Environmental Restrictions: A New Concept Governing HIV-1 Spread Emerging from Integrated Experimental-Computational Analysis of Tissue-Like 3D Cultures
Abstract
:1. Introduction
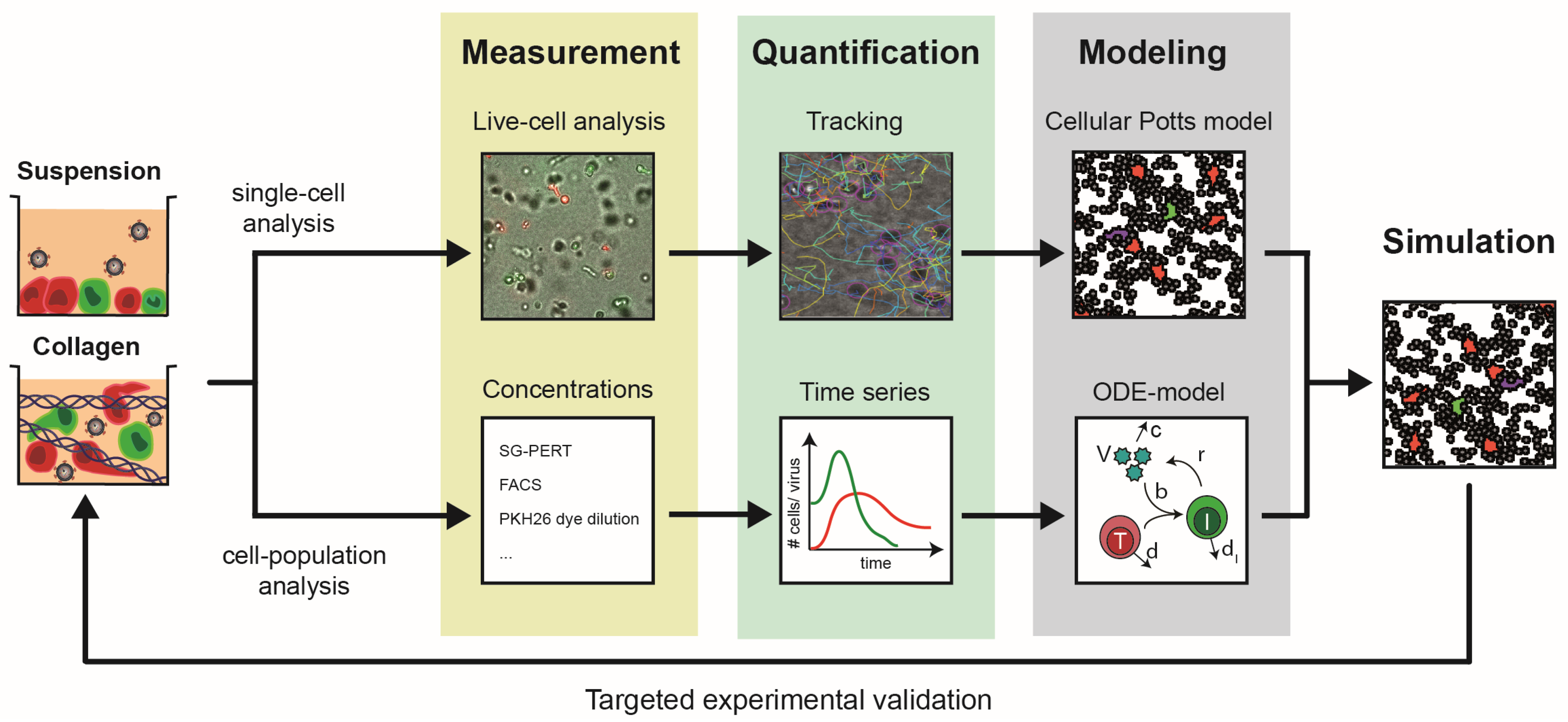
2. 3D Culture Systems
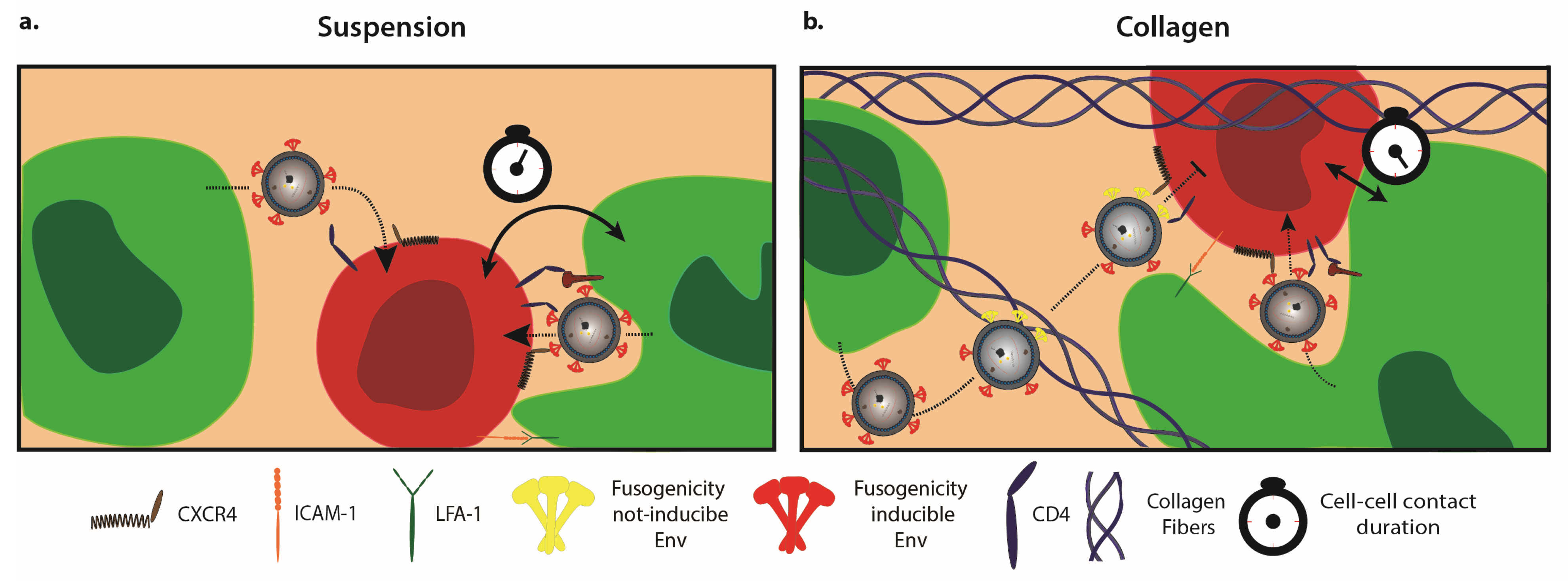
3. New Insights from Combining the Study of HIV-1 Spread in 3D Collagen Cultures with Computation
4. How Well Do Experimental and Computational INSPECT-3D Data Match In Vivo?
5. Requirements and Limitations of Current Computational Approaches to Simulating HIV-1 Spread in Tissue
6. Environmental Restriction to Cell-Free HIV-1 Infection
7. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Zhong, P.; Agosto, L.M.; Munro, J.B.; Mothes, W. Cell-to-cell transmission of viruses. Curr. Opin. Virol. 2013, 3, 44–50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sattentau, Q. Avoiding the void: Cell-to-cell spread of human viruses. Nat. Rev. Microbiol. 2008, 6, 815–826. [Google Scholar] [CrossRef] [PubMed]
- Roberts, K.L.; Smith, G.L. Vaccinia virus morphogenesis and dissemination. Trends Microbiol. 2008, 16, 472–479. [Google Scholar] [CrossRef] [PubMed]
- Graw, F.; Martin, D.N.; Perelson, A.S.; Uprichard, S.L.; Dahari, H. Quantification of Hepatitis C Virus Cell-to-Cell Spread Using a Stochastic Modeling Approach. J. Virol. 2015, 89, 6551–6561. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Polcicova, K.; Goldsmith, K.; Rainish, B.L.; Wisner, T.W.; Johnson, D.C. The extracellular domain of herpes simplex virus gE is indispensable for efficient cell-to-cell spread: Evidence for gE/gI receptors. J. Virol. 2005, 79, 11990–12001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Imai, S.; Nishikawa, J.; Takada, K. Cell-to-cell contact as an efficient mode of Epstein-Barr virus infection of diverse human epithelial cells. J. Virol. 1998, 72, 4371–4378. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, C.F.; Tu, C.H.; Lo, Y.P.; Cheng, C.C.; Chen, W.J. Involvement of Tetraspanin C189 in Cell-to-Cell Spreading of the Dengue Virus in C6/36 Cells. PLoS Negl. Trop. Dis. 2015, 9, e0003885. [Google Scholar] [CrossRef] [Green Version]
- Jolly, C.; Kashefi, K.; Hollinshead, M.; Sattentau, Q.J. HIV-1 cell to cell transfer across an Env-induced, actin-dependent synapse. J. Exp. Med. 2004, 199, 283–293. [Google Scholar] [CrossRef]
- Bracq, L.; Xie, M.; Benichou, S.; Bouchet, J. Mechanisms for Cell-to-Cell Transmission of HIV-1. Front. Immunol. 2018, 9, 260. [Google Scholar] [CrossRef]
- Igakura, T.; Stinchcombe, J.C.; Goon, P.K.; Taylor, G.P.; Weber, J.N.; Griffiths, G.M.; Tanaka, Y.; Osame, M.; Bangham, C.R. Spread of HTLV-I between lymphocytes by virus-induced polarization of the cytoskeleton. Science 2003, 299, 1713–1716. [Google Scholar] [CrossRef] [Green Version]
- Pais-Correia, A.M.; Sachse, M.; Guadagnini, S.; Robbiati, V.; Lasserre, R.; Gessain, A.; Gout, O.; Alcover, A.; Thoulouze, M.I. Biofilm-like extracellular viral assemblies mediate HTLV-1 cell-to-cell transmission at virological synapses. Nat. Med. 2010, 16, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Gross, C.; Thoma-Kress, A.K. Molecular Mechanisms of HTLV-1 Cell-to-Cell Transmission. Viruses 2016, 8, 74. [Google Scholar] [CrossRef] [PubMed]
- Dimitrov, D.S.; Willey, R.L.; Sato, H.; Chang, L.J.; Blumenthal, R.; Martin, M.A. Quantitation of human immunodeficiency virus type 1 infection kinetics. J. Virol. 1993, 67, 2182–2190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sourisseau, M.; Sol-Foulon, N.; Porrot, F.; Blanchet, F.; Schwartz, O. Inefficient human immunodeficiency virus replication in mobile lymphocytes. J. Virol. 2007, 81, 1000–1012. [Google Scholar] [CrossRef] [Green Version]
- Orlandi, C.; Flinko, R.; Lewis, G.K. A new cell line for high throughput HIV-specific antibody-dependent cellular cytotoxicity (ADCC) and cell-to-cell virus transmission studies. J. Immunol. Methods 2016, 433, 51–58. [Google Scholar] [CrossRef] [Green Version]
- Xie, M.; Leroy, H.; Mascarau, R.; Woottum, M.; Dupont, M.; Ciccone, C.; Schmitt, A.; Raynaud-Messina, B.; Verollet, C.; Bouchet, J.; et al. Cell-to-Cell Spreading of HIV-1 in Myeloid Target Cells Escapes SAMHD1 Restriction. Mbio 2019, 10. [Google Scholar] [CrossRef] [Green Version]
- Hubner, W.; McNerney, G.P.; Chen, P.; Dale, B.M.; Gordon, R.E.; Chuang, F.Y.; Li, X.D.; Asmuth, D.M.; Huser, T.; Chen, B.K. Quantitative 3D video microscopy of HIV transfer across T cell virological synapses. Science 2009, 323, 1743–1747. [Google Scholar] [CrossRef] [Green Version]
- Sowinski, S.; Jolly, C.; Berninghausen, O.; Purbhoo, M.A.; Chauveau, A.; Kohler, K.; Oddos, S.; Eissmann, P.; Brodsky, F.M.; Hopkins, C.; et al. Membrane nanotubes physically connect T cells over long distances presenting a novel route for HIV-1 transmission. Nat. Cell Biol. 2008, 10, 211–219. [Google Scholar] [CrossRef]
- Felts, R.L.; Narayan, K.; Estes, J.D.; Shi, D.; Trubey, C.M.; Fu, J.; Hartnell, L.M.; Ruthel, G.T.; Schneider, D.K.; Nagashima, K.; et al. 3D visualization of HIV transfer at the virological synapse between dendritic cells and T cells. Proc. Natl. Acad. Sci. USA 2010. [Google Scholar] [CrossRef] [Green Version]
- Real, F.; Sennepin, A.; Ganor, Y.; Schmitt, A.; Bomsel, M. Live Imaging of HIV-1 Transfer across T Cell Virological Synapse to Epithelial Cells that Promotes Stromal Macrophage Infection. Cell Rep. 2018, 23, 1794–1805. [Google Scholar] [CrossRef]
- McDonald, D.; Wu, L.; Bohks, S.M.; KewalRamani, V.N.; Unutmaz, D.; Hope, T.J. Recruitment of HIV and its receptors to dendritic cell-T cell junctions. Science 2003, 300, 1295–1297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Earl, L.A.; Lifson, J.D.; Subramaniam, S. Catching HIV ‘in the act’ with 3D electron microscopy. Trends Microbiol. 2013, 21, 397–404. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, P.; Hubner, W.; Spinelli, M.A.; Chen, B.K. Predominant mode of human immunodeficiency virus transfer between T cells is mediated by sustained Env-dependent neutralization-resistant virological synapses. J. Virol. 2007, 81, 12582–12595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martin, N.; Welsch, S.; Jolly, C.; Briggs, J.A.; Vaux, D.; Sattentau, Q.J. Virological synapse-mediated spread of human immunodeficiency virus type 1 between T cells is sensitive to entry inhibition. J. Virol. 2010, 84, 3516–3527. [Google Scholar] [CrossRef] [Green Version]
- Kolodkin-Gal, D.; Hulot, S.L.; Korioth-Schmitz, B.; Gombos, R.B.; Zheng, Y.; Owuor, J.; Lifton, M.A.; Ayeni, C.; Najarian, R.M.; Yeh, W.W.; et al. Efficiency of cell-free and cell-associated virus in mucosal transmission of human immunodeficiency virus type 1 and simian immunodeficiency virus. J. Virol. 2013, 87, 13589–13597. [Google Scholar] [CrossRef] [Green Version]
- Sewald, X.; Gonzalez, D.G.; Haberman, A.M.; Mothes, W. In vivo imaging of virological synapses. Nat. Commun. 2012, 3, 1320. [Google Scholar] [CrossRef]
- Murooka, T.T.; Deruaz, M.; Marangoni, F.; Vrbanac, V.D.; Seung, E.; von Andrian, U.H.; Tager, A.M.; Luster, A.D.; Mempel, T.R. HIV-infected T cells are migratory vehicles for viral dissemination. Nature 2012, 490, 283–287. [Google Scholar] [CrossRef] [Green Version]
- Ladinsky, M.S.; Khamaikawin, W.; Jung, Y.; Lin, S.; Lam, J.; An, D.S.; Bjorkman, P.J.; Kieffer, C. Mechanisms of virus dissemination in bone marrow of HIV-1-infected humanized BLT mice. ELife 2019, 8. [Google Scholar] [CrossRef]
- Sewald, X.; Ladinsky, M.S.; Uchil, P.D.; Beloor, J.; Pi, R.; Herrmann, C.; Motamedi, N.; Murooka, T.T.; Brehm, M.A.; Greiner, D.L.; et al. Retroviruses use CD169-mediated trans-infection of permissive lymphocytes to establish infection. Science 2015, 350, 563–567. [Google Scholar] [CrossRef] [Green Version]
- Miyauchi, K.; Kim, Y.; Latinovic, O.; Morozov, V.; Melikyan, G.B. HIV enters cells via endocytosis and dynamin-dependent fusion with endosomes. Cell 2009, 137, 433–444. [Google Scholar] [CrossRef] [Green Version]
- Dale, B.M.; McNerney, G.P.; Thompson, D.L.; Hubner, W.; de Los Reyes, K.; Chuang, F.Y.; Huser, T.; Chen, B.K. Cell-to-cell transfer of HIV-1 via virological synapses leads to endosomal virion maturation that activates viral membrane fusion. Cell Host Microbe 2011, 10, 551–562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melikyan, G.B. HIV entry: A game of hide-and-fuse? Curr. Opin. Virol. 2014, 4, 1–7. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fackler, O.T.; Murooka, T.T.; Imle, A.; Mempel, T.R. Adding new dimensions: Towards an integrative understanding of HIV-1 spread. Nat. Rev. Microbiol. 2014, 12, 563–574. [Google Scholar] [CrossRef] [PubMed]
- Marsden, M.D.; Zack, J.A. Humanized Mouse Models for Human Immunodeficiency Virus Infection. Annu. Rev. Virol. 2017, 4, 393–412. [Google Scholar] [CrossRef] [PubMed]
- Rossi, G.; Manfrin, A.; Lutolf, M.P. Progress and potential in organoid research. Nat. Rev. Genet. 2018, 19, 671–687. [Google Scholar] [CrossRef] [PubMed]
- Bar-Ephraim, Y.E.; Kretzschmar, K.; Clevers, H. Organoids in immunological research. Nat. Rev. Immunol. 2019. [Google Scholar] [CrossRef]
- Merbah, M.; Introini, A.; Fitzgerald, W.; Grivel, J.C.; Lisco, A.; Vanpouille, C.; Margolis, L. Cervico-vaginal tissue ex vivo as a model to study early events in HIV-1 infection. Am. J. Reprod. Immunol. 2011, 65, 268–278. [Google Scholar] [CrossRef] [Green Version]
- Sixt, M.; Lammermann, T. In vitro analysis of chemotactic leukocyte migration in 3D environments. Methods Mol. Biol. 2011, 769, 149–165. [Google Scholar] [CrossRef]
- Wolf, K.; Alexander, S.; Schacht, V.; Coussens, L.M.; von Andrian, U.H.; van Rheenen, J.; Deryugina, E.; Friedl, P. Collagen-based cell migration models in vitro and in vivo. Semin. Cell Dev. Biol. 2009, 20, 931–941. [Google Scholar] [CrossRef] [Green Version]
- Lammermann, T.; Bader, B.L.; Monkley, S.J.; Worbs, T.; Wedlich-Soldner, R.; Hirsch, K.; Keller, M.; Forster, R.; Critchley, D.R.; Fassler, R.; et al. Rapid leukocyte migration by integrin-independent flowing and squeezing. Nature 2008, 453, 51–55. [Google Scholar] [CrossRef]
- Miron-Mendoza, M.; Seemann, J.; Grinnell, F. The differential regulation of cell motile activity through matrix stiffness and porosity in three dimensional collagen matrices. Biomaterials 2010, 31, 6425–6435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olivares, V.; Condor, M.; Del Amo, C.; Asin, J.; Borau, C.; Garcia-Aznar, J.M. Image-based Characterization of 3D Collagen Networks and the Effect of Embedded Cells. Microsc. Microanal. 2019, 25, 971–981. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jansen, K.A.; Licup, A.J.; Sharma, A.; Rens, R.; MacKintosh, F.C.; Koenderink, G.H. The Role of Network Architecture in Collagen Mechanics. Biophys. J. 2018, 114, 2665–2678. [Google Scholar] [CrossRef] [PubMed]
- Friedl, P.; Brocker, E.B. The biology of cell locomotion within three-dimensional extracellular matrix. Cell. Mol. Life Sci. 2000, 57, 41–64. [Google Scholar] [CrossRef]
- Thippabhotla, S.; Zhong, C.; He, M. 3D cell culture stimulates the secretion of in vivo like extracellular vesicles. Sci. Rep. 2019, 9, 13012. [Google Scholar] [CrossRef] [Green Version]
- Abu-Shah, E.; Demetriou, P.; Balint, S.; Mayya, V.; Kutuzov, M.A.; Dushek, O.; Dustin, M.L. A tissue-like platform for studying engineered quiescent human T-cells’ interactions with dendritic cells. ELife 2019, 8. [Google Scholar] [CrossRef]
- Imle, A.; Kumberger, P.; Schnellbacher, N.D.; Fehr, J.; Carrillo-Bustamante, P.; Ales, J.; Schmidt, P.; Ritter, C.; Godinez, W.J.; Muller, B.; et al. Experimental and computational analyses reveal that environmental restrictions shape HIV-1 spread in 3D cultures. Nat. Commun. 2019, 10, 2144. [Google Scholar] [CrossRef] [Green Version]
- Wolf, K.; Te Lindert, M.; Krause, M.; Alexander, S.; Te Riet, J.; Willis, A.L.; Hoffman, R.M.; Figdor, C.G.; Weiss, S.J.; Friedl, P. Physical limits of cell migration: Control by ECM space and nuclear deformation and tuning by proteolysis and traction force. J. Cell Biol. 2013, 201, 1069–1084. [Google Scholar] [CrossRef] [Green Version]
- Graw, F.; Perelson, A.S. Modeling Viral Spread. Annu. Rev. Virol. 2016, 3, 555–572. [Google Scholar] [CrossRef] [Green Version]
- Perelson, A.S. Modelling viral and immune system dynamics. Nat. Rev. Immunol. 2002, 2, 28–36. [Google Scholar] [CrossRef]
- Perelson, A.S.; Neumann, A.U.; Markowitz, M.; Leonard, J.M.; Ho, D.D. HIV-1 dynamics in vivo: Virion clearance rate, infected cell life-span, and viral generation time. Science 1996, 271, 1582–1586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramratnam, B.; Bonhoeffer, S.; Binley, J.; Hurley, A.; Zhang, L.; Mittler, J.E.; Markowitz, M.; Moore, J.P.; Perelson, A.S.; Ho, D.D. Rapid production and clearance of HIV-1 and hepatitis C virus assessed by large volume plasma apheresis. Lancet 1999, 354, 1782–1785. [Google Scholar] [CrossRef]
- Perelson, A.S.; Essunger, P.; Cao, Y.; Vesanen, M.; Hurley, A.; Saksela, K.; Markowitz, M.; Ho, D.D. Decay characteristics of HIV-1-infected compartments during combination therapy. Nature 1997, 387, 188–191. [Google Scholar] [CrossRef] [PubMed]
- Markowitz, M.; Louie, M.; Hurley, A.; Sun, E.; Di Mascio, M.; Perelson, A.S.; Ho, D.D. A novel antiviral intervention results in more accurate assessment of human immunodeficiency virus type 1 replication dynamics and T-cell decay in vivo. J. Virol. 2003, 77, 5037–5038. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iwami, S.; Takeuchi, J.S.; Nakaoka, S.; Mammano, F.; Clavel, F.; Inaba, H.; Kobayashi, T.; Misawa, N.; Aihara, K.; Koyanagi, Y.; et al. Cell-to-cell infection by HIV contributes over half of virus infection. ELife 2015, 4. [Google Scholar] [CrossRef]
- De Boer, R.J.; Ribeiro, R.M.; Perelson, A.S. Current estimates for HIV-1 production imply rapid viral clearance in lymphoid tissues. PLoS Comput. Biol. 2010, 6, e1000906. [Google Scholar] [CrossRef] [Green Version]
- Timpe, J.M.; Stamataki, Z.; Jennings, A.; Hu, K.; Farquhar, M.J.; Harris, H.J.; Schwarz, A.; Desombere, I.; Roels, G.L.; Balfe, P.; et al. Hepatitis C virus cell-cell transmission in hepatoma cells in the presence of neutralizing antibodies. Hepatology 2008, 47, 17–24. [Google Scholar] [CrossRef]
- Barretto, N.; Sainz, B., Jr.; Hussain, S.; Uprichard, S.L. Determining the involvement and therapeutic implications of host cellular factors in hepatitis C virus cell-to-cell spread. J. Virol. 2014, 88, 5050–5061. [Google Scholar] [CrossRef] [Green Version]
- Barretto, N.; Uprichard, S.L. Hepatitis C virus Cell-to-cell Spread Assay. Bio Protoc. 2014, 4. [Google Scholar] [CrossRef] [Green Version]
- Iwami, S.; Holder, B.P.; Beauchemin, C.A.; Morita, S.; Tada, T.; Sato, K.; Igarashi, T.; Miura, T. Quantification system for the viral dynamics of a highly pathogenic simian/human immunodeficiency virus based on an in vitro experiment and a mathematical model. Retrovirology 2012, 9, 18. [Google Scholar] [CrossRef]
- Chen, H.Y.; Di Mascio, M.; Perelson, A.S.; Ho, D.D.; Zhang, L. Determination of virus burst size in vivo using a single-cycle SIV in rhesus macaques. Proc. Natl. Acad. Sci. USA 2007, 104, 19079–19084. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reilly, C.; Wietgrefe, S.; Sedgewick, G.; Haase, A. Determination of simian immunodeficiency virus production by infected activated and resting cells. AIDS (London, England) 2007, 21, 163–168. [Google Scholar] [CrossRef] [PubMed]
- Althaus, C.L.; De Vos, A.S.; De Boer, R.J. Reassessing the human immunodeficiency virus type 1 life cycle through age-structured modeling: Life span of infected cells, viral generation time, and basic reproductive number, R0. J. Virol. 2009, 83, 7659–7667. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumberger, P.; Durso-Cain, K.; Uprichard, S.L.; Dahari, H.; Graw, F. Accounting for Space-Quantification of Cell-To-Cell Transmission Kinetics Using Virus Dynamics Models. Viruses 2018, 10, 200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pilyugin, S.S.; Antia, R. Modeling immune responses with handling time. Bull. Math. Biol. 2000, 62, 869–890. [Google Scholar] [CrossRef] [PubMed]
- Bauer, A.L.; Beauchemin, C.A.; Perelson, A.S. Agent-based modeling of host-pathogen systems: The successes and challenges. Infor. Sci. 2009, 179, 1379–1389. [Google Scholar] [CrossRef]
- Strain, M.C.; Richman, D.D.; Wong, J.K.; Levine, H. Spatiotemporal dynamics of HIV propagation. J. Theor. Biol. 2002, 218, 85–96. [Google Scholar] [CrossRef] [Green Version]
- dos Santos, R.M.Z.; Coutinho, S. Dynamics of HIV Infection: A cellular automata approach. Phys. Rev. Lett. 2001, 87, 168102. [Google Scholar] [CrossRef] [Green Version]
- Coombes, J.L.; Robey, E.A. Dynamic imaging of host-pathogen interactions in vivo. Nat. Rev. Immunol. 2010, 10, 353–364. [Google Scholar] [CrossRef]
- Germain, R.N.; Miller, M.J.; Dustin, M.L.; Nussenzweig, M.C. Dynamic imaging of the immune system: Progress, pitfalls and promise. Nat. Rev. Immunol. 2006, 6, 497–507. [Google Scholar] [CrossRef]
- Starruss, J.; de Back, W.; Brusch, L.; Deutsch, A. Morpheus: A user-friendly modeling environment for multiscale and multicellular systems biology. Bioinformatics 2014, 30, 1331–1332. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Klinger, E.; Rickert, D.; Hasenauer, J. pyABC: Distributed, likelihood-free inference. Bioinformatics 2018, 34, 3591–3593. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Novkovic, M.; Onder, L.; Cheng, H.W.; Bocharov, G.; Ludewig, B. Integrative Computational Modeling of the Lymph Node Stromal Cell Landscape. Front. Immunol. 2018, 9, 2428. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Novkovic, M.; Onder, L.; Bocharov, G.; Ludewig, B. Graph Theory-Based Analysis of the Lymph Node Fibroblastic Reticular Cell Network. Meth. Mol. Biol. 2017, 1591, 43–57. [Google Scholar] [CrossRef]
- Novkovic, M.; Onder, L.; Cupovic, J.; Abe, J.; Bomze, D.; Cremasco, V.; Scandella, E.; Stein, J.V.; Bocharov, G.; Turley, S.J.; et al. Topological Small-World Organization of the Fibroblastic Reticular Cell Network Determines Lymph Node Functionality. PLoS Biol. 2016, 14, e1002515. [Google Scholar] [CrossRef] [Green Version]
- Novkovic, M.; Onder, L.; Bocharov, G.; Ludewig, B. Topological Structure and Robustness of the Lymph Node Conduit System. Cell Rep. 2020, 30, 893–904. [Google Scholar] [CrossRef] [Green Version]
- Licup, A.J.; Munster, S.; Sharma, A.; Sheinman, M.; Jawerth, L.M.; Fabry, B.; Weitz, D.A.; MacKintosh, F.C. Stress controls the mechanics of collagen networks. Proc. Natl. Acad. Sci. USA 2015, 112, 9573–9578. [Google Scholar] [CrossRef] [Green Version]
- Lindstrom, S.B.; Vader, D.A.; Kulachenko, A.; Weitz, D.A. Biopolymer network geometries: Characterization, regeneration, and elastic properties. Phys. Rev. E. 2010, 82, 051905. [Google Scholar] [CrossRef]
- Stein, A.M.; Vader, D.A.; Jawerth, L.M.; Weitz, D.A.; Sander, L.M. An algorithm for extracting the network geometry of three-dimensional collagen gels. J. Microsc. 2008, 232, 463–475. [Google Scholar] [CrossRef]
- Harjanto, D.; Zaman, M.H. Modeling extracellular matrix reorganization in 3D environments. PLoS ONE 2013, 8, e52509. [Google Scholar] [CrossRef] [Green Version]
- Graw, F.; Balagopal, A.; Kandathil, A.J.; Ray, S.C.; Thomas, D.L.; Ribeiro, R.M.; Perelson, A.S. Inferring viral dynamics in chronically HCV infected patients from the spatial distribution of infected hepatocytes. PLoS Computat. Biol. 2014, 10, e1003934. [Google Scholar] [CrossRef] [Green Version]
- Beltman, J.B.; Henrickson, S.E.; von Andrian, U.H.; de Boer, R.J.; Maree, A.F. Towards estimating the true duration of dendritic cell interactions with T cells. J. Immunol. Methods 2009, 347, 54–69. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beltman, J.B.; Maree, A.F.; de Boer, R.J. Analysing immune cell migration. Nat. Rev. Immunol. 2009, 9, 789–798. [Google Scholar] [CrossRef] [PubMed]
- Howat, T.J.; Barreca, C.; O’Hare, P.; Gog, J.R.; Grenfell, B.T. Modelling dynamics of the type I interferon response to in vitro viral infection. J. R. Soc. Interface 2006, 3, 699–709. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nelson, C.A.; Wilen, C.B.; Dai, Y.N.; Orchard, R.C.; Kim, A.S.; Stegeman, R.A.; Hsieh, L.L.; Smith, T.J.; Virgin, H.W.; Fremont, D.H. Structural basis for murine norovirus engagement of bile acids and the CD300lf receptor. Proc. Natl. Acad. Sci. USA 2018, 115, E9201–E9210. [Google Scholar] [CrossRef] [Green Version]
- Schupp, A.K.; Trilling, M.; Rattay, S.; Le-Trilling, V.T.K.; Haselow, K.; Stindt, J.; Zimmermann, A.; Haussinger, D.; Hengel, H.; Graf, D. Bile Acids Act as Soluble Host Restriction Factors Limiting Cytomegalovirus Replication in Hepatocytes. J. Virol. 2016, 90, 6686–6698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Veloso Alves Pereira, I.; Buchmann, B.; Sandmann, L.; Sprinzl, K.; Schlaphoff, V.; Dohner, K.; Vondran, F.; Sarrazin, C.; Manns, M.P.; Pinto Marques Souza de Oliveira, C.; et al. Primary biliary acids inhibit hepatitis D virus (HDV) entry into human hepatoma cells expressing the sodium-taurocholate cotransporting polypeptide (NTCP). PLoS ONE 2015, 10, e0117152. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Munch, J.; Rucker, E.; Standker, L.; Adermann, K.; Goffinet, C.; Schindler, M.; Wildum, S.; Chinnadurai, R.; Rajan, D.; Specht, A.; et al. Semen-derived amyloid fibrils drastically enhance HIV infection. Cell 2007, 131, 1059–1071. [Google Scholar] [CrossRef] [Green Version]
- Bart, S.M.; Cohen, C.; Dye, J.M.; Shorter, J.; Bates, P. Enhancement of Ebola virus infection by seminal amyloid fibrils. Proc. Natl. Acad. Sci. USA 2018, 115, 7410–7415. [Google Scholar] [CrossRef] [Green Version]
- Sanjuan, R. Collective Infectious Units in Viruses. Trends Microbiol. 2017, 25, 402–412. [Google Scholar] [CrossRef]
- Andreu-Moreno, I.; Sanjuan, R. Collective Infection of Cells by Viral Aggregates Promotes Early Viral Proliferation and Reveals a Cellular-L0evel Allee Effect. Curr. Biol. 2018, 28, 3212–3219. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zotova, A.; Atemasova, A.; Pichugin, A.; Filatov, A.; Mazurov, D. Distinct Requirements for HIV-1 Accessory Proteins during Cell Coculture and Cell-Free Infection. Viruses 2019, 11, 390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malbec, M.; Porrot, F.; Rua, R.; Horwitz, J.; Klein, F.; Halper-Stromberg, A.; Scheid, J.F.; Eden, C.; Mouquet, H.; Nussenzweig, M.C.; et al. Broadly neutralizing antibodies that inhibit HIV-1 cell to cell transmission. J. Exp. Med. 2013, 210, 2813–2821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhong, P.; Agosto, L.M.; Ilinskaya, A.; Dorjbal, B.; Truong, R.; Derse, D.; Uchil, P.D.; Heidecker, G.; Mothes, W. Cell-to-Cell Transmission Can Overcome Multiple Donor and Target Cell Barriers Imposed on Cell-Free HIV. PLoS ONE 2013, 8, e53138. [Google Scholar] [CrossRef] [Green Version]
- Jolly, C.; Booth, N.J.; Neil, S.J. Cell-cell spread of human immunodeficiency virus type 1 overcomes tetherin/BST-2-mediated restriction in T cells. J. Virol. 2010, 84, 12185–12199. [Google Scholar] [CrossRef] [Green Version]
- Dufloo, J.; Bruel, T.; Schwartz, O. HIV-1 cell-to-cell transmission and broadly neutralizing antibodies. Retrovirology 2018, 15, 51. [Google Scholar] [CrossRef] [Green Version]
- Chen, P.; Chen, B.K.; Mosoian, A.; Hays, T.; Ross, M.J.; Klotman, P.E.; Klotman, M.E. Virological synapses allow HIV-1 uptake and gene expression in renal tubular epithelial cells. J. Am. Soc. Nephrol. 2011, 22, 496–507. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.H.; Schierer, S.; Blume, K.; Dindorf, J.; Wittki, S.; Xiang, W.; Ostalecki, C.; Koliha, N.; Wild, S.; Schuler, G.; et al. HIV-Nef and ADAM17-Containing Plasma Extracellular Vesicles Induce and Correlate with Immune Pathogenesis in Chronic HIV Infection. EBioMedicine 2016, 6, 103–113. [Google Scholar] [CrossRef] [Green Version]
- Ostalecki, C.; Wittki, S.; Lee, J.H.; Geist, M.M.; Tibroni, N.; Harrer, T.; Schuler, G.; Fackler, O.T.; Baur, A.S. HIV Nef- and Notch1-dependent Endocytosis of ADAM17 Induces Vesicular TNF Secretion in Chronic HIV Infection. EBioMedicine 2016, 13, 294–304. [Google Scholar] [CrossRef] [Green Version]
- Mazurov, D.; Ilinskaya, A.; Heidecker, G.; Lloyd, P.; Derse, D. Quantitative comparison of HTLV-1 and HIV-1 cell-to-cell infection with new replication dependent vectors. PLoS Pathog. 2010, 6, e1000788. [Google Scholar] [CrossRef] [Green Version]
- Shunaeva, A.; Potashnikova, D.; Pichugin, A.; Mishina, A.; Filatov, A.; Nikolaitchik, O.; Hu, W.S.; Mazurov, D. Improvement of HIV-1 and Human T Cell Lymphotropic Virus Type 1 Replication-Dependent Vectors via Optimization of Reporter Gene Reconstitution and Modification with Intronic Short Hairpin RNA. J. Virol. 2015, 89, 10591–10601. [Google Scholar] [CrossRef] [PubMed] [Green Version]



| Parameter | Description | Unit | Cell Lines | Primary Cells [47] | In Vivo | ||
|---|---|---|---|---|---|---|---|
| 2D Suspension | 3D Collagen | ||||||
| Loose | Dense | ||||||
| ρ | Viral production rate | ×104 day−1 | 2.61 (1.55, 3.70) (RNA copies) [60] | 1.02 (0.80, 1.38) (RT cell−1) | 0.48 (0.37, 0.66) (RT cell−1) | 0.43 (0.32, 0.61) (RT cell−1) | 5.0 * (1.3, 12.0) (virion cell−1) [61] |
| 0.07–0.34 [56,62] | |||||||
| ρI | Total viral production rate | ×1010 virions day−1 | 1.03 ± 1.17 [52,53] | ||||
| δI | Death rate of infected cells | day−1 | 0.50 ± 0.10 [55] | 0.42 (0.40, 0.44) | 0.48 (0.46, 0.50) | 0.52 (0.51, 0.56) | 0.48–1.36 ± 0.16 [53,63] |
| 0.7 ± 0.25 [51] | |||||||
| 1.0 ± 0.3 [54] | |||||||
| 0.88 * ± 0.40 [61] | |||||||
| βf | Cell-free infection rate | ×10−5 day−1 | 0.42 ± 0.14 [55] (p24) | 2.3 (0, 3.0) (RT) | 0.14 × suspension | 0.14 × suspension | |
| βc | Cell–cell infection rate | ×10−5 (cell × day)−1 | 0.11 ± 0.03 [55] | 10−6 (0, 7.0) | 4.3 (3.6, 5.4) | 1.7 (1.4, 2.6) | |
| Pf | Predicted percentage of infections by cell-free transmission | % | 57% ± 7% [55] | 99.6% (0.0%, 100%) | 22.0% (0.0%, 27.0%) | 37% (0.0%, 44.2%) | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sid Ahmed, S.; Bundgaard, N.; Graw, F.; Fackler, O.T. Environmental Restrictions: A New Concept Governing HIV-1 Spread Emerging from Integrated Experimental-Computational Analysis of Tissue-Like 3D Cultures. Cells 2020, 9, 1112. https://doi.org/10.3390/cells9051112
Sid Ahmed S, Bundgaard N, Graw F, Fackler OT. Environmental Restrictions: A New Concept Governing HIV-1 Spread Emerging from Integrated Experimental-Computational Analysis of Tissue-Like 3D Cultures. Cells. 2020; 9(5):1112. https://doi.org/10.3390/cells9051112
Chicago/Turabian StyleSid Ahmed, Samy, Nils Bundgaard, Frederik Graw, and Oliver T. Fackler. 2020. "Environmental Restrictions: A New Concept Governing HIV-1 Spread Emerging from Integrated Experimental-Computational Analysis of Tissue-Like 3D Cultures" Cells 9, no. 5: 1112. https://doi.org/10.3390/cells9051112




