Effect of Circadian Clock and Light–Dark Cycles in Onchidium reevesii: Possible Implications for Long-Term Memory
Abstract
:1. Introduction
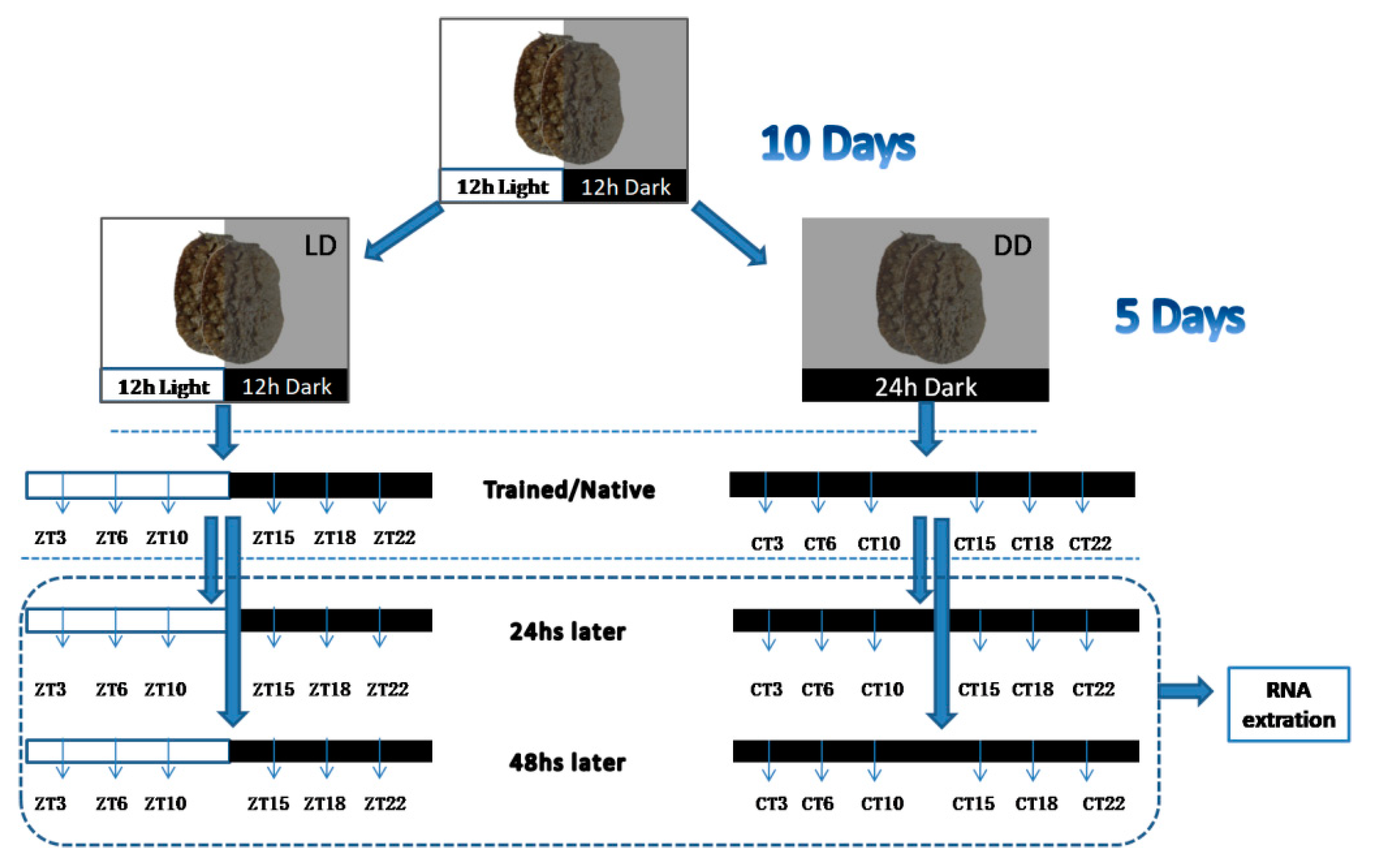
2. Materials and Methods
2.1. Experimental Animals
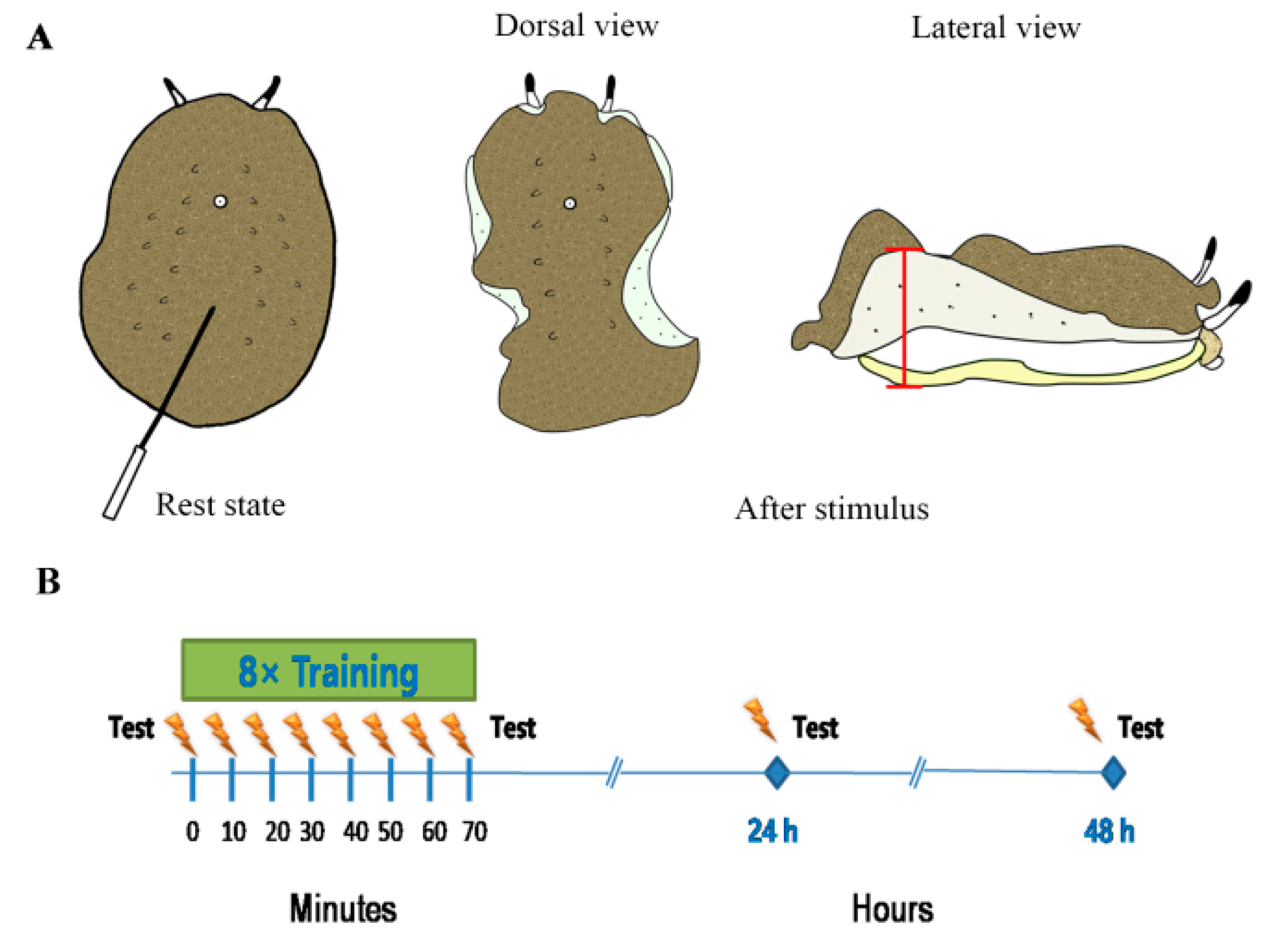
2.2. Training Experiments
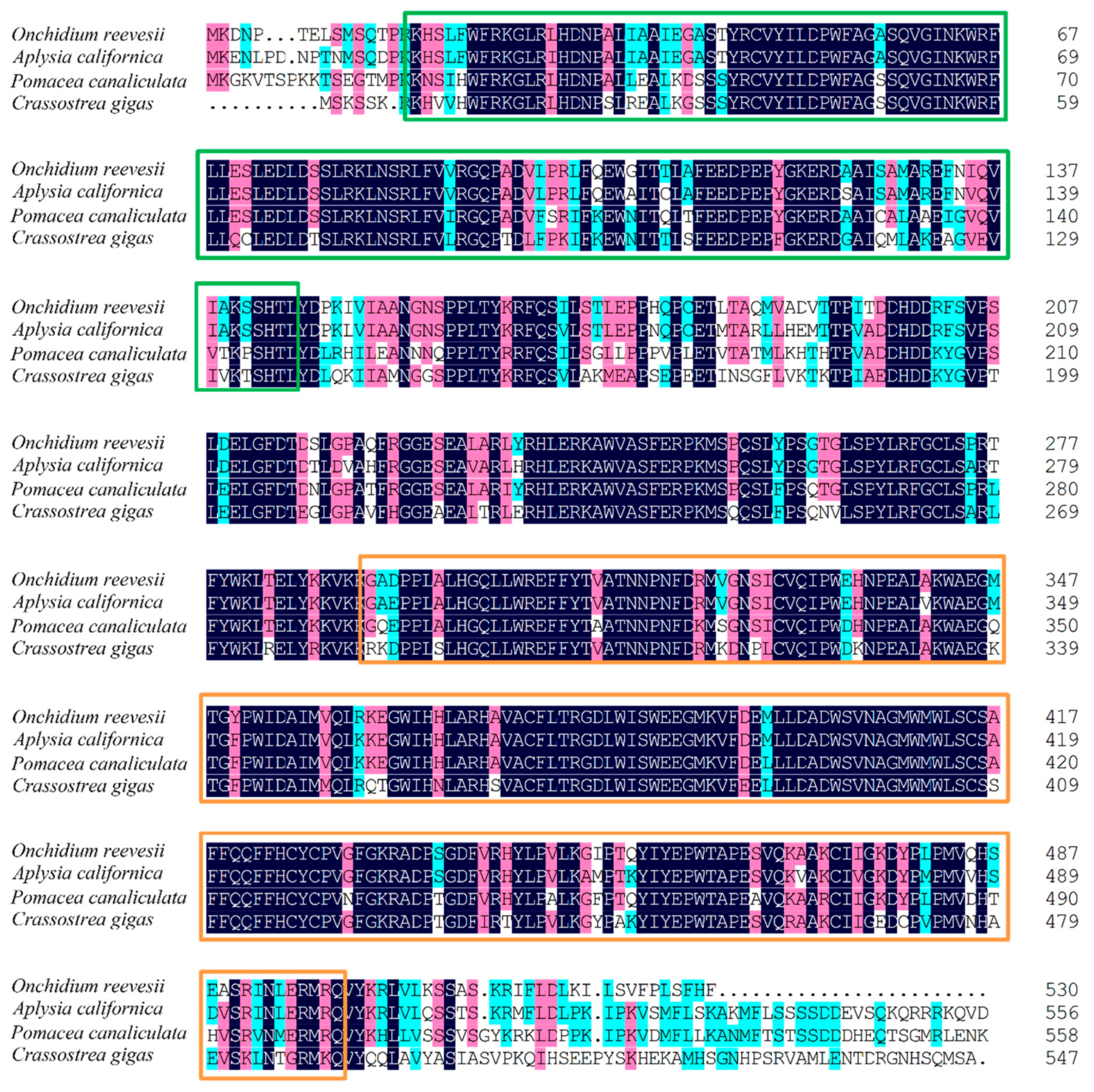
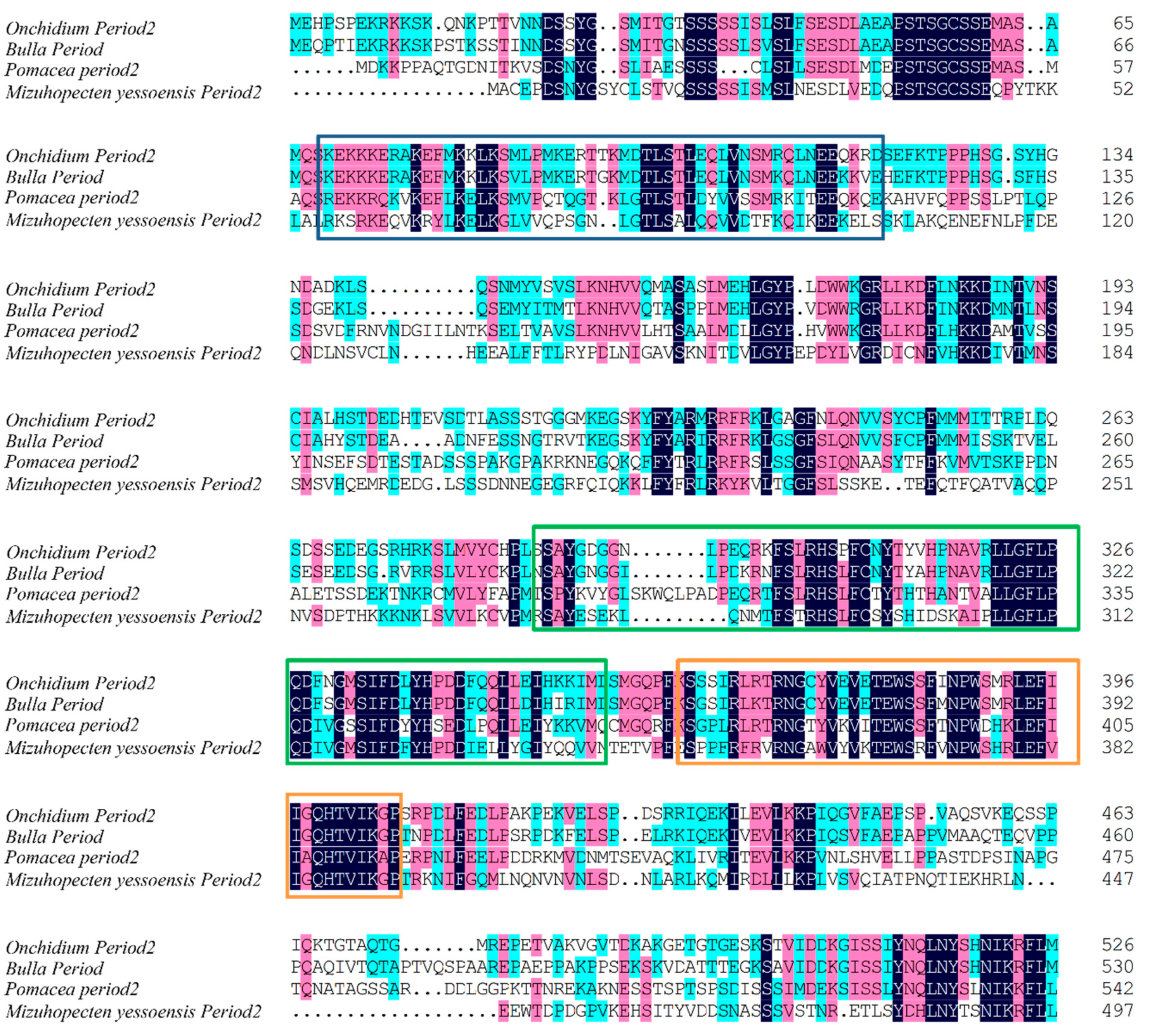
2.3. Cloning of cry1 and per2 Genes and Quantitative Analysis of OnCREB1, cry1, and per2 Genes
3. Results
3.1. Learning/Memory Acquisition and Long-Term Memory Expressed a Certain Rhythm in Different Conditions
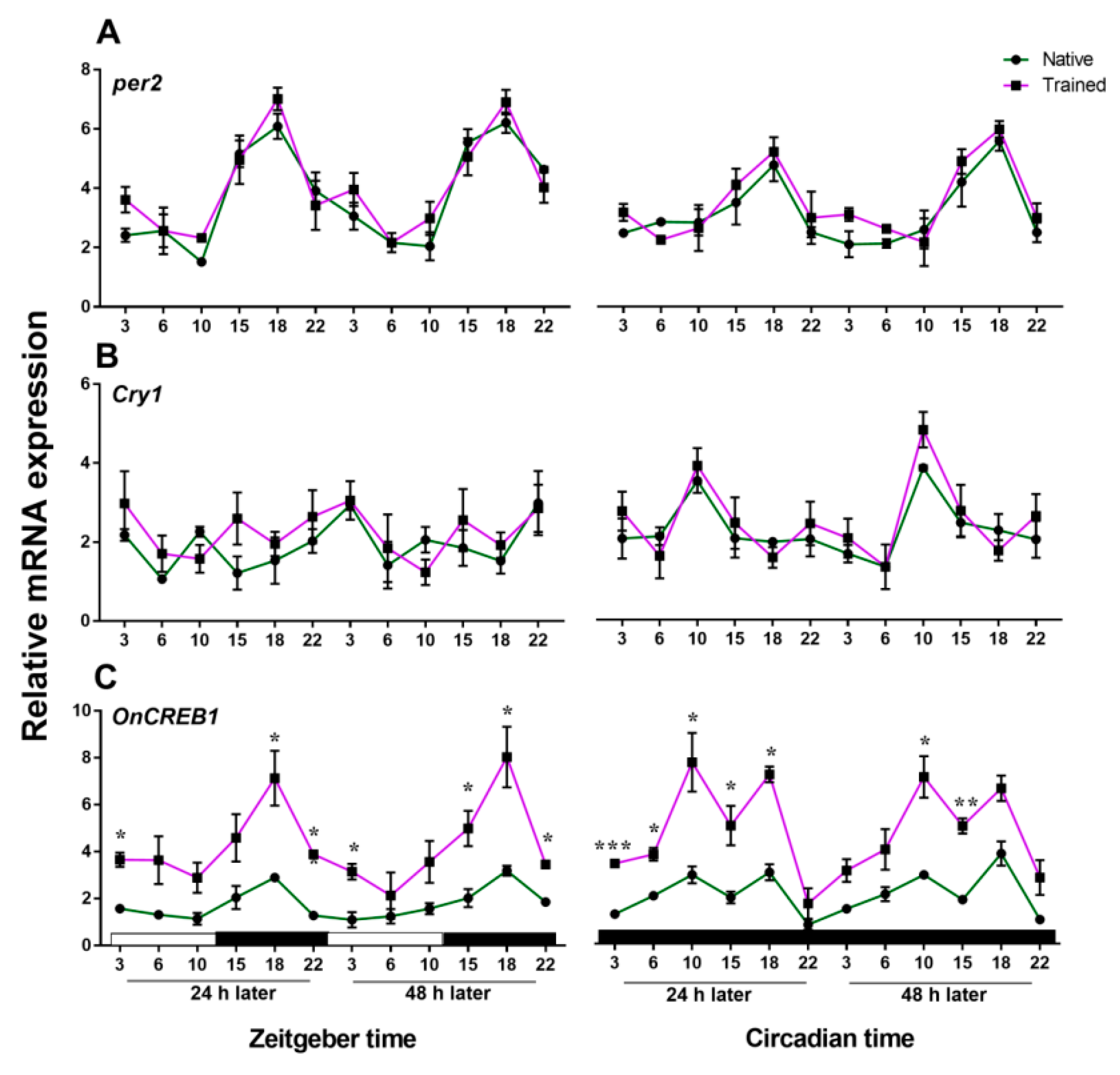
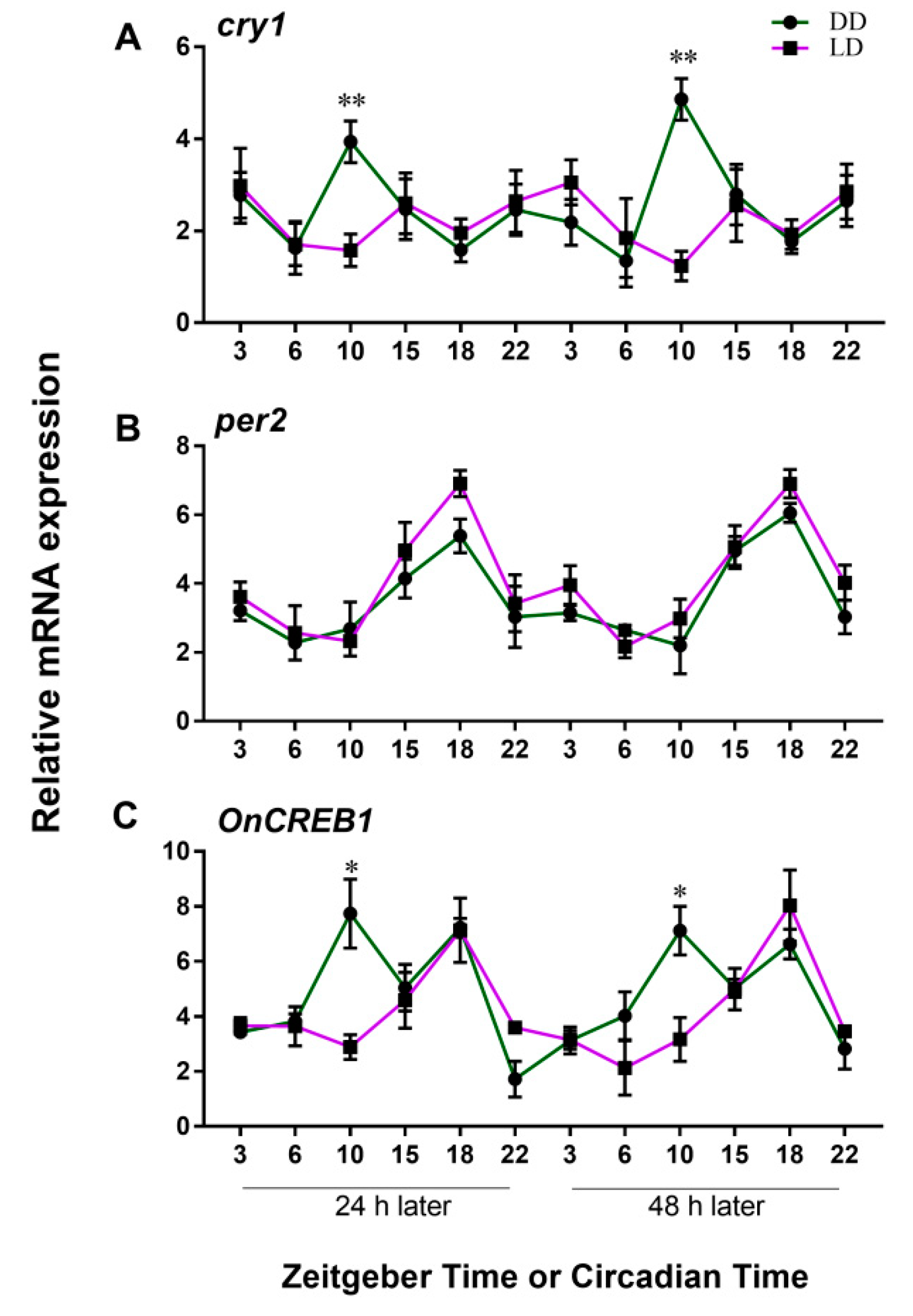
3.2. Expressions of cry1, per2, and OnCREB1 under The Control of Light–Dark (LD) and Constant Darkness (DD) Cycles
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Arey, L.B.; Crozier, W.J. The ‘homing habits’ of the pulmonate mollusk Onchidium. Proc. Natl. Acad. Sci. USA 1918, 4, 319–321. [Google Scholar] [CrossRef] [PubMed]
- Naylor, E. Tidal and diurnal rhythms of locomotory activity in Carcinus maenas (L.). J. Exp. Biol. 1958, 35, 131–138. [Google Scholar]
- Dubofsky, E.A.; Simpson, S.D.; Chabot, C.C. Patterns of activity expressed by juvenile horseshoe crabs. Biol. Bull. 2013, 225, 42–49. [Google Scholar] [CrossRef] [PubMed]
- Forward, R.B.; Diaz, H.; Cohen, J.H. The tidal rhythm in activity of the mole crab Emerita talpoida. J. Mar. Biol. Assoc. UK 2005, 85, 895–901. [Google Scholar] [CrossRef]
- Barker, G.M. Biology of Terrestrial Molluscs; CABI Publishing: New York, NY, USA, 2001. [Google Scholar]
- Naylor, E. Chronobiology of Marine Organisms; Cambridge University Press: Cambridge, UK, 2010; pp. 1–252. [Google Scholar]
- Kristin, T.R.; Florian, R.; Enrique, A. Another place, another timer: Marine species and the rhythms of life. Bioessays News Rev. Mol. Cell. Dev. Biol. 2015, 33, 165–172. [Google Scholar]
- Deborah, B.P.; Cassone, V.M.; Earnest, D.J.; Golden, S.S.; Hardin, P.E.; Thomas, T.L.; Zoran, M.J. Circadian rhythms from multiple oscillators: Lessons from diverse organisms. Nat. Rev. Genet. 2005, 6, 544–556. [Google Scholar]
- Hardin, P.E.; Hall, J.C.; Rosbash, M. Feedback of the Drosophila period gene product on circadian cycling of its messenger RNA levels. Nature 1990, 343, 536–540. [Google Scholar] [CrossRef] [PubMed]
- Mat, A.M.; Perrigault, M.; Massabuau, J.C.; Tran, D. Role and expression of cry1 in the adductor muscle of the oyster Crassostrea gigas during daily and tidal valve activity rhythms. Chronobiol. Int. 2016, 33, 949–963. [Google Scholar] [CrossRef]
- Tei, H.; Okamura, H.; Shigeyoshi, Y.; Fukuhara, C.; Ozawa, R.; Hirose, M.; Sakaki, Y. Circadian oscillation of a mammalian homologue of the Drosophila period gene. Nature 1997, 389, 512–516. [Google Scholar] [CrossRef]
- Padmanabhan, K.; Weitz, C.J. Feedback regulation of transcriptional termination by the mammalian circadian clock PERIOD complex. Science 2012, 337, 599. [Google Scholar] [CrossRef]
- Cyran, S.A.; Buchsbaum, A.M.; Reddy, K.L.; Meng-Chi, L.; Glossop, N.R.J.; Hardin, P.E.; Young, M.W.; Storti, R.V.; Justin, B. vrille, Pdp1, and dClock form a second feedback loop in the Drosophila circadian clock. Cell 2003, 112, 329–341. [Google Scholar] [CrossRef]
- Levy, O.; Appelbaum, L.; Leggat, W.; Gothlif, Y.; Hayward, D.C.; Miller, D.J.; Hoegh-Guldberg, O. Light-responsive cryptochromes from a simple multicellular animal, the coral Acropora millepora. Science 2007, 318, 467–470. [Google Scholar] [CrossRef] [PubMed]
- Todo, T.; Ryo, H.; Yamamoto, K.; Toh, H.; Inui, T.; Ayaki, H.; Nomura, T.; Ikenaga, M. Similarity among the Drosophila (6-4)photolyase, a human photolyase homolog, and the DNA photolyase-blue-light photoreceptor family. Science 1996, 272, 109–112. [Google Scholar] [CrossRef] [PubMed]
- Masato, F.; Takahiro, T.; Yuki, T.; Sung-Pyo, H.; Nozomi, S.; Akihiro, T.; Yoko, K.; Keiko, O.; Toshiyuki, O. Lunar phase-dependent expression of cryptochrome and a photoperiodic mechanism for lunar phase-recognition in a reef fish, goldlined spinefoot. PLoS ONE 2011, 6, e28643. [Google Scholar]
- Feng, D.; Lazar, M.A. Clocks, metabolism, and the epigenome. Mol. Cell 2012, 47, 158–167. [Google Scholar] [CrossRef] [PubMed]
- Dipesh, C.; Christopher, S.C. Circadian modulation of learning and memory in fear-conditioned mice. Behav. Brain Res. 2002, 133, 95–108. [Google Scholar]
- Smarr, B.L.; Jennings, K.J.; Driscoll, J.R.; Kriegsfeld, L.J. A time to remember: The role of circadian clocks in learning and memory. Behav. Neurosci. 2014, 128, 283–303. [Google Scholar] [CrossRef]
- Dash, P.K.; Hochner, B.; Kandel, E.R. Injection of the cAMP-responsive element into the nucleus of Aplysia sensory neurons blocks long-term facilitation. Nature 1990, 345, 718–721. [Google Scholar] [CrossRef]
- Bourtchuladze, R.; Frenguelli, B.; Blendy, J.; Cioffi, D.; Schutz, G.; Silva, A.J. Deficient long-term memory in mice with a targeted mutation of the cAMP-responsive element-binding protein. Cell 1994, 79, 59–68. [Google Scholar] [CrossRef]
- Silva, A.J.; Kogan, J.H.; And, P.W.F.; Kida, S. CREB and Memory. Annu. Rev. Neurosci. 1998, 21, 127–148. [Google Scholar] [CrossRef]
- Gau, D.; Lemberger, T.; Gall, C.V.; Kretz, O.; Minh, N.L.; Gass, P.; Schmid, W.; Schibler, U.; Korf, H.W.; Schütz, G. Phosphorylation of CREB Ser142 regulates light-induced phase shifts of the circadian clock. Neuron 2002, 34, 245–253. [Google Scholar] [CrossRef]
- Reppert, S.M.; Weaver, D.R. Coordination of circadian timing in mammals. Nature 2002, 418, 935–941. [Google Scholar] [CrossRef] [PubMed]
- Mohamed, H.A.; Yao, W.; Fioravante, D.; Smolen, P.D.; Byrne, J.H. cAMP-response elements in Aplysia creb1, creb2, and Ap-uch promoters: Implications for feedback loops modulating long term memory. J. Biol. Chem. 2005, 280, 27035–27043. [Google Scholar] [CrossRef] [PubMed]
- Bartsch, D.; Casadio, A.; Karl, K.A.; Serodio, P.; Kandel, E.R. CREB1 encodes a nuclear activator, a repressor, and a cytoplasmic modulator that form a regulatory unit critical for long-term facilitation. Cell 1998, 95, 211–223. [Google Scholar] [CrossRef]
- Rawashdeh, O.; Jilg, A.; Maronde, E.; Fahrenkrug, J.; Stehle, J.H. Period1 gates the circadian modulation of memory-relevant signaling in mouse hippocampus by regulating the nuclear shuttling of the CREB kinase pP90RSK. J. Neurochem. 2016, 138, 731–745. [Google Scholar] [CrossRef] [PubMed]
- Campos, L.M.; Cruz-Rizzolo, R.J.; Pinato, L. The primate seahorse rhythm. Brain Res. 2015, 1613, 81–91. [Google Scholar] [CrossRef]
- Fernandez, R.I.; Lyons, L.C.; Jonathan, L.; Omar, K.; Arnold, E. Circadian modulation of long-term sensitization in Aplysia. Proc. Natl. Acad. Sci. USA 2003, 100, 14415–14420. [Google Scholar] [CrossRef]
- Mcdonald, R.J.; Hong, N.S.; Ray, C.; Ralph, M.R. No time of day modulation or time stamp on multiple memory tasks in rats. Learn. Motiv. 2002, 33, 230–252. [Google Scholar] [CrossRef]
- Chabot, C.C.; Watson, W.H. Daily and Tidal Rhythms in Intertidal Marine Invertebrates; Springer: Tokyo, Japan, 2014. [Google Scholar]
- Page, T.L. Masking in invertebrates. Chronobiol. Int. 1989, 6, 3–11. [Google Scholar] [CrossRef]
- Shen, H.; Li, K.; Chen, H.; Chen, X.; He, Y.; Shi, Z. Experimental ecology and hibernation of Onchidium struma (Gastropoda: Pulmonata: Systellommatophora). J. Exp. Mar. Biol. Ecol. 2011, 396, 71–76. [Google Scholar]
- Nishi, T.; Gotow, T. A neural mechanism for processing colour information in molluscan extra-ocular photoreceptors. J. Exp. Biol. 1992, 168, 77–91. [Google Scholar]
- Gotow, T.; Tateda, H.; Kuwabara, M. The function of photoexcitive neurones in the central ganglia for behavioral activity of the marine mollusc, Onchidium verruculatum. J. Comp. Physiol. 1973, 83, 361–376. [Google Scholar] [CrossRef]
- Shimotsu, K.; Nishi, T.; Nakagawa, S.; Gotow, T. A new role for photoresponsive neurons called simple photoreceptors in the sea slug Onchidium verruculatum: Potentiation of synaptic transmission and motor response. Comp. Biochem. Physiol. Part A Mol. Integr. Physiol. 2010, 156, 201–210. [Google Scholar] [CrossRef] [PubMed]
- Xu, G.; Yang, T.; Wang, D.; Li, J.; Liu, X.; Wu, X.; Shen, H. A comprehensive comparison of four species of Onchidiidae provides insights on the morphological and molecular adaptations of invertebrates from shallow seas to wetlands. PLoS ONE 2018, 13, e0196252. [Google Scholar] [CrossRef] [PubMed]
- Zantke, J.; Ishikawa-Fujiwara, T.; Arboleda, E.; Lohs, C.; Schipany, K.; Hallay, N.; Straw, A.; Todo, T.; Tessmar-Raible, K. Circadian and circalunar clock interactions in a marine annelid. Cell Rep. 2013, 5, 99–113. [Google Scholar] [CrossRef]
- Goto, S.G.; Takekata, H. Circatidal rhythm and the veiled clockwork. Curr. Opin. Insect Sci. 2015, 7, 92–97. [Google Scholar] [CrossRef]
- Page, T.L. Circadian regulation of learning and memory. Curr. Opin. Insect Sci. 2015, 7, 87–91. [Google Scholar] [CrossRef]
- Garren, M.V.; Sexauer, S.B.; Page, T.L. Effect of circadian phase on memory acquisition and recall: Operant conditioning vs. classical conditioning. PLoS ONE 2013, 8, e58693. [Google Scholar] [CrossRef]
- Vorster, A.P.A.; Krishnan, H.C.; Chiara, C.; Lyons, L.C. Characterization of sleep in Aplysia californica. Sleep 2014, 37, 1453. [Google Scholar] [CrossRef]
- Marques, M.D.; Waterhouse, J.M. Masking and the evolution of circadian rhythmicity. Chronobiol. Int. 1994, 11, 146–155. [Google Scholar] [CrossRef]
- Rietveld, W.J.; Minors, D.S.; Waterhouse, J.M. Circadian rhythms and masking: An overview. Chronobiol. Int. 1993, 10, 306–312. [Google Scholar] [CrossRef] [PubMed]
- Palmer, J.D. The rhythmic lives of crabs. Bioscience 1990, 40, 352–358. [Google Scholar] [CrossRef]
- Chabot, C.C.; Skinner, S.J.; Watson, W.H. Rhythms of locomotion expressed by Limulus polyphemus, the American horseshoe crab: I. synchronization by artificial tides. Biol. Bull. 2008, 215, 34–45. [Google Scholar] [CrossRef] [PubMed]
- Takekata, H.; Goto, S.G.; Satoh, A.; Numata, H. Light masking of the circatidal activity rhythm in the mangrove cricket Apteronemobius asahinai. Biol. Rhythm Res. 2014, 45, 229–233. [Google Scholar] [CrossRef]
- Wang, M.C.; Dragich, J.M.; Kudo, T.; Odom, I.H.; Welsh, D.K.; O’Dell, T.J.; Colwell, C.S. Expression of the circadian clock gene Period2 in the hippocampus: Possible implications for synaptic plasticity and learned behaviour. ASN Neuro 2009, 1, 240–242. [Google Scholar] [CrossRef] [PubMed]
- Bjorn, R.; Jan, B. About sleep’s role in memory. Physiol. Rev. 2013, 93, 681–766. [Google Scholar]
- Takaomi, S.; Takuya, T.; Toshihiro, K.; Yoshiaki, K. A clock gene, period, plays a key role in long-term memory formation in Drosophila. Proc. Natl. Acad. Sci. USA 2004, 101, 16058–16063. [Google Scholar]
- Buhr, E.D.; Takahashi, J.S. Molecular Components of the Mammalian Circadian Clock. In Handbook of Experimental Pharmacology; Springer: Berlin/Heidelberg, Germany, 2013. [Google Scholar]
- Montenegro-Montero, A.; Canessa, P.; Larrondo, L.F. Chapter Four—Around the Fungal Clock: Recent Advances in the Molecular Study of Circadian Clocks in Neurospora and Other Fungi. Adv. Genet. 2015, 92, 107–184. [Google Scholar]
- Mahan, K.L. Circadian Oscillation of MAPK Activity and cAMP in the Hippocampus: Implications for Memory Persistence. Ph.D. Thesis, University of Washington, Seattle, WA, USA, 2008. [Google Scholar]
- Adams, J.P.; Roberson, E.D.; English, J.D.; Selcher, J.C.; Sweatt, J.D. MAPK regulation of gene expression in the central nervous system. Acta Neurobiol. Exp. 2000, 60, 377–394. [Google Scholar]
- Zhang, E.E.; Liu, Y.; Dentin, R.; Pongsawakul, P.Y.; Liu, A.C.; Hirota, T.; Nusinow, D.A.; Sun, X.; Landais, S.; Kodama, Y. Cryptochrome mediates circadian regulation of cAMP signaling and hepatic gluconeogenesis. Nat. Med. 2010, 16, 1152. [Google Scholar] [CrossRef]
- Liu, R.Y.; Shah, S.; Cleary, L.J.; Byrne, J.H. Serotonin- and training-induced dynamic regulation of CREB2 in Aplysia. Learn. Mem. 2011, 18, 245–249. [Google Scholar] [CrossRef] [PubMed]
- Sadamoto, H.; Kitahashi, T.; Fujito, Y.; Ito, E. Learning-dependent gene expression of CREB1 isoforms in the molluscan brain. Front. Behav. Neurosci. 2010, 4, 25. [Google Scholar] [CrossRef] [PubMed]
- Mizuno, M.; Yamada, K.; Maekawa, N.; Saito, K.; Seishima, M.; Nabeshima, T. CREB phosphorylation as a molecular marker of memory processing in the hippocampus for spatial learning. Behav. Brain Res. 2002, 133, 135–141. [Google Scholar] [CrossRef]
- Lyons, L.C.; Oliver, R.; Ayelet, K.; Susswein, A.J.; Arnold, E. Circadian modulation of complex learning in diurnal and nocturnal Aplysia. Proc. Natl. Acad. Sci. USA 2005, 102, 12589–12594. [Google Scholar] [CrossRef] [PubMed]
- Chiang, C.K.; Xu, B.; Mehta, N.; Mayne, J.; Sun, W.Y.L.; Cheng, K.; Ning, Z.; Dong, J.; Zou, H.; Cheng, H.Y.M. Phosphoproteome profiling reveals circadian clock regulation of posttranslational modifications in the murine hippocampus. Front. Neurol. 2017, 8, 110. [Google Scholar] [CrossRef] [PubMed]
- Cain, S.W.; Ko, C.H.; Chalmers, J.A.; Ralph, M.R. Time of day modulation of conditioned place preference in rats depends on the strain of rat used. Neurobiol. Learn. Mem. 2004, 81, 217–220. [Google Scholar] [CrossRef] [PubMed]

| Usage | Primer Name | Primer Sequence (5′-3′) | Description |
|---|---|---|---|
| RT-PCR | Test-1F | GTTTAAAACGCCACCCCCAC | Used to amplify one part of the per2 fragment |
| Test-1R | ATGCTGAACTGAGTGGGTGG | ||
| Test-2F | TTCGAAAACTGGGTGCAGGT | Used to amplify one part of the per2 fragment | |
| Test-2R | GTCTCAGGCTCTCTCATGCC | ||
| Test-3F | ATGAGAGAGCCTGAGACCGT | Used to amplify one part of the per2 fragment | |
| Test-3R | TGCGCATGTTCAGAAGGAGT | ||
| Test-4F | TCGTGAATCAGCGGACAACA | Used to amplify one part of the per2 fragment | |
| Test-4R | GAGGGCGATAGAAGCCAGTC | ||
| Test-5F | ACATCCCACCAGAAACGTCC | Used to amplify one part of the per2 fragment | |
| Test-5R | ATGGGCTCCTGAAACATGGG | ||
| Test-6F | CAGGTGATGACTGGCTTCTATC | Used to amplify one part of the per2 fragment | |
| Test-6R | CTGGTGTGTCGTAGTTCTCATC | ||
| Test-7F | CCCTAAGGATGGGTCCCTCA | Used to amplify one part of the per2 fragment | |
| Test-7R | CACCGTGGCTGTTGTTGATG | ||
| Test-8F | AAACCCTCACTGCCCAGATG | Used to amplify one part of the cry1 fragment | |
| Test-8R | GTGAGAAAGCAAGCCACAGC | ||
| Test-9F | GTGTACAAGAGGCTGGTGCT | Used to amplify one part of the cry1 fragment | |
| Test-9R | AAAAGCCCTCAGGAGCTGAC | ||
| Test-10F | GTCAGCTCCTGAGGGCTTTT | Used to amplify one part of the cry1 fragment | |
| Test-10R | CCTTGTCTGCGTCACAAAGC | ||
| Test-11F | GGTGCCTCACAAGTAGGAATTA | Used to amplify one part of the cry1 fragment | |
| Test-11R | GATTGTGTTCCCACGGTATCT | ||
| Test-12F | GATGCTGACTGGAGCGTAAA | Used to amplify one part of the cry1 fragment | |
| Test-12R | CCCTCAGGAGCTGACTAATAAAC | ||
| Test-13F | AGACTGCAGACCCTGTTAATG | Used to amplify one part of the cry1 fragment | |
| Test-13R | ATGGGCTAGAGAGCTGATACT | ||
| Test-14F | ATGGAAACAGCCCACCACTC | Used to amplify one part of the cry1 fragment | |
| Test-14R | TCCACCACGAAACTGAGCTG | ||
| Test-15F | AGCTCAGTTTCGTGGTGGAG | Used to amplify one part of the cry1 fragment | |
| Test-15R | GGGGATAGTCCTTGCCGATG | ||
| RACE | 3′RACE-F1 | CCCTAAGGATGGGTCCCTCA | Gene-specific outer primer for per2 |
| 3′RACE-F2 | AATGTGTCTCTCAGCCCTGC | Gene-specific inner primer for per2 | |
| 3′RACE-F3 | GTCAGCTCCTGAGGGCTTTT | Gene-specific outer primer for cry1 | |
| 3′RACE-F4 | AGACTGCAGACCCTGTTAATG | Gene-specific inner primer for cry1 | |
| 3′RACE outer primer | TACCGTCGTTCCACTAGTGATTT | Primers from kit | |
| 3′RACE inner primer | CGCGGATCCTCCACTAGTGATTTCACTATAGG | ||
| 5′RACE-R1 | ATGCTGAACTGAGTGGGTGG | Gene-specific outer primer for per2 | |
| 5′RACE-R2 | AGACGAATGCTGCTGCTCTT | Gene-specific inner primer for per2 | |
| 5′RACE-R3 | GATTGTGTTCCCACGGTATCT | Gene-specific outer primer for cry1 | |
| 5′RACE-R4 | TCCACCACGAAACTGAGCTG | Gene-specific inner primer for cry1 | |
| 5′RACE outer primer | CATGGCTACATGCTGACAGCCTA | Primers from kit | |
| 5′RACE inner primer | CGCGGATCCACAGCCTACTGATGATCAGTCGATG | ||
| qRT-PCR | qRT-PCR primer F | TGTTGAGTCCGCCAACCTTT | Used to amplify the per2 fragment for real-time PCR |
| qRT-PCR primer R | AGTGGCTGCTCCTCTGAAAC | ||
| qRT-PCR primer F | GATGCTGACTGGAGCGTAAA | Used to amplify the Cry1 fragment for real-time PCR | |
| qRT-PCR primer R | CCAAAGCCCACAGGACAATA | ||
| qRT-PCR primer F | CCAGTTGGAGGAACCAATGT | Used to amplify the OnCREB1 fragment for real-time PCR | |
| qRT-PCR primer R | CATGTGCTGTGGACTTGAAATAG | ||
| 18S primer F | TCCGCAGGAGTTGCTTCGAT | Used to amplify the 18S fragment for real-time PCR | |
| 18S primer R | ATTAAGCCGCAGGCTCCACT |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, G.; Yang, T.; Shen, H. Effect of Circadian Clock and Light–Dark Cycles in Onchidium reevesii: Possible Implications for Long-Term Memory. Genes 2019, 10, 488. https://doi.org/10.3390/genes10070488
Xu G, Yang T, Shen H. Effect of Circadian Clock and Light–Dark Cycles in Onchidium reevesii: Possible Implications for Long-Term Memory. Genes. 2019; 10(7):488. https://doi.org/10.3390/genes10070488
Chicago/Turabian StyleXu, Guolyu, Tiezhu Yang, and Heding Shen. 2019. "Effect of Circadian Clock and Light–Dark Cycles in Onchidium reevesii: Possible Implications for Long-Term Memory" Genes 10, no. 7: 488. https://doi.org/10.3390/genes10070488
APA StyleXu, G., Yang, T., & Shen, H. (2019). Effect of Circadian Clock and Light–Dark Cycles in Onchidium reevesii: Possible Implications for Long-Term Memory. Genes, 10(7), 488. https://doi.org/10.3390/genes10070488





