, D. Suppression of the deafness and thyroid dysfunction in Thrb-null mice by an independent mutation in the Thra thyroid hormone receptor α gene. Hum. Molec. Genet. 2001, 10, 2701–2708. [Google Scholar] [CrossRef] [Green Version]
- Hodin, R.A.; Lazar, M.; Wintman, B.; Darling, D.; Koenig, R.; Larsen, P.; Moore, D.; Chin, W. Identification of a thyroid hormone receptor that is pituitary-specific. Science 1989, 244, 76–79. [Google Scholar] [CrossRef] [PubMed]
- Koenig, R.J.; Warne, R.L.; Brent, G.A.; Harney, J.W.; Larsen, P.R.; Moore, D.D. Isolation of a cDNA clone encoding a biologically active thyroid hormone receptor. Proc. Natl. Acad. Sci. USA 1988, 85, 5031–5035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodríguez-García, M.; Jolín, T.; Santos, A.; Pérez-Castillo, A. Effect of perinatal hypothyroidism on the developmental regulation of rat pituitary growth hormone and thyrotropin genes. Endocrinology 1995, 136, 4339–4350. [Google Scholar]
- Ercan-Fang, S.; Schwartz, H.L.; Oppenheimer, J.H. Isoform-specific 3,5,3′-triiodothyronine receptor binding capacity and messenger ribonucleic acid content in rat adenohypophysis: Effect of thyroidal state and comparison with extrapituitary tissues. Endocrinology 1996, 137, 3228–3233. [Google Scholar] [CrossRef] [Green Version]
- Yen, P.M.; Sunday, M.E.; Darling, D.S.; Chin, W.W. Isoform-specific thyroid hormone receptor antibodies detect multiple thyroid hormone receptors in rat and human pituitaries. Endocrinology 1992, 130, 1539–1546. [Google Scholar] [PubMed]
- Childs, G.V.; Taub, K.; Jones, K.E.; Chin, W.W. Triiodothyronine Receptor β-2 Messenger Ribonucleic Acid Expression by Somatotropes and Thyrotropes: Effect of Propylthiouracil-Induced Hypothyroidism in Rats. Endocrinology 1991, 129, 2767–2773. [Google Scholar] [CrossRef] [PubMed]
- Martin, N.M.; Small, C.J.; Sajedi, A.; Liao, X.H.; Weiss, R.E.; Gardiner, J.V.; Ghatei, M.A.; Bloom, S.R. Abnormalities of the hypothalamo-pituitary-thyroid axis in the pro-opiomelanocortin deficient mouse. Regul. Pept. 2004, 122, 169–172. [Google Scholar] [CrossRef] [PubMed]
- Chambon, P. A decade of molecular biology of retinoic acid receptors. FASEB J. 1996, 10, 940–954. [Google Scholar] [CrossRef] [PubMed]
- Chlapek, P.; Slavikova, V.; Mazanek, P.; Sterba, J.; Veselska, R. Why Differentiation Therapy Sometimes Fails: Molecular Mechanisms of Resistance to Retinoids. Int. J. Mol. Sci. 2018, 19, 132. [Google Scholar] [CrossRef] [Green Version]
- Huang, Z.; Liu, Y.; Qi, G.; Brand, D.; Zheng, S.G. Role of Vitamin A in the Immune System. J. Clin. Med. 2018, 7, 258. [Google Scholar] [CrossRef] [Green Version]
- Pallet, V.; Touyarot, K. Vitamin A and cognitive processes. Nutr. Aging 2015, 3, 21–31. [Google Scholar] [CrossRef] [Green Version]
- Davey, J.C.; Nomikos, A.P.; Wungjiranirun, M.; Sherman, J.R.; Ingram, L.; Batki, C.; Lariviere, J.P.; Hamilton, J.W. Arsenic as an endocrine disruptor: Arsenic disrupts retinoic acid receptor-and thyroid hormone receptor-mediated gene regulation and thyroid hormone-mediated amphibian tail metamorphosis. Environ. Health Perspect. 2008, 116, 165–172. [Google Scholar] [CrossRef] [Green Version]
- Karadag, A.S.; Takci, Z.; Ertugrul, D.T.; Bilgili, S.G.; Balahoroglu, R.; Takir, M. The Effect of Different Doses of Isotretinoin on Pituitary Hormones. Dermatology 2015, 230, 354–359. [Google Scholar] [CrossRef]
- Vilar, L.; Albuquerque, J.L.; Lyra, R.; Diniz, E.T.; Filho, F.R.; Gadelha, P.; Thé, A.C.; Ibiapina, G.R.; Gomes, B.S.; Santos, V.; et al. The Role of Isotretinoin Therapy for Cushing’s Disease: Results of a Prospective Study. Int. J. Endocrinol. 2016, 2016, 9. [Google Scholar] [CrossRef] [Green Version]
- Pecori Giraldi, F.; Ambrogio, A.G.; Andrioli, M.; Sanguin, F.; Karamouzis, I.; Corsello, S.M.; Scaroni, C.; Arvat, E.; Pontercorvi, A.; Cavagnini, F. Potential Role for Retinoic Acid in Patients with Cushing’s Disease. J. Clin. Endocrinol. Metab. 2012, 97, 3577–3583. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Castillo, V.; Giacomini, D.; Páez-Pereda, M.; Stalla, J.; Labeur, M.; Theodoropoulou, M.; Holsboer, F.; Grossman, A.B.; Stalla, G.K.; Arzt, E. Retinoic Acid as a Novel Medical Therapy for Cushing’s Disease in Dogs. Endocrinology 2006, 147, 4438–4444. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Páez-Pereda, M.; Kovalovsky, D.; Hopfner, U.; Theodoropoulou, M.; Pagotto, U.; Uhl, E.; Losa, M.; Stalla, J.; Grübler, Y.; Missale, C.; et al. Retinoic acid prevents experimental Cushing syndrome. J. Clin. Investig. 2001, 108, 1123–1131. [Google Scholar]
- Tran, P.; Zhang, X.K.; Salbert, G.; Hermann, T.; Lehmann, J.M.; Pfahl, M. COUP orphan receptors are negative regulators of retinoic acid response pathways. Mol. Cell Biol. 1992, 12, 4666–4676. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nudi, M.; Ouimette, J.F.; Drouin, J. Bone Morphogenic Protein (Smad)-Mediated Repression of Proopiomelanocortin Transcription by Interference with Pitx/Tpit Activity. Mol. Endocrinol. 2005, 19, 1329–1342. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ludot, M.; Mouchabac, S.; Ferreri, F. Inter-relationships between isotretinoin treatment and psychiatric disorders: Depression, bipolar disorder, anxiety, psychosis and suicide risks. World J. Psychiatry 2015, 5, 222–227. [Google Scholar] [CrossRef]
- Ghyselinck, N.B.; Dupe, V.; Dierich, A.; Messaddeq, N.; Garnier, J.; Rochette-Egly, C.; Chambon, P.; Mark, M. Role of the retinoic acid receptor beta (RARbeta) during mouse development. Int. J. Dev. Biol. 1997, 41, 425–447. [Google Scholar]
- Metzler, M.A.; Raja, S.; Elliott, K.H.; Friedl, R.M.; Tran, N.Q.H.; Brugmann, S.A.; Larsen, M.; Sandell, L.L. RDH10-mediated retinol metabolism and RARα-mediated retinoic acid signaling are required for submandibular salivary gland initiation. Development 2018, 145, dev164822. [Google Scholar] [CrossRef] [Green Version]
- Labeur, M.; Paez-Pereda, M.; Arzt, E.; Stalla, G.K. Potential of retinoic acid derivatives for the treatment of corticotroph pituitary adenomas. Rev. Endocrinol. Metab. Disord. 2009, 10, 103. [Google Scholar] [CrossRef] [Green Version]
- Renaud, J.P.; Rochel, N.; Ruff, M.; Vivat, V.; Chambon, P.; Gronemeyer, H.; Moras, D. Crystal structure of the RAR-γ ligand-binding domain bound to all-trans retinoic acid. Nature 1995, 378, 681–689. [Google Scholar] [CrossRef]
- Uruno, A.; Saito-Hakoda, A.; Yokoyama, A.; Kogure, N.; Matsuda, K.; Parvin, R.; Shimizu, K.; Sato, I.; Kudo, M.; Yoshikawa, T.; et al. Retinoic acid receptor-alpha up-regulates proopiomelanocortin gene expression in AtT20 corticotroph cells. Endocr. J. 2014, 61, 1105–1114. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Evans, R.M.; Mangelsdorf, D.J. Nuclear Receptors, RXR, and the Big Bang. Cell 2014, 157, 255–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saito-Hakoda, A.; Uruno, A.; Yokoyama, A.; Shimizu, K.; Parvin, R.; Kudo, M.; Saito-Ito, T.; Sato, I.; Kogure, N.; Suzuki, D. Effects of RXR Agonists on Cell Proliferation/Apoptosis and ACTH Secretion/Pomc Expression. PLoS ONE 2015, 10, e0141960. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Issemann, I.; Green, S. Activation of a member of the steroid hormone receptor superfamily by peroxisome proliferators. Nature 1990, 347, 645–650. [Google Scholar] [CrossRef]
- Vitti, M.; Di Emidio, G.; Di Carlo, M.; Carta, G.; Antonosante, A.; Artini, P.G.; Cimini, A.; Tatone, C.; Benedetti, E. Peroxisome Proliferator-Activated Receptors in Female Reproduction and Fertility. PPAR Res. 2016, 2016, 4612306. [Google Scholar] [CrossRef]
- Garcia-Bates, T.M.; Lehmann, G.M.; Simpson-Haidaris, P.J.; Bernstein, S.H.; Sime, P.J.; Phipps, R.P. Role of peroxisome proliferator-activated receptor gamma and its ligands in the treatment of hematological malignancies. PPAR Res. 2008, 2008, 834612. [Google Scholar] [CrossRef] [Green Version]
- Yang, J.; Chen, L.; Zhang, X.; Zhou, Y.; Zhang, D.; Huo, M.; Guan, Y. PPARs and Female Reproduction: Evidence from Genetically Manipulated Mice. PPAR Res. 2008, 2008, 8. [Google Scholar] [CrossRef]
- Borel, V.; Gallot, D.; Marceau, G.; Sapin, V.; Blanchon, L. Placental implications of peroxisome proliferator-activated receptors in gestation and parturition. PPAR Res. 2008, 2008, 758562. [Google Scholar] [CrossRef] [Green Version]
- Mukherjee, S.; Mani, S. Orphan nuclear receptors as targets for drug development. Pharm. Res. 2010, 27, 1439–1468. [Google Scholar] [CrossRef] [Green Version]
- Wieser, F.; Waite, L.; Depoix, C.; Taylor, R.N. PPAR Action in Human Placental Development and Pregnancy and Its Complications. PPAR Res. 2008, 2008, 527048. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.C. The role of peroxisome proliferator-activated receptors in the development and physiology of gametes and preimplantation embryos. PPAR Res. 2008, 2008, 732303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bookout, A.L.; Jeong, Y.; Downes, M.; Yu, R.T.; Evans, R.M.; Mangelsdorf, D.J. Anatomical profiling of nuclear receptor expression reveals a hierarchical transcriptional network. Cell 2006, 126, 789–799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bogazzi, F.; Russo, D.; Locci, M.T.; Chifenti, B.; Ultimieri, F.; Raggi, F.; Viacava, P.; Cecchetti, D.; Cosci, C.; Sardella, C.; et al. Peroxisome proliferator-activated receptor (PPAR)γ is highly expressed in normal human pituitary gland. J. Endocrinol. Investig. 2005, 28, 899–904. [Google Scholar] [CrossRef]
- Occhi, G.; Albiger, N.; Berlucchi, S.; Gardiman, M.; Scanarini, M.; Scienza, R.; Fassina, A.; Mantero, F.; Scaroni, C. Peroxisome Proliferator-Activated Receptor γ in the Human Pituitary Gland: Expression and Splicing Pattern in Adenomas Versus Normal Pituitary. J. Neuroendocrinol. 2007, 19, 552–559. [Google Scholar] [CrossRef] [PubMed]
- Heaney, A.P.; Fernando, M.; Yong, W.H.; Melmed, S. Functional PPAR-γ receptor is a novel therapeutic target for ACTH-secreting pituitary adenomas. Nat. Med. 2002, 8, 1281–1287. [Google Scholar] [CrossRef]
- Emery, M.N.; Leontiou, C.; Bonner, S.E.; Merulli, C.; Nanzer, A.M.; Musat, M.; Galloway, M.; Powell, M.; Nikookam, K.; Korbonits, M.; et al. PPAR-γ expression in pituitary tumours and the functional activity of the glitazones: Evidence that any anti-proliferative effect of the glitazones is independent of the PPAR-γ receptor. Clin. Endocrinol. 2006, 65, 389–395. [Google Scholar] [CrossRef]
- Giraldi, F.P.; Scaroni, C.; Arvat, E.; De Martin, M.; Giordano, R.; Albiger, N.; Leao, A.A.S.; Picu, A.; Mantero, F.; Cavagnini, F. Effect of protracted treatment with rosiglitazone, a PPARγ agonist, in patients with Cushing’s disease. Clin. Endocrinol. 2006, 64, 219–224. [Google Scholar] [CrossRef]
- Ambrosi, B.; Dall’Asta, C.; Cannavo, S.; Libe, R.; Vigo, T.; Epaminonda, P.; Chiodini, I.; Ferrero, S.; Trimarchi, F.; Arosio, M.; et al. Effects of chronic administration of PPAR-gamma ligand rosiglitazone in Cushing’s disease. Eur. J. Endocrinol. 2004, 151, 173–178. [Google Scholar] [CrossRef] [Green Version]
- Morcos, M.; Fohr, B.; Tafel, J.; Pfisterer, F.; Hamann, A.; Humpert, P.; Bode, H.; Schwenger, V.; Zeier, M.; Becker, C.; et al. Long-Term Treatment of Central Cushing’s Syndrome with Rosiglitazone. Exp. Clin. Endocrinol. Diabetes 2007, 115, 292–297. [Google Scholar] [CrossRef]
- Woodcock, J.; Sharfstein, J.M.; Hamburg, M. Regulatory Action on Rosiglitazone by the U.S. Food and Drug Administration. N. Engl. J. Med. 2010, 363, 1489–1491. [Google Scholar] [CrossRef]
- Parvin, R.; Noro, E.; Saito-Hakoda, A.; Shimada, H.; Suzuki, S.; Shimizu, K.; Miyachi, H.; Yokoyama, A.; Sugawara, A. Inhibitory Effects of a Novel PPAR-gamma Agonist MEKT1 on Pomc Expression/ACTH Secretion in AtT20 Cells. PPAR Res. 2018, 2018, 16. [Google Scholar] [CrossRef]
- Solt, L.A.; Wang, Y.; Banerjee, S.; Hughes, T.; Kojetin, D.J.; Lundasen, T.; Shin, Y.; Liu, J.; Cameron, M.D.; Noel, R.; et al. Regulation of circadian behaviour and metabolism by synthetic REV-ERB agonists. Nature 2012, 485, 62–68. [Google Scholar] [CrossRef]
- Chung, S.; Son, G.H.; Kim, K. Circadian rhythm of adrenal glucocorticoid: Its regulation and clinical implications. BBA Mol. Basis Dis. 2011, 1812, 581–591. [Google Scholar] [CrossRef] [Green Version]
- Tsigos, C.; Kyrou, I.; Kassi, E.; Chrousos, G.P. Stress, endocrine physiology and pathophysiology. In Endotext [Internet]; MDText.com, Inc.: South Dartmouth, MA, USA, 2016. [Google Scholar]
- Solt, L.A.; Kojetin, D.J.; Burris, T.P. The REV-ERBs and RORs: Molecular links between circadian rhythms and lipid homeostasis. Future Med. Chem. 2011, 3, 623–638. [Google Scholar] [CrossRef] [Green Version]
- Preitner, N.; Damiola, F.; Luis Lopez, M.; Zakany, J.; Duboule, D.; Albrecht, U.; Schibler, U. The Orphan Nuclear Receptor REV-ERBa Controls Circadian Transcription within the Positive Limb of the Mammalian Circadian Oscillator. Cell 2002, 110, 251–260. [Google Scholar] [CrossRef]
- Angelousi, A.; Kassi, E.; Ansari-Nasiri, N.; Randeva, H.; Kaltsas, G.; Chrousos, G. Clock genes and cancer development in particular in endocrine tissues. Endocr. Relat. Cancer 2019, 26, R305–R317. [Google Scholar] [CrossRef] [Green Version]
- Delezie, J.; Dumont, S.; Dardente, H.; Oudart, H.; Gréchez-Cassiau, A.; Klosen, P.; Teboul, M.; Delaunay, F.; Pévet, P.; Challet, E. The nuclear receptor REV-ERBα is required for the daily balance of carbohydrate and lipid metabolism. FASEB J. 2012, 26, 3321–3335. [Google Scholar] [CrossRef] [Green Version]
- Tsukamoto-Yamauchi, N.; Terasaka, T.; Iwasaki, Y.; Otsuka, F. Interaction of pituitary hormones and expression of clock genes modulated by bone morphogenetic protein-4 and melatonin. Biochem. Biophys. Res. Commun. 2015, 459, 172–177. [Google Scholar] [CrossRef]
- Walker, J.J.; Terry, J.R.; Lightman, S.L. Origin of ultradian pulsatility in the hypothalamic–pituitary–adrenal axis. Proc. Biol. Sci. 2010, 277, 1627–1633. [Google Scholar] [CrossRef] [Green Version]
- Le Tissier, P.; Fiordelisio Coll, T.; Mollard, P. The Processes of Anterior Pituitary Hormone Pulse Generation. Endocrinology 2018, 159, 3524–3535. [Google Scholar] [CrossRef] [Green Version]
- Kalafatakis, K.; Russell, G.M.; Zarros, A.; Lightman, S.L. Temporal control of glucocorticoid neurodynamics and its relevance for brain homeostasis, neuropathology and glucocorticoid-based therapeutics. Neurosci. Biobehav. Rev. 2016, 61, 12–25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bur, I.M.; Zouaoui, S.; Fontanaud, P.; Coutry, N.; Molino, F.; Martin, A.O.; Mollard, P.; Bonnefont, X. The Comparison between Circadian Oscillators in Mouse Liver and Pituitary Gland Reveals Different Integration of Feeding and Light Schedules. PLoS ONE 2010, 5, e15316. [Google Scholar] [CrossRef] [PubMed]
- Abe, M.; Herzog, E.D.; Yamazaki, S.; Straume, M.; Tei, H.; Sakaki, Y.; Menaker, M.; Block, G.D. Circadian Rhythms in Isolated Brain Regions. J. Neurosci. 2002, 22, 350–356. [Google Scholar] [CrossRef]
- Yoo, S.H.; Yamazaki, S.; Lowrey, P.L.; Shimomura, K.; Ko, C.H.; Buhr, E.D.; Siepka, S.M.; Hong, H.K.; Oh, W.J.; Yoo, O.J.; et al. PERIOD2::LUCIFERASE real-time reporting of circadian dynamics reveals persistent circadian oscillations in mouse peripheral tissues. Proc. Natl. Acad. Sci. USA 2004, 101, 5339–5346. [Google Scholar] [CrossRef] [Green Version]
- Chu, A.; Zhu, L.; Blum, I.D.; Mai, O.; Leliavski, A.; Fahrenkrug, J.; Oster, H.; Boehm, U.; Storch, K.F. Global But Not Gonadotrope-Specific Disruption of Bmal1 Abolishes the Luteinizing Hormone Surge Without Affecting Ovulation. Endocrinology 2013, 154, 2924–2935. [Google Scholar] [CrossRef] [Green Version]
- Becquet, D.; Boyer, B.; Rasolonjanahary, R.; Brue, T.; Guillen, S.; Moreno, M.; Franc, J.L.; François-Bellan, A.M. Evidence for an internal and functional circadian clock in rat pituitary cells. Mol. Cell. Endocrinol. 2014, 382, 888–898. [Google Scholar] [CrossRef]
- Lin, X.W.; Blum, I.D.; Storch, K.F. Clocks within the Master Gland:Hypophyseal Rhythms and Their Physiological Significance. J. Biol. Rhythms 2015, 30, 263–276. [Google Scholar] [CrossRef]
- Wunderer, F.; Kühne, S.; Jilg, A.; Ackermann, K.; Sebesteny, T.; Maronde, E.; Stehle, J.H. Clock Gene Expression in the Human Pituitary Gland. Endocrinology 2013, 154, 2046–2057. [Google Scholar] [CrossRef] [Green Version]
- Giguère, V.; Tini, M.; Flock, G.; Ong, E.; Evans, R.M.; Otulakowski, G. Isoform-specific amino-terminal domains dictate DNA-binding properties of ROR alpha, a novel family of orphan hormone nuclear receptors. Genes Dev. 1994, 8, 538–553. [Google Scholar] [CrossRef] [Green Version]
- Carlberg, C.; Hooft van Huijsduijnen, R.; Staple, J.K.; DeLamarter, J.F.; Becker-André, M. RZRs, a new family of retinoid-related orphan receptors that function as both monomers and homodimers. Mol. Endocrinol. 1994, 8, 757–770. [Google Scholar]
- Hirose, T.; Smith, R.J.; Jetten, A.M. ROR-γ: The Third Member of ROR/RZR Orphan Receptor Subfamily That Is Highly Expressed in Skeletal Muscle. Biochem. Biophys. Res. Com. 1994, 205, 1976–1983. [Google Scholar] [CrossRef] [Green Version]
- Moretti, R.M.; Marelli, M.M.; Motta, M.; Polizzi, D.; Monestiroli, S.; Pratesi, G.; Limonta, P. Activation of the orphan nuclear receptor RORα induces growth arrest in androgen-independent DU 145 prostate cancer cells. Prostate 2001, 46, 327–335. [Google Scholar] [CrossRef]
- Harding, H.P.; Atkins, G.B.; Jaffe, A.B.; Seo, W.J.; Lazar, M.A. Transcriptional Activation and Repression by RORα, an Orphan Nuclear Receptor Required for Cerebellar Development. Mol. Endocrinol. 1997, 11, 1737–1746. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.; Cho, H.; Yu, R.T.; Atkins, A.R.; Downes, M.; Evans, R.M. Nuclear receptors rock around the clock. EMBO Rep. 2014, 15, 518–528. [Google Scholar] [CrossRef]
- Early, J.O.; Curtis, A.M. Immunometabolism: Is it under the eye of the clock? Semin. Immunol. 2016, 28, 478–490. [Google Scholar] [CrossRef]
- Carter, S.J.; Durrington, H.J.; Gibbs, J.E.; Blaikley, J.; Loudon, A.S.; Ray, D.W.; Sabroe, I. A matter of time: Study of circadian clocks and their role in inflammation. J. Leukoc. Biol. 2016, 99, 549–560. [Google Scholar] [CrossRef] [Green Version]
- Hamilton, B.A.; Frankel, W.N.; Kerrebrock, A.W.; Hawkins, T.L.; FitzHugh, W.; Kusumi, K.; Russell, L.B.; Mueller, K.L.; van Berkel, V.; Birren, B.W.; et al. Disruption of the nuclear hormone receptor RORα in staggerer mice. Nature 1996, 379, 736–739. [Google Scholar] [CrossRef]
- Klar, J.; Åsling, B.; Carlsson, B.; Ulvsbäck, M.; Dellsén, A.; Ström, C.; Rhedin, M.; Forslund, A.; Annerén, G.; Ludvigsson, J.F.; et al. RAR-related orphan receptor A isoform 1 (RORa1) is disrupted by a balanced translocation t(4;15)(q22.3;q21.3) associated with severe obesity. Eur. J. Hum. Genet. 2005, 13, 928–934. [Google Scholar] [CrossRef] [Green Version]
- Guissart, C.; Latypova, X.; Rollier, P.; Khan, T.N.; Stamberger, H.; McWalter, K.; Cho, M.T.; Kjaergaard, S.; Weckhuysen, S.; Lesca, G.; et al. Dual Molecular Effects of Dominant RORA Mutations Cause Two Variants of Syndromic Intellectual Disability with Either Autism or Cerebellar Ataxia. Am. J. Hum. Genet. 2018, 102, 744–759. [Google Scholar] [CrossRef] [Green Version]
- Giguère, V.; Beatty, B.; Squire, J.; Copeland, N.G.; Jenkins, N.A. The Orphan Nuclear Receptor RORα (RORA) Maps to a Conserved Region of Homology on Human Chromosome 15q21-q22 and Mouse Chromosome 9. Genomics 1995, 28, 596–598. [Google Scholar]
- Dussault, I.; Fawcett, D.; Matthyssen, A.; Bader, J.A.; Giguère, V. Orphan nuclear receptor RORα-deficient mice display the cerebellar defects of staggerer. Mech. Dev. 1998, 70, 147–153. [Google Scholar] [CrossRef]
- Steinmayr, M.; André, E.; Conquet, F.; Rondi-Reig, L.; Delhaye-Bouchaud, N.; Auclair, N.; Daniel, H.; Crépel, F.; Mariani, J.; Sotelo, C.; et al. Staggerer phenotype in retinoid-related orphan receptor α-deficient mice. Proc. Natl. Acad. Sci. USA 1998, 95, 3960–3965. [Google Scholar] [CrossRef] [Green Version]
- Mamontova, A.; Séguret-Macé, S.; Esposito, B.; Chaniale, C.; Bouly, M.; Delhaye-Bouchaud, N.; Luc, G.; Staels, B.; Duverger, N.; Mariani, J.; et al. Severe Atherosclerosis and Hypoalphalipoproteinemia in the Staggerer Mouse, a Mutant of the Nuclear Receptor RORα. Circulation 1998, 98, 2738–2743. [Google Scholar] [CrossRef]
- Jarvis, C.I.; Staels, B.; Brugg, B.; Lemaigre-Dubreuil, Y.; Tedgui, A.; Mariani, J. Age-related phenotypes in the staggerer mouse expand the RORα nuclear receptor’s role beyond the cerebellum. Mol. Cell. Endocrinol. 2002, 186, 1–5. [Google Scholar] [CrossRef]
- Akashi, M.; Takumi, T. The orphan nuclear receptor RORα regulates circadian transcription of the mammalian core-clock Bmal1. Nat. Struct. Mol. Biol. 2005, 12, 441–448. [Google Scholar] [CrossRef]
- Ino, H. Immunohistochemical Characterization of the Orphan Nuclear Receptor RORα in the Mouse Nervous System. J. Histochem. Cytochem. 2004, 52, 311–323. [Google Scholar] [CrossRef] [Green Version]
- Hazlerigg, D.G.; Barrett, P.; Hastings, M.H.; Morgan, P.J. Are nuclear receptors involved in pituitary responsiveness to melatonin? Mol. Cell. Endocrinol. 1996, 123, 53–59. [Google Scholar] [CrossRef]
- Frédéric, F.; Chianale, C.; Oliver, C.; Mariani, J. Enhanced endocrine response to novel environment stress and lack of corticosterone circadian rhythm in staggerer (Rora sg/sg) mutant mice. J. Neurosci. Res. 2006, 83, 1525–1532. [Google Scholar] [CrossRef]
- Teboul, M.; Enmark, E.; Li, Q.; Wikström, A.C.; Pelto-Huikko, M.; Gustafsson, J.A. OR-1, a member of the nuclear receptor superfamily that interacts with the 9-cis-retinoic acid receptor. Proc. Natl. Acad. Sci. USA 1995, 92, 2096–2100. [Google Scholar] [CrossRef] [Green Version]
- Apfel, R.; Benbrook, D.; Lernhardt, E.; Ortiz, M.A.; Salbert, G.; Pfahl, M. A novel orphan receptor specific for a subset of thyroid hormone-responsive elements and its interaction with the retinoid/thyroid hormone receptor subfamily. Mol. Cell. Biol. 1994, 14, 7025–7035. [Google Scholar] [CrossRef] [Green Version]
- Lu, T.T.; Repa, J.J.; Mangelsdorf, D.J. Orphan Nuclear Receptors as eLiXiRs and FiXeRs of Sterol Metabolism. J. Biol. Chem. 2001, 276, 37735–37738. [Google Scholar] [CrossRef] [Green Version]
- Hu, X.; Li, S.; Wu, J.; Xia, C.; Lala, D.S. Liver X Receptors Interact with Corepressors to Regulate Gene Expression. Mol. Endocrinol. 2003, 17, 1019–1026. [Google Scholar] [CrossRef] [Green Version]
- Komati, R.; Spadoni, D.; Zheng, S.; Sridhar, J.; Riley, K.E.; Wang, G. Ligands of Therapeutic Utility for the Liver X Receptors. Molecules 2017, 22, 88. [Google Scholar] [CrossRef]
- Edwards, P.A.; Ericsson, J. Sterols and Isoprenoids: Signaling Molecules Derived from the Cholesterol Biosynthetic Pathway. Ann. Rev. Biochem. 1999, 68, 157–185. [Google Scholar] [CrossRef]
- Yoshikawa, T.; Shimano, H.; Yahagi, N.; Ide, T.; Amemiya-Kudo, M.; Matsuzaka, T.; Nakakuki, M.; Tomita, S.; Okazaki, H.; Tamura, Y.; et al. Polyunsaturated Fatty Acids Suppress Sterol Regulatory Element-binding Protein 1c Promoter Activity by Inhibition of Liver X Receptor (LXR) Binding to LXR Response Elements. J. Biol. Chem. 2002, 277, 1705–1711. [Google Scholar] [CrossRef] [Green Version]
- Matsumoto, S.; Hashimoto, K.; Yamada, M.; Satoh, T.; Hirato, J.; Mori, M. Liver X receptor-alpha regulates proopiomelanocortin (POMC) gene transcription in the pituitary. Mol. Endocrinol. 2009, 23, 47–60. [Google Scholar] [CrossRef] [Green Version]
- Hashimoto, K.; Matsumoto, S.; Ishida, E.; Miura, A.; Horiguchi, K.; Ozawa, A.; Shibusawa, N.; Satoh, T.; Yamada, M.; Yamada, S.; et al. Liver X receptor-α/β expression ratio is increased in ACTH-secreting pituitary adenomas. Neurosci. Lett. 2011, 494, 34–37. [Google Scholar] [CrossRef]
- Nilsson, M.; Stulnig, T.M.; Lin, C.Y.; Yeo, A.L.; Nowotny, P.; Liu, E.T.; Steffensen, K.R. Liver X Receptors Regulate Adrenal Steroidogenesis and Hypothalamic-Pituitary-Adrenal Feedback. Mol. Endocrinol. 2007, 21, 126–137. [Google Scholar] [CrossRef] [Green Version]
- Kliewer, S.A.; Moore, J.T.; Wade, L.; Staudinger, J.L.; Watson, M.A.; Jones, S.A.; McKee, D.D.; Oliver, B.B.; Willson, T.M.; Zetterström, R.H.; et al. An Orphan Nuclear Receptor Activated by Pregnanes Defines a Novel Steroid Signaling Pathway. Cell 1998, 92, 73–82. [Google Scholar] [CrossRef] [Green Version]
- Lehmann, J.M.; McKee, D.D.; Watson, M.A.; Willson, T.M.; Moore, J.T.; Kliewer, S.A. The human orphan nuclear receptor PXR is activated by compounds that regulate CYP3A4 gene expression and cause drug interactions. J. Clin. Investig. 1998, 102, 1016–1023. [Google Scholar] [CrossRef]
- Bertilsson, G.; Heidrich, J.; Svensson, K.; Åsman, M.; Jendeberg, L.; Sydow-Bäckman, M.; Ohlsson, R.; Postlind, H.; Blomquist, P.; Berkenstam, A. Identification of a human nuclear receptor defines a new signaling pathway for CYP3A induction. Proc. Natl. Acad. Sci. USA 1998, 95, 12208–12213. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Masi, A.; Marinis, E.D.; Ascenzi, P.; Marino, M. Nuclear receptors CAR and PXR: Molecular, functional, and biomedical aspects. Mol. Aspects Med. 2009, 30, 297–343. [Google Scholar] [CrossRef]
- Prakash, C.; Zuniga, B.; Song, C.S.; Jiang, S.; Cropper, J.; Park, S.; Chatterjee, B. Nuclear Receptors in Drug Metabolism, Drug Response and Drug Interactions. Nucl. Receptor Res. 2015, 2, 101178. [Google Scholar] [CrossRef] [PubMed]
- Iyer, M.; Reschly, E.J.; Krasowski, M.D. Functional evolution of the pregnane X receptor. Expert Opin. Drug. Metab. Toxicol. 2006, 2, 381–397. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.; Xie, W. Targeting xenobiotic receptors PXR and CAR for metabolic diseases. Trends Pharmacol. Sci. 2012, 33, 552–558. [Google Scholar] [CrossRef] [Green Version]
- Xue, Y.; Moore, L.B.; Orans, J.; Peng, L.; Bencharit, S.; Kliewer, S.A.; Redinbo, M.R. Crystal Structure of the Pregnane X Receptor-Estradiol Complex Provides Insights into Endobiotic Recognition. Mol. Endocrinol. 2007, 21, 1028–1038. [Google Scholar] [CrossRef] [Green Version]
- Jones, S.A.; Moore, L.B.; Shenk, J.L.; Wisely, G.B.; Hamilton, G.A.; McKee, D.D.; Tomkinson, N.C.O.; LeCluyse, E.L.; Lambert, M.H.; Willson, T.M.; et al. The Pregnane X Receptor: A Promiscuous Xenobiotic Receptor That Has Diverged during Evolution. Mol. Endocrinol. 2000, 14, 27–39. [Google Scholar] [CrossRef]
- He, J.; Cheng, Q.; Xie, W. Minireview: Nuclear receptor-controlled steroid hormone synthesis and metabolism. Mol. Endocrinol. 2010, 24, 11–21. [Google Scholar] [CrossRef] [Green Version]
- Xie, W.; Yeuh, M.F.; Radominska-Pandya, A.; Saini, S.P.S.; Negishi, Y.; Bottroff, B.S.; Cabrera, G.Y.; Tukey, R.H.; Evans, R.M. Control of steroid, heme, and carcinogen metabolism by nuclear pregnane X receptor and constitutive androstane receptor. Proc. Natl. Acad. Sci. USA 2003, 100, 4150–4155. [Google Scholar] [CrossRef] [Green Version]
- Zhai, Y.; Pai, H.V.; Zhou, J.; Amico, J.A.; Vollmer, R.R.; Xie, W. Activation of Pregnane X Receptor Disrupts Glucocorticoid and Mineralocorticoid Homeostasis. Mol. Endocrinol. 2007, 21, 138–147. [Google Scholar] [CrossRef]
- Giguère, V. Orphan Nuclear Receptors: From Gene to Function*. Endocr. Rev. 1999, 20, 689–725. [Google Scholar] [CrossRef]
- Heaney, A.P. Novel Pituitary Ligands: Peroxisome Proliferator Activating Receptor-γ. Pituitary 2003, 6, 153–159. [Google Scholar] [CrossRef]
- Heaney, A.P.; Fernando, M.; Melmed, S. PPAR-gamma receptor ligands: Novel therapy for pituitary adenomas. J. Clin. Investig. 2003, 111, 1381–1388. [Google Scholar] [CrossRef]
- Chang, C.; Kokontis, J.; Acakpo-Satchivi, L.; Liao, S.; Takeda, H.; Chang, Y. Molecular cloning of new human TR2 receptors: A class of steroid receptor with multiple ligand-binding domains. Biochem. Biophys. Res. Commun. 1989, 165, 735–741. [Google Scholar] [CrossRef]
- Mullican, S.E.; DiSpirito, J.R.; Lazar, M.A. The Orphan Nuclear Receptors at Their 25th Year Reunion. J. Mol. Endocrinol. 2013, 51, T115–T140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, Y.F.; Lee, H.J.; Chang, C. Recent advances in the TR2 and TR4 orphan receptors of the nuclear receptor superfamily. J. Steroid Biochem. Mol. Biol. 2002, 81, 291–308. [Google Scholar] [CrossRef]
- Lin, S.J.; Zhang, Y.; Liu, N.C.; Yang, D.R.; Li, G.; Chang, C. Minireview: Pathophysiological Roles of the TR4 Nuclear Receptor: Lessons Learned From Mice Lacking TR4. Mol. Endocrinol. 2014, 28, 805–821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shyr, C.R.; Kang, H.Y.; Tsai, M.Y.; Liu, N.C.; Ku, P.Y.; Huang, K.E.; Chang, C. Roles of Testicular Orphan Nuclear Receptors 2 and 4 in Early Embryonic Development and Embryonic Stem Cells. Endocrinology 2009, 150, 2454–2462. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.E.; Suino-Powell, K.M.; Xu, Y.; Chan, C.W.; Tanabe, O.; Kruse, S.W.; Reynolds, R.; Engel, J.D.; Xu, H.E. The Orphan Nuclear Receptor TR4 Is a Vitamin A-activated Nuclear Receptor. J. Biol. Chem. 2011, 286, 2877–2885. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.F.; Young, W.J.; Burbach, J.P.H.; Chang, C. Negative Feedback Control of the Retinoid-Retinoic Acid/Retinoid X Receptor Pathway by the Human TR4 Orphan Receptor, a Member of the Steroid Receptor Superfamily. J. Biol. Chem. 1998, 273, 13437–13443. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.F.; Bao, B.Y.; Chang, C. Modulation of the retinoic acid-induced cell apoptosis and differentiation by the human TR4 orphan nuclear receptor. Biochem. Biophys. Res. Commun. 2004, 323, 876–883. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.T.; Collins, L.L.; Uno, H.; Chang, C. Deficits in Motor Coordination with Aberrant Cerebellar Development in Mice Lacking Testicular Orphan Nuclear Receptor 4. Mol. Cell. Biol. 2005, 25, 2722–2732. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Du, L.; Bergsneider, M.; Mirsadraei, L.; Young, S.H.; Jonker, J.W.; Downes, M.; Yong, W.H.; Evans, R.M.; Heaney, A.P. Evidence for orphan nuclear receptor TR4 in the etiology of Cushing disease. Proc. Natl. Acad. Sci. USA 2013, 110, 8555–8560. [Google Scholar] [CrossRef] [Green Version]
- Zhang, D.; Du, L.; Heaney, A.P. Testicular Receptor-4: Novel Regulator of Glucocorticoid Resistance. J. Clin. Endocrinol. Metab. 2016, 101, 3123–3133. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, D.; Bergsneider, M.; Wang, M.B.; Heaney, A.P. Targeting the ERK pathway for the treatment of Cushing’s disease. Oncotarget 2016, 7, 69149–69158. [Google Scholar] [PubMed] [Green Version]
- Raccurt, M.; Smallwood, S.; Mertani, H.C.; Devost, D.; Abbaci, K.; Boutin, J.M.; Morel, G. Cloning, expression and regulation of chicken ovalbumin upstream promoter transcription factors (COUP-TFII and EAR-2) in the rat anterior pituitary gland. Neuroendocrinology 2005, 82, 233–244. [Google Scholar] [CrossRef]
- Cooney, A.J.; Tsai, S.Y.; O’Malley, B.W.; Tsai, M.J. Chicken ovalbumin upstream promoter transcription factor (COUP-TF) dimers bind to different GGTCA response elements, allowing COUP-TF to repress hormonal induction of the vitamin D3, thyroid hormone, and retinoic acid receptors. Mol. Cell Biol. 1992, 12, 4153–4163. [Google Scholar] [CrossRef]
- Qiu, Y.; Tsai, S.Y.; Tsai, M.J. COUP-TF an orphan member of the steroid/thyroid hormone receptor superfamily. Trends Endocrinol. Metab. 1994, 5, 234–239. [Google Scholar] [CrossRef]
- De Souza, F.S.J.; Nasif, S.; López-Leal, R.; Levi, D.H.; Low, M.J.; Rubinsten, M. The estrogen receptor α colocalizes with proopiomelanocortin in hypothalamic neurons and binds to a conserved motif present in the neuron-specific enhancer nPE2. Eur. J. Pharmacol. 2011, 660, 181–187. [Google Scholar] [CrossRef] [Green Version]
- Friend, K.E.; McCutcheon, I.E. Estrogen Receptor Expression in the Pituitary Gland. In Sex-Steroid Interactions with Growth Hormone; Veldhuis, J.D., Giustina, A., Eds.; Springer New York: New York, NY, USA, 1999; pp. 301–307. [Google Scholar]
- Friend, K.E.; Chiou, Y.K.; Lopes, M.B.; Laws, E.R.; Hughes, K.M.; Shupnik, M.A. Estrogen receptor expression in human pituitary: Correlation with immunohistochemistry in normal tissue, and immunohistochemistry and morphology in macroadenomas. J. Clin. Endocrinol. Metabol. 1994, 78, 1497–1504. [Google Scholar]
- Nakao, H.; Koga, M.; Arao, M.; Nakao, M.; Sato, B.; Kishimoto, S.; Saitoh, Y.; Arita, N.; Mori, S. Enzyme-immunoassay for estrogen receptors in human pituitary adenomas. Eur. J. Endocrinol. 1989, 120, 233–238. [Google Scholar] [CrossRef] [PubMed]
- Pichon, M.; Bression, D.; Peillon, F.; Milgrom, E. Estrogen Receptors in Human Pituitary Adenomas. J. Clin. Endocrinol. Metab. 1980, 51, 897–902. [Google Scholar] [CrossRef] [PubMed]
- Lloyd, R.V.; Osamura, R.Y. Transcription factors in normal and neoplastic pituitary tissues. Microsc. Res. Tech. 1997, 39, 168–181. [Google Scholar] [CrossRef]
- Manoranjan, B.; Salehi, F.; Scheithauer, B.; Rotondo, F.; Kovacs, K.; Cusimano, M. Estrogen Receptors α and β Immunohistochemical Expression: Clinicopathological Correlations in Pituitary Adenomas. Anticancer Res. 2010, 30, 2897–2904. [Google Scholar] [PubMed]
- Waterman, M.L.; Adlerf, S.; Nelson, C.; Greene, G.L.; Evans, R.M.; Rosenfeld, M.G. A Single Domain of the Estrogen Receptor Confers Deoxyribonucleic Acid Binding and Transcriptional Activation of the Rat Prolactin Gene. Mol. Endocrinol. 1988, 2, 14–21. [Google Scholar] [CrossRef]
- Maurer, R.A.; Notides, A.C. Identification of an estrogen-responsive element from the 5′-flanking region of the rat prolactin gene. Mol. Cell Biol. 1987, 7, 4247–4254. [Google Scholar] [CrossRef]
- Nowakowski, B.E.; Maurer, R.A. Multiple Pit-1-binding sites facilitate estrogen responsiveness of the prolactin gene. Mol. Endocrinol. 1994, 8, 1742–1749. [Google Scholar]
- Holloway, J.M.; Szeto, D.P.; Scully, K.M.; Glass, C.K.; Rosenfeld, M.G. Pit-1 binding to specific DNA sites as a monomer or dimer determines gene-specific use of a tyrosine-dependent synergy domain. Genes Dev. 1995, 9, 1992–2006. [Google Scholar] [CrossRef] [Green Version]
- Scully, K.M.; Gleiberman, A.S.; Lindzey, J.; Lubahn, D.B.; Korach, K.S.; Rosenfeld, M.G. Role of Estrogen Receptor-α in the Anterior Pituitary Gland. Mol. Endocrinol. 1997, 11, 674–681. [Google Scholar]
- Chaidarun, S.S.; Swearingen, B.; Alexander, J.M. Differential Expression of Estrogen Receptor-β (ERβ) in Human Pituitary Tumors: Functional Interactions with ERα and a Tumor-Specific Splice Variant1. J. Clin. Endocrinol. Metab. 1998, 83, 3308–3315. [Google Scholar] [CrossRef]
- Ramamoorthy, S.; Cidlowski, J.A. Exploring the Molecular Mechanisms of Glucocorticoid Receptor Action from Sensitivity to Resistance. Endocr. Dev. 2013, 24, 41–56. [Google Scholar] [PubMed] [Green Version]
- Eberwine, J.H.; Roberts, J.L. Glucocorticoid regulation of pro-opiomelanocortin gene transcription in the rat pituitary. J. Biol. Chem. 1984, 259, 2166–2170. [Google Scholar] [PubMed]
- Drouin, J.; Lin, S.Y.; Nemer, M. Glucocorticoid repression of pro-opiomelanocortin gene transcription. J. Steroid Biochem. 1989, 34, 63–69. [Google Scholar] [CrossRef]
- Drouin, J.; Trifiro, M.A.; Plante, R.K.; Nemer, M.; Eriksson, P.; Wrange, O. Glucocorticoid receptor binding to a specific DNA sequence is required for hormone-dependent repression of pro-opiomelanocortin gene transcription. Mol. Cell Biol. 1989, 9, 5305–5314. [Google Scholar] [CrossRef] [Green Version]
- Drouin, J.; Sun, Y.L.; Nemer, M. Regulatory elements of the pro-opiomelanocortin gene pituitary specificity and glucocorticoid repression. Trends Endocrinol. Metab. 1990, 1, 219–225. [Google Scholar] [CrossRef]
- Drouin, J.; Sun, Y.L.; Chamberland, M.; Gauthier, Y.; De Léan, A.; Nemer, M.; Schmidt, T.J. Novel glucocorticoid receptor complex with DNA element of the hormone-repressed POMC gene. EMBO J. 1993, 12, 145–156. [Google Scholar] [CrossRef]
- Oakley, R.H.; Cidlowski, J.A. The Biology of the Glucocorticoid Receptor: New Signaling Mechanisms in Health and Disease. J. Allergy Clin. Immunol. 2013, 132, 1033–1044. [Google Scholar] [CrossRef] [Green Version]
- Panek, M.; Pietras, T.; Fabijan, A.; MiŁAnowski, M.; Wieteska, Ł.; GÓRski, P.; Kuna, P.; Szemraj, J. Effect of glucocorticoid receptor gene polymorphisms on asthma phenotypes. Exp.Ther. Med. 2013, 5, 572–580. [Google Scholar] [CrossRef] [Green Version]
- Sánchez-Vega, B.; Krett, N.; Rosen, S.T.; Gandhi, V. Glucocorticoid receptor transcriptional isoforms and resistance in multiple myeloma cells. Mol. Cancer Ther. 2006, 5, 3062–3070. [Google Scholar] [CrossRef] [Green Version]
- Smith, L.K.; Cidlowski, J.A. Glucocorticoid-Induced Apoptosis of Healthy and Malignant Lymphocytes. Prog. Brain Res. 2010, 182, 1–30. [Google Scholar]
- John, K.; Marino, J.S.; Sanchez, E.R.; Hinds, T.D. The glucocorticoid receptor: Cause of or cure for obesity? Am. J. Physiol. Endocrinol. Metabol. 2016, 310, E249–E257. [Google Scholar] [CrossRef] [Green Version]
- Oakley, R.H.; Cidlowski, J.A. Cellular Processing of the Glucocorticoid Receptor Gene and Protein: New Mechanisms for Generating Tissue-specific Actions of Glucocorticoids. J. Biol. Chem. 2011, 286, 3177–3184. [Google Scholar] [CrossRef] [Green Version]
- Rivers, C.; Levy, A.; Hancock, J.; Lightman, S.; Norman, M. Insertion of an Amino Acid in the DNA-Binding Domain of the Glucocorticoid Receptor as a Result of Alternative Splicing. J. Clin. Endocrinol. Metab. 1999, 84, 4283–4286. [Google Scholar] [CrossRef] [PubMed]
- Kadmiel, M.; Cidlowski, J.A. Glucocorticoid receptor signaling in health and disease. Trends Pharmacol. Sci. 2013, 34, 518–530. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kino, T. Glucocorticoid receptor. In Endotext [Internet], 2017; Feingold, K.R., Anawalt, B., Boyce, A., Chrousos, G., Dungan, K., Grossman, A., Hershman, J.M., Kaltsas, G., Koch, C., Kopp, P., et al., Eds.; MDText.com, Inc.: South Dartmouth, MA, USA, 2017. [Google Scholar]
- Charmandari, E.; Kino, T.; Chrousos, G.P. Familial/sporadic glucocorticoid resistance: Clinical phenotype and molecular mechanisms. Ann. N. Y. Acad. Sci. 2004, 1024, 168–181. [Google Scholar] [CrossRef] [PubMed]
- Nicolaides, N.C.; Charmandari, E.; Chrousos, G.P.; Kino, T. Recent advances in the molecular mechanisms determining tissue sensitivity to glucocorticoids: Novel mutations, circadian rhythm and ligand-induced repression of the human glucocorticoid receptor. BMC Endocr. Disord. 2014, 14, 71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Charmandari, E.; Kino, T.; Ichijo, T.; Chrousos, G.P. Generalized Glucocorticoid Resistance: Clinical Aspects, Molecular Mechanisms, and Implications of a Rare Genetic Disorder. The J. Clin. Endocrinol. Metab. 2008, 93, 1563–1572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Charmandari, E.; Ichijo, T.; Jubiz, W.; Baid, S.; Zachman, K.; Chrousos, G.P.; Kino, T. A Novel Point Mutation in the Amino Terminal Domain of the Human Glucocorticoid Receptor (hGR) Gene Enhancing hGR-Mediated Gene Expression. J. Clin. Endocrinol. Metab. 2008, 93, 4963–4968. [Google Scholar] [CrossRef] [Green Version]
- Van Rossum, E.F.C.; Koper, J.W.; Huizenga, N.A.T.M.; Uitterlinden, A.G.; Janssen, J.A.M.J.L.; Brinkmann, A.O.; Grobbee, D.E.; de Jong, F.H.; van Duyn, C.M.; Pols, H.A.P.; et al. A Polymorphism in the Glucocorticoid Receptor Gene, Which Decreases Sensitivity to Glucocorticoids In Vivo, Is Associated With Low Insulin and Cholesterol Levels. Diabetes 2002, 51, 3128–3134. [Google Scholar] [CrossRef] [Green Version]
- Karl, M.; Lamberts, S.W.; Detera-Wadleigh, S.D.; Encio, I.J.; Stratakis, C.A.; Hurley, D.M.; Accili, D.; Chrousos, G.P. Familial glucocorticoid resistance caused by a splice site deletion in the human glucocorticoid receptor gene. J. Clin. Endocrinol. Metab. 1993, 76, 683–689. [Google Scholar]
- Syed, A.A.; Irving, J.A.E.; Redfern, C.P.F.; Hall, A.G.; Unwin, N.C.; White, M.; Bhopal, R.S.; Weaver, J.U. Association of Glucocorticoid Receptor Polymorphism A3669G in Exon 9β with Reduced Central Adiposity in Women. Obesity 2006, 14, 759–764. [Google Scholar] [CrossRef] [PubMed]
- Liberman, A.C.; Antunica-Noguerol, M.; Arzt, E. Modulation of the glucocorticoid receptor activity by post-translational modifications. Nucl. Recept. Res. 2014, 1, 1–15. [Google Scholar] [CrossRef]
- Galliher-Beckley, A.J.; Cidlowski, J.A. Emerging roles of glucocorticoid receptor phosphorylation in modulating glucocorticoid hormone action in health and disease. IUBMB Life 2009, 61, 979–986. [Google Scholar] [CrossRef] [PubMed]
- Helzer, K.T.; Hooper, C.; Miyamoto, S.; Alarid, E.T. Ubiquitylation of nuclear receptors: New linkages and therapeutic implications. J. Mol. Endocrinol. 2015, 54, R151. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le Drean, Y.; Mincheneau, N.; Le Goff, P.; Michel, D. Potentiation of Glucocorticoid Receptor Transcriptional Activity by Sumoylation. Endocrinology 2002, 143, 3482–3489. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Druker, J.; Liberman, A.C.; Antunica-Noguerol, M.; Gerez, J.; Paez-Pereda, M.; Rein, T.; Iñiguez-Lluhí, J.A.; Holsboer, F.; Arzt, E. RSUME Enhances Glucocorticoid Receptor SUMOylation and Transcriptional Activity. Mol. Cell. Biol. 2013, 33, 2116–2127. [Google Scholar] [CrossRef] [Green Version]
- Murphy, E.P.; Conneely, O.M. Neuroendocrine Regulation of the Hypothalamic Pituitary Adrenal Axis by the nurr1/nur77 Subfamily of Nuclear Receptors. Mol. Endocrinol. 1997, 11, 39–47. [Google Scholar] [CrossRef]
- Milbrandt, J. Nerve growth factor induces a gene homologous to the glucocorticoid receptor gene. Neuron 1988, 1, 183–188. [Google Scholar] [CrossRef]
- Campos-Melo, D.; Galleguillos, D.; Sánchez, N.; Gysling, K.; Andrés, M. Nur transcription factors in stress and addiction. Front. Mol. Neurosci. 2013, 6, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Wilson, T.; Fahrner, T.; Johnston, M.; Milbrandt, J. Identification of the DNA binding site for NGFI-B by genetic selection in yeast. Science 1991, 252, 1296–1300. [Google Scholar] [CrossRef]
- Perlmann, T.; Jansson, L. A novel pathway for vitamin A signaling mediated by RXR heterodimerization with NGFI-B and NURR1. Genes Dev. 1995, 9, 769–782. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Okabe, T.; Takayanagi, R.; Adachi, M.; Imasaki, K.; Nawata, H. Nur77, a member of the steroid receptor superfamily, antagonizes negative feedback of ACTH synthesis and secretion by glucocorticoid in pituitary corticotrope cells. J. Endocrinol. 1998, 156, 169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Philips, A.; Lesage, S.; Gingras, R.; Maira, M.H.; Gauthier, Y.; Hugo, P.; Drouin, J. Novel dimeric Nur77 signaling mechanism in endocrine and lymphoid cells. Mol. Cell Biol. 1997, 17, 5946–5951. [Google Scholar] [CrossRef] [Green Version]
- Kovalovsky, D.; Paez Pereda, M.; Labeur, M.; Renner, U.; Holsboer, F.; Stalla, G.K.; Arzt, E. Nur77 induction and activation are necessary for interleukin-1 stimulation of proopiomelanocortin in AtT-20 corticotrophs. FEBS Lett. 2004, 563, 229–233. [Google Scholar] [CrossRef] [Green Version]
- Făgăraşan, M.O.; Eskay, R.; Axelrod, J. Interleukin 1 potentiates the secretion of beta-endorphin induced by secretagogues in a mouse pituitary cell line (AtT-20). Proc. Natl. Acad. Sci. USA 1989, 86, 2070–2073. [Google Scholar] [CrossRef] [Green Version]
- Webster, E.L.; Tracey, D.E.; De Souza, E.B. Upregulation of Interleukin-1 Receptors in Mouse AtT-20 Pituitary Tumor Cells Following Treatment with Corticotropin-Releasing Factor. Endocrinology 1991, 129, 2796–2798. [Google Scholar] [CrossRef]
- Philips, A.; Maira, M.; Mullick, A.; Chamberland, M.; Lesage, S.; Hugo, P.; Drouin, J. Antagonism between Nur77 and glucocorticoid receptor for control of transcription. Mol. Cell Biol. 1997, 17, 5952–5959. [Google Scholar] [CrossRef] [Green Version]
- Martens, C.; Bilodeau, S.; Maira, M.; Gauthier, Y.; Drouin, J. Protein-Protein Interactions and Transcriptional Antagonism between the Subfamily of NGFI-B/Nur77 Orphan Nuclear Receptors and Glucocorticoid Receptor. Mol. Endocrinology 2005, 19, 885–897. [Google Scholar] [CrossRef]
- Maxwell, M.A.; Muscat, G.E.O. The NR4A subgroup: Immediate early response genes with pleiotropic physiological roles. Nucl. Recept. Signal 2006, 4, e002. [Google Scholar] [CrossRef] [Green Version]
- Kovalovsky, D.; Refojo, D.; Liberman, A.C.; Hochbaum, D.; Pereda, M.P.; Coso, O.A.; Stalla, G.K.; Holsboer, F.; Arzt, E. Activation and Induction of NUR77/NURR1 in Corticotrophs by CRH/cAMP: Involvement of Calcium, Protein Kinase, A., and MAPK Pathways. Mol. Endocrinol. 2002, 16, 1638–1651. [Google Scholar] [CrossRef]
- Arredondo, C.; Orellana, M.; Vecchiola, A.; Pereira, L.A.; Galdames, L.; Andrés, M.E. PIASγ Enhanced SUMO-2 Modification of Nurr1 Activation-Function-1 Domain Limits Nurr1 Transcriptional Synergy. PLoS ONE 2013, 8, e55035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maira, M.; Martens, C.; Batsché, É.; Gauthier, Y.; Drouin, J. Dimer-Specific Potentiation of NGFI-B (Nur77) Transcriptional Activity by the Protein Kinase A Pathway and AF-1-Dependent Coactivator Recruitment. Mol. Cell Biol. 2003, 23, 763–776. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wansa, K.D.S.A.; Harris, J.M.; Muscat, G.E.O. The Activation Function-1 Domain of Nur77/NR4A1 Mediates Trans-activation, Cell Specificity, and Coactivator Recruitment. J. Biol. Chem. 2002, 277, 33001–33011. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, J.; Li, Q. Review of the in Vivo Functions of the p160 Steroid Receptor Coactivator Family. Mol. Endocrinol. 2003, 17, 1681–1692. [Google Scholar] [CrossRef]
- Rambaud, J.; Desroches, J.; Balsalobre, A.; Drouin, J. TIF1β/KAP-1 Is a Coactivator of the Orphan Nuclear Receptor NGFI-B/Nur77. J. Biol. Chem. 2009, 284, 14147–14156. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Benoit, G.; Liu, J.; Prasad, S.; Aarnisalo, P.; Liu, X.; Xu, H.; Walker, N.P.C.; Perlmann, T. Structure and function of Nurr1 identifies a class of ligand-independent nuclear receptors. Nature 2003, 423, 555–560. [Google Scholar] [CrossRef]
- Flaig, R.; Greschik, H.; Peluso-Iltis, C.; Moras, D. Structural Basis for the Cell-specific Activities of the NGFI-B and the Nurr1 Ligand-binding Domain. J. Biol. Chem. 2005, 280, 19250–19258. [Google Scholar] [CrossRef] [Green Version]
- Chintharlapalli, S.; Burghardt, R.; Papineni, S.; Ramaiah, S.; Yoon, K.; Safe, S. Activation of Nur77 by Selected 1,1-Bis(3′-indolyl)-1-(p-substituted phenyl)methanes Induces Apoptosis through Nuclear Pathways. J. Biol. Chem. 2005, 280, 24903–24914. [Google Scholar] [CrossRef] [Green Version]
- Cho, S.D.; Yoon, K.; Chintharlapalli, S.; Abdelrahim, M.; Lei, P.; Hamilton, S.; Khan, S.; Ramaiah, S.K.; Safe, S. Nur77 Agonists Induce Proapoptotic Genes and Responses in Colon Cancer Cells through Nuclear Receptor–Dependent and Nuclear Receptor–Independent Pathways. Cancer Res. 2007, 67, 674–683. [Google Scholar] [CrossRef] [Green Version]
- Mohankumar, K.; Li, X.; Sung, N.; Cho, Y.J.; Han, S.J.; Safe, S. Bis-indole derived nuclear receptor 4A1 (NR4A1, Nur77) ligands as inhibitors of endometriosis. Endocrinology 2020, 161, bqaa027. [Google Scholar] [CrossRef]
- Zhan, Y.; Du, X.; Chen, H.; Liu, J.; Zhao, B.; Huang, D.; Li, G.; Xu, Q.; Zhang, M.; Weimer, B.C.; et al. Cytosporone B is an agonist for nuclear orphan receptor Nur77. Nat. Chem. Biol. 2008, 4, 548–556. [Google Scholar] [CrossRef] [PubMed]
- Vinayavekhin, N.; Saghatelian, A. Discovery of a protein-metabolite interaction between unsaturated fatty acids and the nuclear receptor Nur77 using a metabolomics approach. J. Am. Chem. Soc. 2011, 133, 17168–17171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lakshmi, S.P.; Reddy, A.T.; Banno, A.; Reddy, R.C. Molecular, chemical, and structural characterization of prostaglandin A2 as a novel agonist for Nur77. Biochem. J. 2019, 476, 2757–2767. [Google Scholar] [CrossRef] [PubMed]
- Jakaria, M.; Haque, M.E.; Cho, D.Y.; Azam, S.; Kim, I.S.; Choi, D.K. Molecular Insights into NR4A2(Nurr1): An Emerging Target for Neuroprotective Therapy Against Neuroinflammation and Neuronal Cell Death. Mol. Neurobiol. 2019, 56, 5799–5814. [Google Scholar] [CrossRef]
- Medzikovic, L.; de Vries, C.J.M.; de Waard, V. NR4A nuclear receptors in cardiac remodeling and neurohormonal regulation. Trends Cardiovasc. Med. 2019, 29, 429–437. [Google Scholar] [CrossRef]
- Hedrick, E.; Lee, S.O.; Doddapaneni, R.; Singh, M.; Safe, S. Nuclear receptor 4A1 as a drug target for breast cancer chemotherapy. Endocr. Relat. Cancer. 2015, 22, 831–840. [Google Scholar] [CrossRef]
- Banno, A.; Lakshmi, S.P.; Reddy, A.T.; Kim, S.C.; Reddy, R.C. Key Functions and Therapeutic Prospects of Nur77 in Inflammation Related Lung Diseases. Am. J. Pathol. 2019, 189, 482–491. [Google Scholar] [CrossRef] [Green Version]
- Gervais, J.; Soghomonian, J.J.; Richard, D.; Rouillard, C. Dopamine and serotonin interactions in the modulation of the expression of the immediate-early transcription factor, nerve growth factor-inducible B, in the striatum. Neuroscience 1999, 91, 1045–1054. [Google Scholar] [CrossRef]
- Pivonello, R.; Ferone, D.; de Herder, W.W.; Kros, J.M.; De Caro, M.L.; Arvigo, M.; Annunziato, L.; Lombardi, G.; Colao, A.; Hofland, L.J.; et al. Dopamine Receptor Expression and Function in Corticotroph Pituitary Tumors. J. Clin. Endocrinol. Metabol. 2004, 89, 2452–2462. [Google Scholar] [CrossRef] [Green Version]
- Crawford, P.A.; Sadovsky, Y.; Woodson, K.; Lee, S.L.; Milbrandt, J. Adrenocortical function and regulation of the steroid 21-hydroxylase gene in NGFI-B-deficient mice. Mol. Cell Biol. 1995, 15, 4331–4336. [Google Scholar] [CrossRef] [Green Version]
- Law, S.W.; Conneely, O.M.; DeMayo, F.J.; O’Malley, B.W. Identification of a new brain-specific transcription factor, NURR1. Mol. Endocrinol. 1992, 6, 2129–2135. [Google Scholar] [PubMed] [Green Version]
- Maira, M.; Martens, C.; Philips, A.; Drouin, J. Heterodimerization between members of the Nur subfamily of orphan nuclear receptors as a novel mechanism for gene activation. Mol. Cell Biol. 1999, 19, 7549–7557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jankovic, J.; Chen, S.; Le, W.D. The role of Nurr1 in the development of dopaminergic neurons and Parkinson’s disease. Prog. Neurobiol. 2005, 77, 128–138. [Google Scholar] [CrossRef] [PubMed]
- Carpentier, R.; Sacchetti, P.; Ségard, P.; Staels, B.; Lefebvre, P. The glucocorticoid receptor is a co-regulator of the orphan nuclear receptor Nurr1. J. Neurochem. 2008, 104, 777–789. [Google Scholar] [CrossRef] [PubMed]
- Sacchetti, P.; Carpentier, R.; Ségard, P.; Olivé-Cren, C.; Lefebvre, P. Multiple signaling pathways regulate the transcriptional activity of the orphan nuclear receptor NURR1. Nucleic Acids Res. 2006, 34, 5515–5527. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galleguillos, D.; Vecchiola, A.; Fuentealba, J.A.; Ojeda, V.; Alvarez, K.; Gómez, A.; Andrés, M.E. PIASγ Represses the Transcriptional Activation Induced by the Nuclear Receptor Nurr1. J. Biol. Chem. 2004, 279, 2005–2011. [Google Scholar] [CrossRef] [Green Version]
- Sacchetti, P.; Dwornik, H.; Formstecher, P.; Rachez, C.; Lefebvre, P. Requirements for Heterodimerization between the Orphan Nuclear Receptor Nurr1 and Retinoid X Receptors. J. Biol. Chem. 2002, 277, 35088–35096. [Google Scholar] [CrossRef] [Green Version]
- Takayasu, S.; Iwasaki, Y.; Nigawara, T.; Asai, M.; Yoshida, M.; Kageyama, K.; Suda, T. Involvement of Nuclear Factor-ĸB and Nurr-1 in Cytokine-Induced Transcription of Proopiomelanocortin Gene in AtT20 Corticotroph Cells. Neuroimmunomodulation 2010, 17, 88–96. [Google Scholar] [CrossRef]
- De Vera, I.M.S.; Giri, P.K.; Munoz-Tello, P.; Brust, R.; Fuhrmann, J.; Matta-Camacho, E.; Shang, J.; Campbell, S.; Wilson, H.D.; Granados, J.; et al. Identification of a Binding Site for Unsaturated Fatty Acids in the Orphan Nuclear Receptor Nurr1. ACS Chem. Biol. 2016, 11, 1795–1799. [Google Scholar] [CrossRef]
- De Vera, I.M.S.; Munoz-Tello, P.; Zheng, J.; Dharmarajan, V.; Marciano, D.P.; Matta-Camacho, E.; Giri, P.K.; Shang, J.; Hughes, T.S.; Rance, M.; et al. Defining a Canonical Ligand-Binding Pocket in the Orphan Nuclear Receptor Nurr1. Structure 2019, 27, 66–77.e65. [Google Scholar] [CrossRef] [Green Version]
- Windshügel, B. Structural insights into ligand-binding pocket formation in Nurr1 by molecular dynamics simulations. J. Biomol. Struct. Dyn. 2019, 37, 4651–4657. [Google Scholar] [CrossRef] [PubMed]
- DeYoung, R.A.; Baker, J.C.; Cado, D.; Winoto, A. The Orphan Steroid Receptor Nur77 Family Member Nor-1 Is Essential for Early Mouse Embryogenesis. J. Biol. Chem. 2003, 278, 47104–47109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mullican, S.E.; Zhang, S.; Konopleva, M.; Ruvolo, V.; Andreeff, M.; Milbrandt, J.; Conneely, O.M. Abrogation of nuclear receptors Nr4a3 andNr4a1 leads to development of acute myeloid leukemia. Nat. Med. 2007, 13, 730–735. [Google Scholar] [CrossRef] [PubMed]
- Winoto, A.; Littman, D.R. Nuclear Hormone Receptors in T Lymphocytes. Cell 2002, 109, S57–S66. [Google Scholar] [CrossRef] [Green Version]
- Winoto, A. Genes involved in T-cell receptor-mediated apoptosis of thymocytes and T-cell hybridomas. Semin. Immunol. 1997, 9, 51–58. [Google Scholar] [CrossRef] [PubMed]
- Banta, K.L.; Wang, X.; Das, P.; Winoto, A. B cell lymphoma 2 (Bcl-2) residues essential for Bcl-2′s apoptosis-inducing interaction with Nur77/Nor-1 orphan steroid receptors. J. Biol. Chem. 2018, 293, 4724–4734. [Google Scholar] [CrossRef] [Green Version]
- Wansa, K.D.S.A.; Harris, J.M.; Yan, G.; Ordentlich, P.; Muscat, G.E.O. The AF-1 Domain of the Orphan Nuclear Receptor NOR-1 Mediates Trans-activation, Coactivator Recruitment, and Activation by the Purine Anti-metabolite 6-Mercaptopurine. J. Biol. Chem. 2003, 278, 24776–24790. [Google Scholar] [CrossRef] [Green Version]
- Shiota, M.; Fujimoto, N.; Kashiwagi, E.; Eto, M. The Role of Nuclear Receptors in Prostate Cancer. Cells 2019, 8, 602. [Google Scholar] [CrossRef] [Green Version]
- Zhao, L.; Bakke, M.; Krimkevich, Y.; Cushman, L.J.; Parlow, A.F.; Camper, S.A.; Parker, K.L. Steroidogenic factor 1 (SF1) is essential for pituitary gonadotrope function. Development 2001, 128, 147–154. [Google Scholar]
- Fortin, J.; Kumar, V.; Zhou, X.; Wang, Y.; Auwerx, J.; Schoonjans, K.; Boehm, U.; Boerboom, D.; Bernard, D.J. NR5A2 regulates Lhb and Fshb transcription in gonadotrope-like cells in vitro, but is dispensable for gonadotropin synthesis and fertility in vivo. PLoS ONE 2013, 8, e59058. [Google Scholar] [CrossRef] [Green Version]
- Ciato, D.; Albani, A. Molecular Mechanisms of Glucocorticoid Resistance in Corticotropinomas: New Developments and Drug Targets. Front. Endocrinol. (Lausanne) 2020, 11, 21. [Google Scholar] [CrossRef] [PubMed]




| Family | Gene Name | Gene Symbol | Abbreviation | Ligand | Effect on POMC Transcription | POMC Promoter Binding Site |
|---|---|---|---|---|---|---|
| 0B | Dosage-sensitive sex reversal-adrenal hypoplasia congenital critical region on the Xchromosome, Gene 1 | NR0B1 | DAX1 | |||
| Short heterodimeric partner | NR0B2 | SHP | ||||
| 1A | Thyroid hormone receptor-α | NR1A1 | TRα | T3, T4 | ||
| Thyroid hormone receptor-β | NR1A2 | TRβ | T3, T4 | |||
| 1B | Retinoic acid receptor-α | NR1B1 | RARα | Retinoic Acids | RAs↓ [41,42,43,44,45,46,47] /Am80↑ [53] | |
| Retinoic acid receptor-β | NR1B2 | RARβ | Retinoic Acids | |||
| Retinoic acid receptor-γ | NR1B3 | RARγ | Retinoic Acids | |||
| 1C | Peroxisome proliferator-activated receptor-α | NR1C1 | PPARα | Fatty Acids | ||
| Peroxisome proliferator-activated receptor-β | NR1C2 | PPARβ | Fatty Acids | |||
| Peroxisome proliferator-activated receptor-γ | NR1C3 | PPARγ | Fatty Acids | Rosiglitazone↓ [67,68,69,70,71] | ||
| 1D | Reverse-Erb-α | NR1D1 | REV-ERBα | Heme | ||
| Reverse-Erb-β | NR1D2 | REV-ERBβ | Heme | |||
| 1F | Retinoic acid receptor-related orphan receptor-α | NR1F1 | RORα | Cholesterol | ||
| Retinoic acid receptor-related orphan receptor-β | NR1F2 | RORβ | Cholesterol | |||
| Retinoic acid receptor-related orphan receptor-γ | NR1F3 | RORγ | Cholesterol | |||
| 1H | Liver X receptor-α | NR1H3 | LXRα | Oxysterols | TO901317↑ [119,121] | (-73 AGGAAGGTCA CGTC CAAGGCTCA -52) [119] |
| Liver X receptor-β | NR1H2 | LXRβ | Oxysterols | |||
| Farnesoid X receptor-α | NR1H4 | FXRα | Bile acids | |||
| Farnesoid X receptor-β | NR1H5P | FXRβ | /Farnesoids | |||
| 1I | Vitamin D receptor | VDR | VDR | Vitamin D | ||
| Pregnane X receptor | NR1I2 | PXR | Endobiotics | |||
| Constitutive androstane receptor | NR1I3 | CAR | /Xenobiotics | |||
| 2A | Hepatocyte nuclear factor-4-α | HNF4A | HNF4α | Fatty Acids | ||
| Hepatocyte nuclear factor-4-γ | HNF4G | HNF4γ | Fatty Acids | |||
| 2B | Retinoid X receptor-α | RXRA | RXRα | 9cisRA | HX630 ↓ [55] | |
| Retinoid X receptor-β | RXRB | RXRβ | 9cisRA | |||
| Retinoid X receptor-γ | RXRG | RXRγ | 9cisRA | |||
| 2C | Testicular orphan nuclear receptor 2 | NR2C1 | TR2 | |||
| Testicular orphan nuclear receptor 4 | NR2C2 | TR4 | MEK-162 ↓ [148] | |||
| 2E | Tailless homolog orphan receptor | NR2E1 | TLX | |||
| Photoreceptor-cell-specific nuclear receptor | NR2E3 | PNR | ||||
| 2F | Chicken ovalbumin upstream promoter-transcription factor-α | NR2F1 | COUP-TFα | |||
| Chicken ovalbumin upstream promoter-transcription factor-β | NR2F2 | COUP-TFβ | ||||
| Chicken ovalbumin upstream promoter-transcription factor-γ | NR2F6 | COUP-TFγ | ||||
| 3A | Estrogen receptor-α | ESR1 | ERα | Estrogen | hypothalamic POMC enhancer ↑ [152] | |
| Estrogen receptor-β | ESR2 | ERβ | Estrogen | |||
| 3B | Estrogen-related receptor-α | ESRRA | ERRα | |||
| Estrogen-related receptor-β | ESRRB | ERRβ | ||||
| Estrogen-related receptor-γ | ESRRG | ERRγ | ||||
| 3C | Glucocorticoid receptor | NR3C1 | GR | GC | GC ↓ [167,168,169,170] | (-63 CGTCCA -58) [167,168,169,170] |
| Mineralocorticoid receptor | NR3C2 | MR | GC/MC | |||
| Progesterone receptor | PGR | PR | Progesterone | |||
| Androgen receptor | AR | AR | Androgen | |||
| 4A | Nerve growth factor 1B | NR4A1 | Nur77 | UFAs | CRH, IL-1 ↑ [197,198,199,200,201] | (-71 GAAGGTCA -63) [197] |
| Nurr-related factor 1 | NR4A2 | Nurr1 | UFAs | (-405 TGATATTT ACCTCC AAATGCCA -384) [198] | ||
| Neuron-derived orphan receptor-1 | NR4A3 | NOR-1 | 6-MP | |||
| 5A | Steroidogenic factor-1 | NR5A1 | SF1 | Phospholipids | ||
| Liver receptor homolog-1 | NR5A2 | LRH-1 | Phospholipids | |||
| 6A | Germ cell nuclear factor | NR6A1 | GCNF |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, D.; Heaney, A.P. Nuclear Receptors as Regulators of Pituitary Corticotroph Pro-Opiomelanocortin Transcription. Cells 2020, 9, 900. https://doi.org/10.3390/cells9040900
Zhang D, Heaney AP. Nuclear Receptors as Regulators of Pituitary Corticotroph Pro-Opiomelanocortin Transcription. Cells. 2020; 9(4):900. https://doi.org/10.3390/cells9040900
Chicago/Turabian StyleZhang, Dongyun, and Anthony P. Heaney. 2020. "Nuclear Receptors as Regulators of Pituitary Corticotroph Pro-Opiomelanocortin Transcription" Cells 9, no. 4: 900. https://doi.org/10.3390/cells9040900




