Evidence Implicating CCNB1IP1, a RING Domain-Containing Protein Required for Meiotic Crossing Over in Mice, as an E3 SUMO Ligase
Abstract
:1. Introduction
2. Results and Discussion
2.1. Expression of CCNB1IP1 and CCNB1IP1mei4 During Spermatogenesis

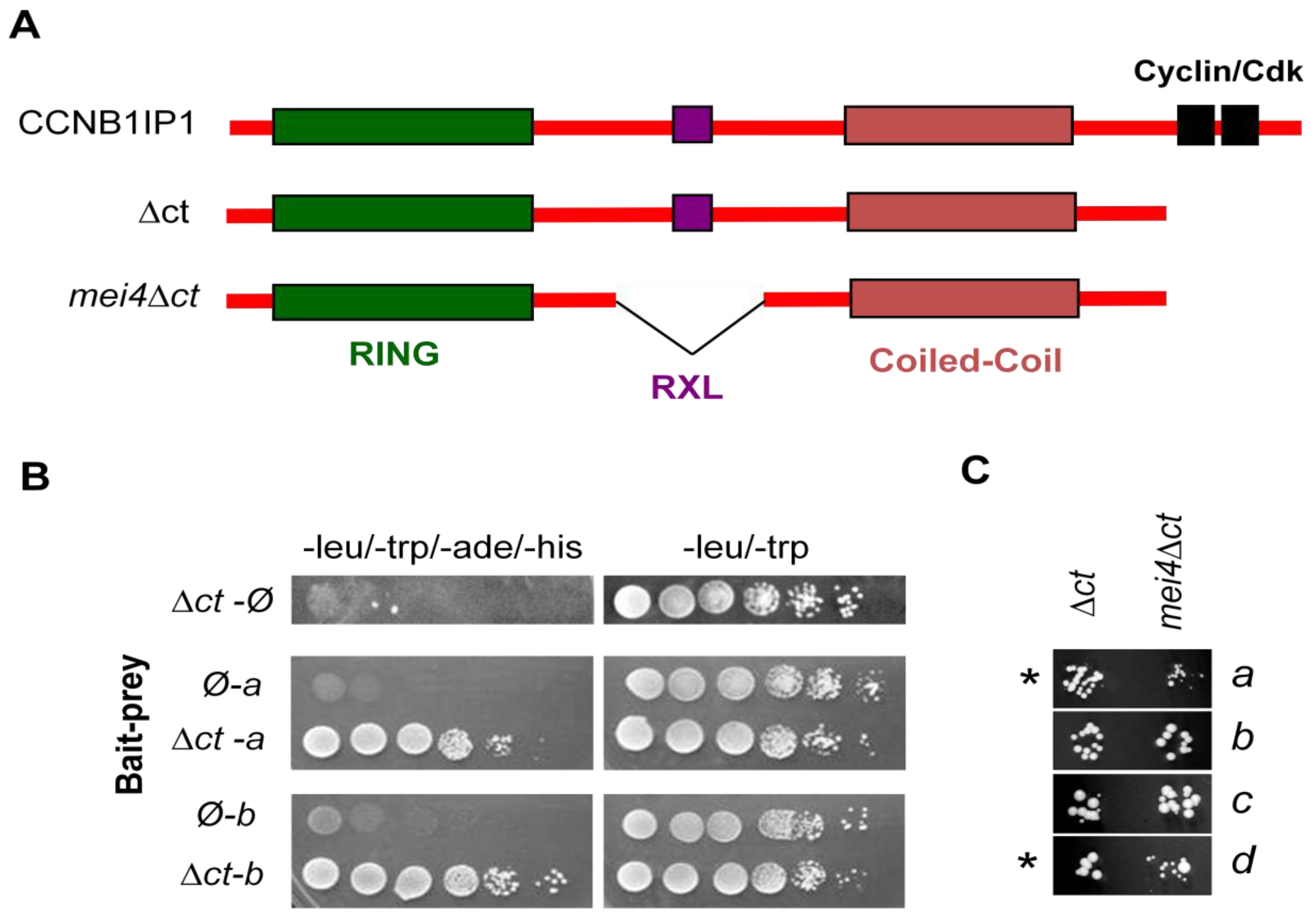
2.2. Identification of CCNB1IP1 Interacting Proteins
| Kinetically Normal with mei4Δct | Kinetically Defective with mei4Δct |
|---|---|
| SUMO2 | 4930455F23RIK (3) |
| AKAP9 | YPEL2 |
| SPINK10 | 1700006A11RIK |
| 1700019N19RIK (2) | EP400 |
| POLR2B | POMP (4) |
| ENAH | MSL1 |
| H3F3B (3) | DDC8 (2) |
| 5730469M10RIK | HOOK1 |
| PHF12 | GGN (4) |
| MRRF | FHL5 |
| OCIAD1 (2) | 4930503B20RIK (2) |
| B9D1 | 1700021F07RIK (3) |
| MORN2 | SPATA3 (3) |
| ATOH8 | |
| SRGN | |
| PENK1 | |
| BRP44 (3) | |
| INSL3 (2) | |
| EMX1 | |
| GSG1 (3) | |
| OAZ3 (3) | |
| MIIP |
2.3. Motif Analysis of CCNB1IP1 Implicates It Is a SUMO E3 Ligase

3. Experimental Section
3.1. Recombinant Expression of CCNB1IP1 and Anti-CCNB1IP1 Production
3.2. SDS-PAGE and Western Blotting
3.3. Yeast Two-Hybrid Screen for CCNB1IP1 Interactors
4. Conclusions
Acknowledgements
References
- Munroe, R.J.; Bergstrom, R.A.; Zheng, Q.Y.; Libby, B.; Smith, R.; John, S.W.M.; Schimenti, K.J.; Browning, V.L.; Schimenti, J.C. Mouse mutants from chemically mutagenized embryonic stem cells. Nat. Genet. 2000, 24, 318–321. [Google Scholar] [CrossRef]
- Ward, J.O.; Reinholdt, L.G.; Hartford, S.A.; Wilson, L.A.; Munroe, R.J.; Schimenti, K.J.; Libby, B.J.; O’Brien, M.; Pendola, J.K.; Eppig, J.; Schimenti, J.C. Toward the genetics of mammalian reproduction: induction and mapping of gametogenesis mutants in mice. Biol. Reprod. 2003, 69, 1615–1625. [Google Scholar] [CrossRef]
- Ward, J.O.; Reinholdt, L.G.; Motley, W.W.; Niswander, L.M.; Deacon, D.C.; Griffin, L.B.; Langlais, K.K.; Backus, V.L.; Schimenti, K.J.; O’Brien, M.J.; Eppig, J.J.; Schimenti, J.C. Mutation in mouse Hei10, an e3 ubiquitin ligase, disrupts meiotic crossing over. PLoS Genet. 2007, 3, e139. [Google Scholar] [CrossRef]
- Marcon, E.; Moens, P. MLH1p and MLH3p localize to precociously induced chiasmata of okadaic-acid-treated mouse spermatocytes. Genetics 2003, 165, 2283–2287. [Google Scholar]
- Gronholm, M.; Muranen, T.; Toby, G.G.; Utermark, T.; Hanemann, C.O.; Golemis, E.A.; Carpen, O. A functional association between merlin and HEI10, a cell cycle regulator. Oncogene 2006, 25, 4389–4398. [Google Scholar] [CrossRef]
- Singh, M.K.; Nicolas, E.; Gherraby, W.; Dadke, D.; Lessin, S.; Golemis, E.A. HEI10 negatively regulates cell invasion by inhibiting cyclin B/Cdk1 and other promotility proteins. Oncogene 2007, 26, 4825–4832. [Google Scholar] [CrossRef]
- Geiss-Friedlander, R.; Melchior, F. Concepts in sumoylation: A decade on. Nat. Rev. 2007, 8, 947–956. [Google Scholar] [CrossRef]
- Ulrich, H.D. The SUMO system: An overview. Methods Mol. Biol. 2009, 497, 3–16. [Google Scholar] [CrossRef]
- Galanty, Y.; Belotserkovskaya, R.; Coates, J.; Polo, S.; Miller, K.M.; Jackson, S.P. Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature 2009, 462, 935–939. [Google Scholar] [CrossRef]
- Rogers, R.S.; Inselman, A.; Handel, M.A.; Matunis, M.J. SUMO modified proteins localize to the XY body of pachytene spermatocytes. Chromosoma 2004, 113, 233–243. [Google Scholar] [CrossRef]
- Shrivastava, V.; Pekar, M.; Grosser, E.; Im, J.; Vigodner, M. SUMO proteins are involved in the stress response during spermatogenesis and are localized to DNA double-strand breaks in germ cells. Reproduction 2010, 139, 999–1010. [Google Scholar] [CrossRef]
- Vigodner, M.; Morris, P.L. Testicular expression of small ubiquitin-related modifier-1 (SUMO-1) supports multiple roles in spermatogenesis: Silencing of sex chromosomes in spermatocytes, spermatid microtubule nucleation, and nuclear reshaping. Dev. Biol. 2005, 282, 480–492. [Google Scholar] [CrossRef]
- Shrivastava, V.; Pekar, M.; Grosser, E.; Im, J.; Vigodner, M. SUMO proteins are involved in the stress response during spermatogenesis and are localized to DNA double-strand breaks in germ cells. Reproduction 2010, 139, 999–1010. [Google Scholar] [CrossRef]
- La Salle, S.; Sun, F.; Zhang, X.D.; Matunis, M.J.; Handel, M.A. Developmental control of sumoylation pathway proteins in mouse male germ cells. Dev. Biol. 2008, 321, 227–237. [Google Scholar] [CrossRef]
- Hunter, N.; Kleckner, N. The single-end invasion: an asymmetric intermediate at the double-strand break to double-holliday junction transition of meiotic recombination. Cell 2001, 106, 59–70. [Google Scholar] [CrossRef]
- Borner, G.V.; Kleckner, N.; Hunter, N. Crossover/noncrossover differentiation, synaptonemal complex formation, and regulatory surveillance at the leptotene/zygotene transition of meiosis. Cell 2004, 117, 29–45. [Google Scholar] [CrossRef]
- Guillon, H.; Baudat, F.; Grey, C.; Liskay, R.M.; de Massy, B. Crossover and noncrossover pathways in mouse meiosis. Mol. Cell 2005, 20, 563–573. [Google Scholar] [CrossRef]
- Bannister, L.; Schimenti, J; Cornell University, Ithaca, NY 14850, USA. Personal communication, 2010.
- Libby, B.J.; de La Fuente, R.; O’Brien, M.J.; Wigglesworth, K.; Cobb, J.; Inselman, A.; Eaker, S.; Handel, M.A.; Eppig, J.J.; Schimenti, J.C. The Mouse Meiotic Mutation mei1 Disrupts Chromosome Synapsis with Sexually Dimorphic Consequences for Meiotic Progression. Dev. Biol. 2002, 242, 174–187. [Google Scholar] [CrossRef]
- Libby, B.J.; Reinholdt, L.G.; Schimenti, J.C. Positional cloning and characterization of Mei1, a vertebrate-specific gene required for normal meiotic chromosome synapsis in mice. Proc. Natl. Acad. Sci. U. S. A. 2003, 100, 15706–15711. [Google Scholar] [CrossRef]
- Nebel, B.R.; Amarose, A.P.; Hacket, E.M. Calendar of gametogenic development in the prepuberal male mouse. Science 1961, 134, 832–833. [Google Scholar]
- Bellve, A.; Cavicchia, J.; Millette, C.; O’Brien, D.; Bhatnagar, Y.; Dym, M. Spermatogenic cells of the prepubertal mouse. Isolation and morphological characterization. J. Cell. Biol. 1977, 74, 68–85. [Google Scholar] [CrossRef]
- Toby, G.G.; Gherraby, W.; Coleman, T.R.; Golemis, E.A. A novel RING finger protein, human enhancer of invasion 10, alters mitotic progression through regulation of cyclin B levels. Mol. Cell Biol. 2003, 23, 2109–2122. [Google Scholar] [CrossRef]
- Lu, B.; Bishop, C.E. Mouse GGN1 and GGN3, two germ cell-specific proteins from the single gene Ggn, interact with mouse POG and play a role in spermatogenesis. J. Biol. Chem. 2003, 278, 16289–16296. [Google Scholar] [CrossRef]
- Mendoza-Lujambio, I.; Burfeind, P.; Dixkens, C.; Meinhardt, A.; Hoyer-Fender, S.; Engel, W.; Neesen, J. The Hook1 gene is non-functional in the abnormal spermatozoon head shape (azh) mutant mouse. Hum. Mol. Genet. 2002, 11, 1647–1658. [Google Scholar] [CrossRef]
- Melchior, F.; Schergaut, M.; Pichler, A. SUMO: ligases, isopeptidases and nuclear pores. Trends Biochem. Sci. 2003, 28, 612–618. [Google Scholar] [CrossRef]
- Denison, C.; Rudner, A.D.; Gerber, S.A.; Bakalarski, C.E.; Moazed, D.; Gygi, S.P. A proteomic strategy for gaining insights into protein sumoylation in yeast. Mol. Cell. Proteomics 2005, 4, 246–254. [Google Scholar] [CrossRef]
- Zhou, F.; Xue, Y.; Lu, H.; Chen, G.; Yao, X. A genome-wide analysis of sumoylation-related biological processes and functions in human nucleus. FEBS Lett. 2005, 579, 3369–3375. [Google Scholar] [CrossRef]
- Weger, S.; Hammer, E.; Engstler, M. The DNA topoisomerase I binding protein topors as a novel cellular target for SUMO-1 modification: characterization of domains necessary for subcellular localization and sumolation. Exp. Cell Res. 2003, 290, 13–27. [Google Scholar] [CrossRef]
- Kahyo, T.; Nishida, T.; Yasuda, H. Involvement of PIAS1 in the sumoylation of tumor suppressor p53. Mol. Cell 2001, 8, 713–718. [Google Scholar] [CrossRef]
- Goehler, H.; Lalowski, M.; Stelzl, U.; Waelter, S.; Stroedicke, M.; Worm, U.; Droege, A.; Lindenberg, K.S.; Knoblich, M.; Haenig, C.; et al. A protein interaction network links GIT1, an enhancer of huntingtin aggregation, to Huntington’s disease. Mol. Cell 2004, 15, 853–865. [Google Scholar] [CrossRef]
- Johnson, E.S.; Gupta, A.A. An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell 2001, 106, 735–744. [Google Scholar]
- Cheng, C.H.; Lo, Y.H.; Liang, S.S.; Ti, S.C.; Lin, F.M.; Yeh, C.H.; Huang, H.Y.; Wang, T.F. SUMO modifications control assembly of synaptonemal complex and polycomplex in meiosis of Saccharomyces cerevisiae. Genes Dev. 2006, 20, 2067–2081. [Google Scholar] [CrossRef]
- Song, J.; Durrin, L.K.; Wilkinson, T.A.; Krontiris, T.G.; Chen, Y. Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc. Natl. Acad. Sci. U. S. A. 2004, 101, 14373–14378. [Google Scholar] [CrossRef]
- Merrill, J.C.; Melhuish, T.A.; Kagey, M.H.; Yang, S.H.; Sharrocks, A.D.; Wotton, D. A role for non-covalent SUMO interaction motifs in Pc2/CBX4 E3 activity. PLoS ONE 2010, 5, e8794. [Google Scholar]
- Song, J.; Zhang, Z.; Hu, W.; Chen, Y. Small ubiquitin-like modifier (SUMO) recognition of a SUMO binding motif: a reversal of the bound orientation. J. Biol. Chem. 2005, 280, 40122–40129. [Google Scholar] [CrossRef]
- Takahashi, Y.; Toh-e, A.; Kikuchi, Y. A novel factor required for the SUMO1/Smt3 conjugation of yeast septins. Gene 2001, 275, 223–231. [Google Scholar]
- Zhu, J.; Zhu, S.; Guzzo, C.M.; Ellis, N.A.; Sung, K.S.; Choi, C.Y.; Matunis, M.J. Small ubiquitin-related modifier (SUMO) binding determines substrate recognition and paralog-selective SUMO modification. J. Biol. Chem. 2008, 283, 29405–29415. [Google Scholar] [CrossRef]
- Lin, D.Y.; Huang, Y.S.; Jeng, J.C.; Kuo, H.Y.; Chang, C.C.; Chao, T.T.; Ho, C.C.; Chen, Y.C.; Lin, T.P.; Fang, H.I.; Hung, C.C.; Suen, C.S.; Hwang, M.J.; Chang, K.S.; Maul, G.G.; Shih, H.M. Role of SUMO-interacting motif in Daxx SUMO modification, subnuclear localization, and repression of sumoylated transcription factors. Mol. Cell 2006, 24, 341–354. [Google Scholar] [CrossRef]
- Macqueen, A.J.; Roeder, G.S. Fpr3 and Zip3 ensure that initiation of meiotic recombination precedes chromosome synapsis in budding yeast. Curr. Biol. 2009, 19, 1519–1526. [Google Scholar] [CrossRef]
- Agarwal, S.; Roeder, G.S. Zip3 provides a link between recombination enzymes and synaptonemal complex proteins. Cell 2000, 102, 245–255. [Google Scholar] [CrossRef]
- Bhalla, N.; Wynne, D.J.; Jantsch, V.; Dernburg, A.F. ZHP-3 acts at crossovers to couple meiotic recombination with synaptonemal complex disassembly and bivalent formation in C. elegans. PLoS Genet. 2008, 4, e1000235. [Google Scholar] [CrossRef]
- Chowdhury, R.; Bois, P.R.; Feingold, E.; Sherman, S.L.; Cheung, V.G. Genetic analysis of variation in human meiotic recombination. PLoS Genet. 2009, 5, e1000648. [Google Scholar] [CrossRef] [Green Version]
- Kong, A.; Thorleifsson, G.; Stefansson, H.; Masson, G.; Helgason, A.; Gudbjartsson, D.F.; Jonsdottir, G.M.; Gudjonsson, S.A.; Sverrisson, S.; Thorlacius, T.; Jonasdottir, A.; Hardarson, G.A.; Palsson, S.T.; Frigge, M.L.; Gulcher, J.R.; Thorsteinsdottir, U.; Stefansson, K. Sequence variants in the RNF212 gene associate with genome-wide recombination rate. Science 2008, 319, 1398–1401. [Google Scholar] [CrossRef]
- Lin, F.M.; Lai, Y.J.; Shen, H.J.; Cheng, Y.H.; Wang, T.F. Yeast axial-element protein, Red1, binds SUMO chains to promote meiotic interhomologue recombination and chromosome synapsis. EMBO J. 2010, 29, 586–596. [Google Scholar] [CrossRef]
- Morris, J.R.; Boutell, C.; Keppler, M.; Densham, R.; Weekes, D.; Alamshah, A.; Butler, L.; Galanty, Y.; Pangon, L.; Kiuchi, T.; Ng, T.; Solomon, E. The SUMO modification pathway is involved in the BRCA1 response to genotoxic stress. Nature 2009, 462, 886–890. [Google Scholar] [CrossRef]
© 2010 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Strong, E.R.; Schimenti, J.C. Evidence Implicating CCNB1IP1, a RING Domain-Containing Protein Required for Meiotic Crossing Over in Mice, as an E3 SUMO Ligase. Genes 2010, 1, 440-451. https://doi.org/10.3390/genes1030440
Strong ER, Schimenti JC. Evidence Implicating CCNB1IP1, a RING Domain-Containing Protein Required for Meiotic Crossing Over in Mice, as an E3 SUMO Ligase. Genes. 2010; 1(3):440-451. https://doi.org/10.3390/genes1030440
Chicago/Turabian StyleStrong, Edward R., and John C. Schimenti. 2010. "Evidence Implicating CCNB1IP1, a RING Domain-Containing Protein Required for Meiotic Crossing Over in Mice, as an E3 SUMO Ligase" Genes 1, no. 3: 440-451. https://doi.org/10.3390/genes1030440
APA StyleStrong, E. R., & Schimenti, J. C. (2010). Evidence Implicating CCNB1IP1, a RING Domain-Containing Protein Required for Meiotic Crossing Over in Mice, as an E3 SUMO Ligase. Genes, 1(3), 440-451. https://doi.org/10.3390/genes1030440




