Synthetic Adrenocorticotropic Peptides Modulate the Expression Pattern of Immune Genes in Rat Brain following the Early Post-Stroke Period
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Rat transient Middle Cerebral Artery Occlusion (tMCAO) Model
2.2.1. Operation
2.2.2. Peptide Administration
2.3. Sample Collection and RNA Isolation
2.4. RNA-Seq
2.5. cDNA Synthesis and Real-Time Reverse Transcription Polymerase Chain Reaction (RT-PCR)
2.6. RNA-Seq Data Analysis
2.7. Real-Time RT-PCR Data Analysis
2.8. Functional Analysis
2.9. Availability of Data and Material
3. Results
3.1. Magnetic Resonance Imaging (MRI)
3.2. RNA-Seq Analysis of the Effect of IR on the Dorsolateral Areas of the Frontal Cortex of Rats 4.5 h after tMCAO
3.3. RNA-Seq Analysis of the Effect of Semax and ACTH(6-9)PGP on the Effect of IR on the Dorsolateral Areas of the Frontal Cortex of Rats 4.5 h after tMCAO
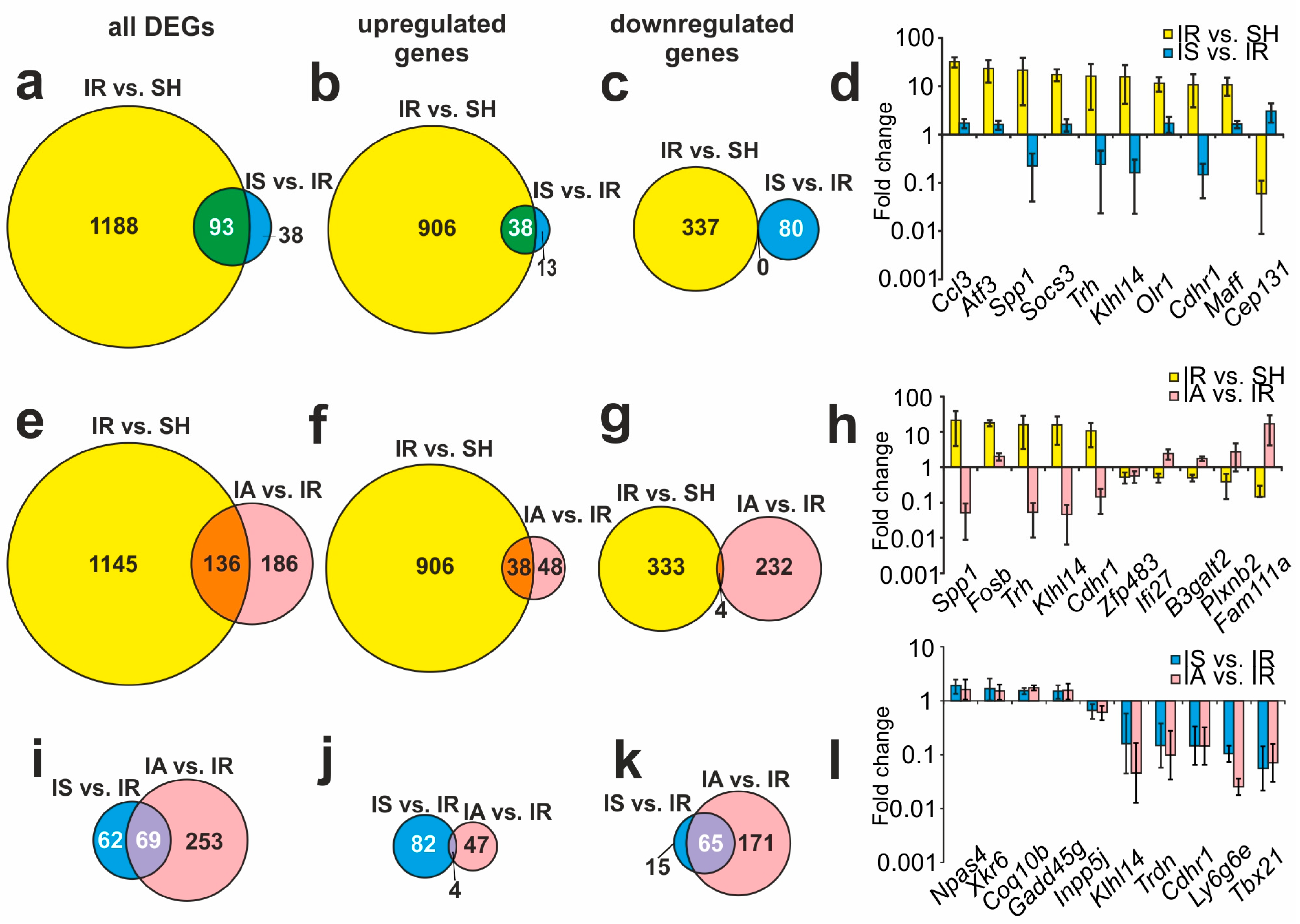
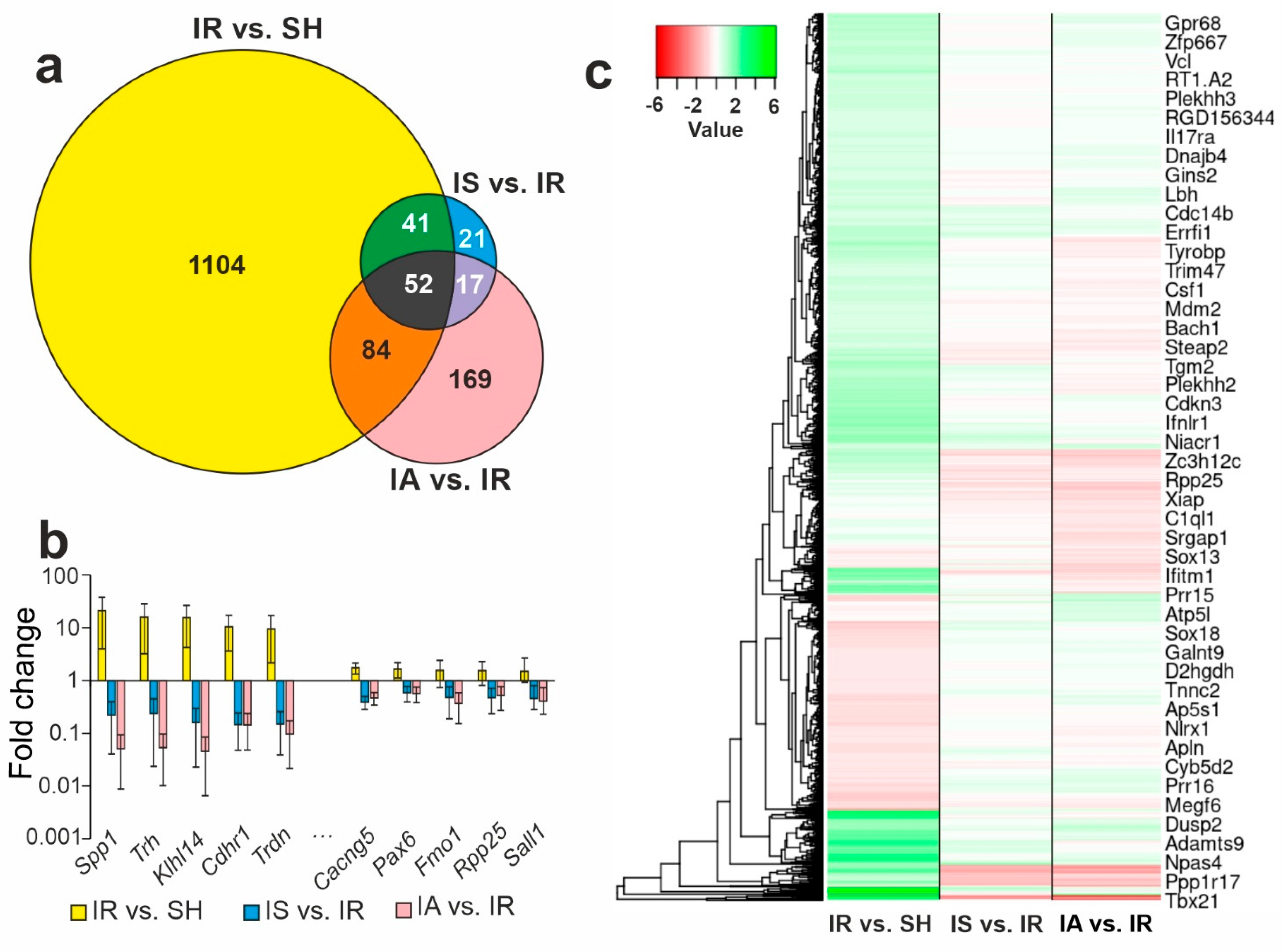
3.4. Comparison of RNA-Seq Results for Different Groups
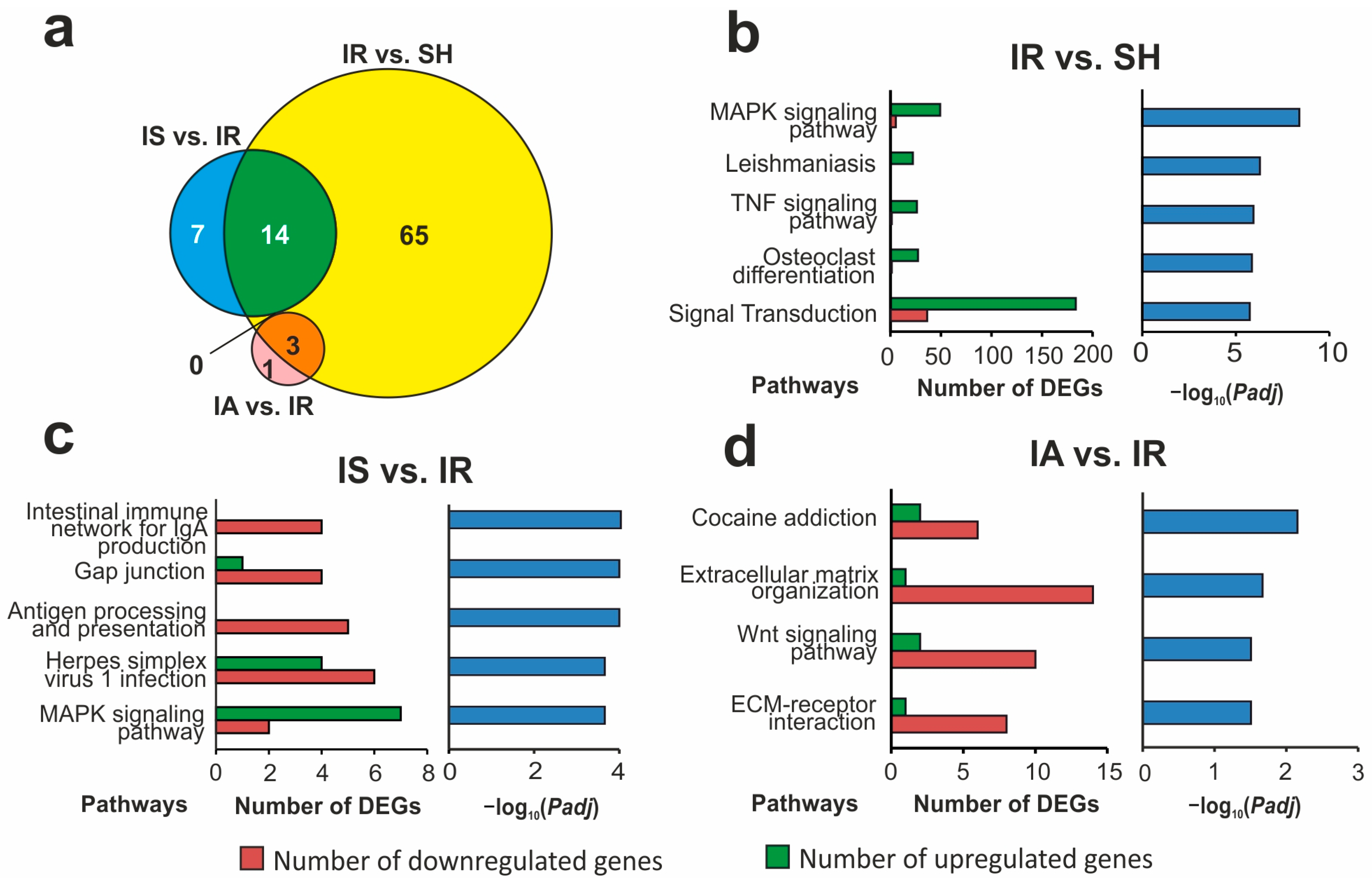
3.5. Functional Annotations of DEGs Altered in Different Comparison Groups
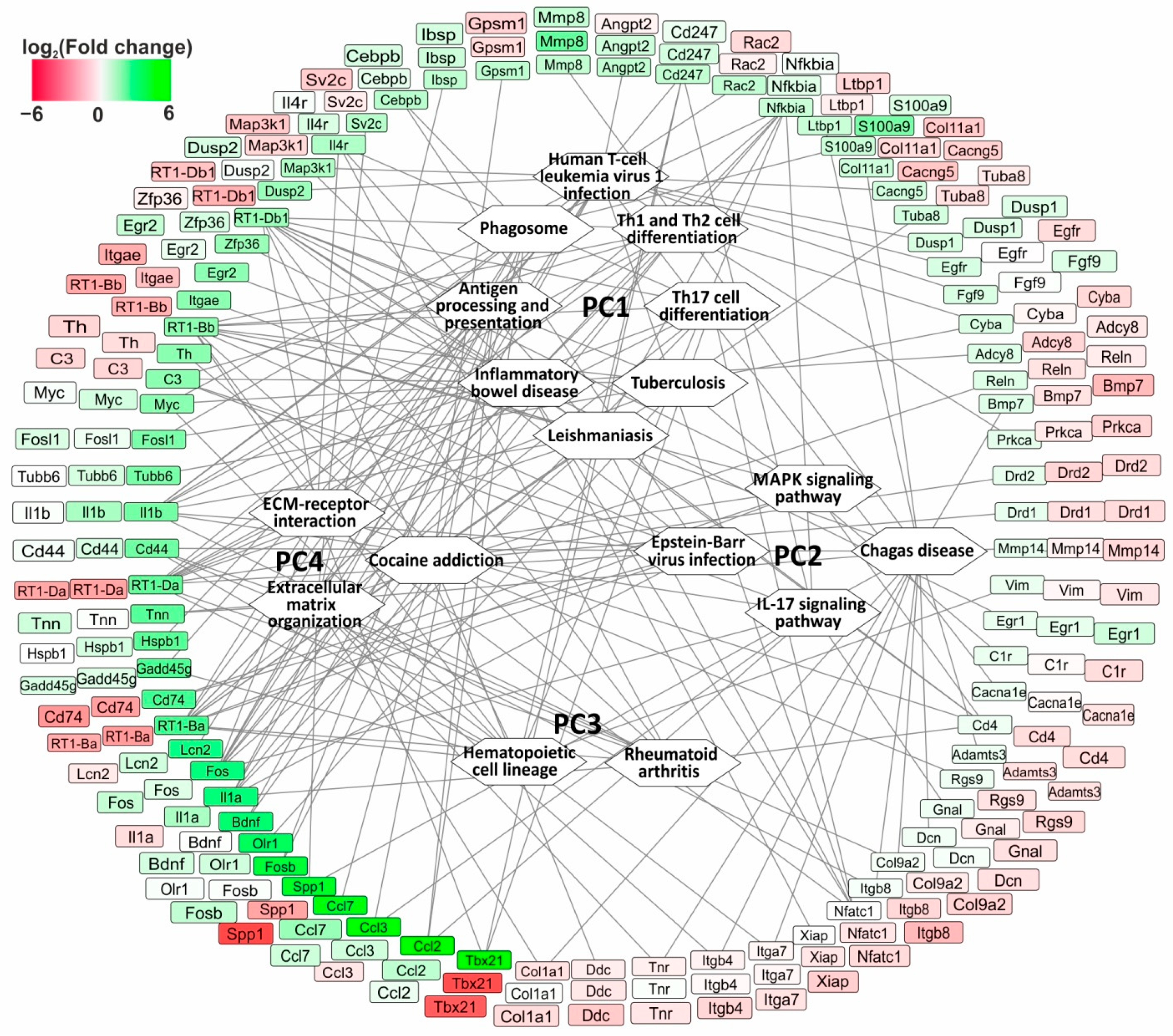
3.6. The Search for Pathways That Reflect Common and Unique Gene Expression Effects of Semax and ACTH(6-9)PGP Peptides in the Early Hours of IR Conditions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Feske, S.K. Ischemic Stroke. Am. J. Med. 2021, 134, 1457–1464. [Google Scholar] [CrossRef]
- Yang, Q.; Tong, X.; Schieb, L.; Vaughan, A.; Gillespie, C.; Wiltz, J.L.; King, S.C.; Odom, E.; Merritt, R.; Hong, Y.; et al. Vital Signs: Recent Trends in Stroke Death Rates-United States, 2000–2015. Morb. Mortal. Wkly. Rep. 2017, 66, 933. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paul, S.; Candelario-Jalil, E. Emerging neuroprotective strategies for the treatment of ischemic stroke: An overview of clinical and preclinical studies. Exp. Neurol. 2021, 335, 113518. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Zheng, Y.; Wu, J.; Chen, Y.; Wu, X.; Zhou, Y.; Yuan, Y.; Lu, S.; Jiang, L.; Qin, Z.; et al. PARK2-dependent mitophagy induced by acidic postconditioning protects against focal cerebral ischemia and extends the reperfusion window. Autophagy 2017, 13, 473–485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuo, D.-P.; Lu, C.-F.; Liou, M.; Chen, Y.-C.; Chung, H.-W.; Chen, C.-Y. Differentiation of the Infarct Core from Ischemic Penumbra within the First 4.5 h, Using Diffusion Tensor Imaging-Derived Metrics: A Rat Model. Korean J. Radiol. 2017, 18, 269–278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, S.; Xu, H.; Chi, X.; Wei, L.; Cheng, Q.; Yang, Y.; Zhou, C.; Ding, F. 2-(4-Methoxyphenyl)ethyl-2-Acetamido-2-deoxy-β-d-pyranoside (A Salidroside Analog) Confers Neuroprotection with a Wide Therapeutic Window by Regulating Local Glucose Metabolism in a Rat Model of Cerebral Ischemic Injury. Neuroscience 2018, 391, 60–72. [Google Scholar] [CrossRef]
- Ma, Y.; Li, L.; Kong, L.; Zhu, Z.; Zhang, W.; Song, J.; Chang, J.; Du, G. Pinocembrin Protects Blood-Brain Barrier Function and Expands the Therapeutic Time Window for Tissue-Type Plasminogen Activator Treatment in a Rat Thromboembolic Stroke Model. BioMed Res. Int. 2018, 2018, 8943210. [Google Scholar] [CrossRef]
- Yousuf, S.; Sayeed, I.; Atif, F.; Tang, H.; Wang, J.; Stein, D.G. Delayed progesterone treatment reduces brain infarction and improves functional outcomes after ischemic stroke: A time-window study in middle-aged rats. J. Cereb. Blood Flow Metab. 2014, 34, 297–306. [Google Scholar] [CrossRef]
- Hori, M.; Nakamachi, T.; Shibato, J.; Rakwal, R.; Shioda, S.; Numazawa, S. Unraveling the Specific Ischemic Core and Penumbra Transcriptome in the Permanent Middle Cerebral Artery Occlusion Mouse Model Brain Treated with the Neuropeptide PACAP38. Microarrays 2015, 4, 2–24. [Google Scholar] [CrossRef] [Green Version]
- Liu, B.H.; Jobichen, C.; Chia, C.S.B.; Chan, T.H.M.; Tang, J.P.; Chung, T.X.Y.; Li, J.; Poulsen, A.; Hung, A.W.; Koh-Stenta, X.; et al. Targeting cancer addiction for SALL4 by shifting its transcriptome with a pharmacologic peptide. Proc. Natl. Acad. Sci. USA 2018, 115, E7119–E7128. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.-M.; Pan, Y.-Y.; Liu, M.-H.; Cheng, B.-H.; Bai, B.; Chen, J. RNA-seq expression profiling of rat MCAO model following reperfusion Orexin-A. Oncotarget 2017, 8, 113066–113081. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Starowicz, K.; Przewłocka, B. The role of melanocortins and their receptors in inflammatory processes, nerve regeneration and nociception. Life Sci. 2003, 73, 823–847. [Google Scholar] [CrossRef] [PubMed]
- Adan, R.A.H.; Gispen, W.H. Brain Melanocortin Receptors: From Cloning to Function. Peptides 1997, 18, 1279–1287. [Google Scholar] [CrossRef]
- Catania, A.; Gatti, S.; Colombo, G.; Lipton, J.M. Targeting Melanocortin Receptors as a Novel Strategy to Control Inflammation. Pharmacol. Rev. 2004, 56, 1–29. [Google Scholar] [CrossRef] [PubMed]
- Cone, R.D. Anatomy and regulation of the central melanocortin system. Nat. Neurosci. 2005, 8, 571–578. [Google Scholar] [CrossRef] [PubMed]
- Giuliani, D.; Minutoli, L.; Ottani, A.; Spaccapelo, L.; Bitto, A.; Galantucci, M.; Altavilla, D.; Squadrito, F.; Guarini, S. Melanocortins as potential therapeutic agents in severe hypoxic conditions. Front. Neuroendocrinol. 2012, 33, 179–193. [Google Scholar] [CrossRef]
- Wikberg, J. Melanocortin receptors: New opportunities in drug discovery. Exp. Opin. Ther. Pat. 2001, 11, 61. [Google Scholar] [CrossRef]
- Gusev, E.I.; Martynov, M.Y.; Kostenko, E.V.; Petrova, L.V.; Bobyreva, S.N. The efficacy of semax in the tretament of patients at different stages of ischemic stroke. Zhurnal Nevrol. Psihiatr. Im. S.S. Korsakova 2018, 118, 61–68. [Google Scholar] [CrossRef]
- Potaman, V.N.; Alfeeva, L.Y.; Kamensky, A.A.; Levitzkaya, N.G.; Nezavibatko, V.N. N-terminal degradation of ACTH(4–10) and its synthetic analog semax by the rat blood enzymes. Biochem. Biophys. Res. Commun. 1991, 176, 741–746. [Google Scholar] [CrossRef]
- Stavchansky, V.V.; Yuzhakov, V.V.; Botsina, A.Y.; Skvortsova, V.I.; Bondurko, L.N.; Tsyganova, M.G.; Limborska, S.A.; Myasoedov, N.F.; Dergunova, L.V. The effect of Semax and its C-end peptide PGP on the morphology and proliferative activity of rat brain cells during experimental ischemia: A pilot study. J. Mol. Neurosci. 2011, 45, 177–185. [Google Scholar] [CrossRef]
- Dmitrieva, V.G.; Povarova, O.V.; Skvortsova, V.I.; Limborska, S.A.; Myasoedov, N.F.; Dergunova, L.V. Semax and Pro-Gly-Pro activate the transcription of neurotrophins and their receptor genes after cerebral ischemia. Cell. Mol. Neurobiol. 2010, 30, 71–79. [Google Scholar] [CrossRef]
- Medvedeva, E.V.; Dmitrieva, V.G.; Povarova, O.V.; Limborska, S.A.; Skvortsova, V.I.; Myasoedov, N.F.; Dergunova, L.V. The peptide semax affects the expression of genes related to the immune and vascular systems in rat brain focal ischemia: Genome-wide transcriptional analysis. BMC Genom. 2014, 15, 228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Medvedeva, E.V.; Dmitrieva, V.G.; Limborska, S.A.; Myasoedov, N.F.; Dergunova, L.V. Semax, an analog of ACTH(4-7), regulates expression of immune response genes during ischemic brain injury in rats. Mol. Genet. Genom. 2017, 292, 635–653. [Google Scholar] [CrossRef] [PubMed]
- Filippenkov, I.B.; Stavchansky, V.V.; Denisova, A.E.; Yuzhakov, V.V.; Sevan’kaeva, L.E.; Sudarkina, O.Y.; Dmitrieva, V.G.; Gubsky, L.V.; Myasoedov, N.F.; Limborska, S.A.; et al. Novel Insights into the Protective Properties of ACTH (4–7) PGP (Semax) Peptide at the Transcriptome Level Following Cerebral Ischaemia-Reperfusion in Rats. Genes 2020, 11, 681. [Google Scholar] [CrossRef]
- Stavchansky, V.V.; Filippenkov, I.B.; Remizova, J.A.; Denisova, A.E.; Mozgovoy, I.V.; Gubsky, L.V.; Myasoedov, N.F.; Andreeva, L.A.; Limborska, S.A.; Dergunova, L.V. Insight into Glyproline Peptides’ Activity through the Modulation of the Inflammatory and Neurosignaling Genetic Response following Cerebral Ischemia–Reperfusion. Genes 2022, 13, 2380. [Google Scholar] [CrossRef]
- Sudarkina, O.Y.; Filippenkov, I.B.; Stavchansky, V.V.; Denisova, A.E.; Yuzhakov, V.V.; Sevan’kaeva, L.E.; Valieva, L.V.; Remizova, J.A.; Dmitrieva, V.G.; Gubsky, L.V.; et al. Brain Protein Expression Profile Confirms the Protective Effect of the ACTH(4–7)PGP Peptide (Semax) in a Rat Model of Cerebral Ischemia–Reperfusion. Int. J. Mol. Sci. 2021, 22, 6179. [Google Scholar] [CrossRef]
- Filippenkov, I.B.; Stavchansky, V.V.; Glazova, N.Y.; Sebentsova, E.A.; Remizova, J.A.; Valieva, L.V.; Levitskaya, N.G.; Myasoedov, N.F.; Limborska, S.A.; Dergunova, L.V. Antistress action of melanocortin derivatives associated with correction of gene expression patterns in the hippocampus of male rats following acute stress. Int. J. Mol. Sci. 2021, 22, 10054. [Google Scholar] [CrossRef]
- Vorvul, A.O.; Bobyntsev, I.I.; Medvedeva, O.A.; Mukhina, A.Y.; Svishcheva, M.V.; Azarova, I.E.; Andreeva, L.A.; Myasoedov, N.F. ACTH(6-9)-Pro-Gly-Pro ameliorates anxiety-like and depressive-like behaviour and gut mucosal microbiota composition in rats under conditions of chronic restraint stress. Neuropeptides 2022, 93, 35487169. [Google Scholar] [CrossRef]
- Bakaeva, Z.V.; Surin, A.M.; Lizunova, N.V.; Zgodova, A.E.; Krasilnikova, I.A.; Fisenko, A.P.; Frolov, D.A.; Andreeva, L.A.; Myasoedov, N.F.; Pinelis, V.G. Neuroprotective Potential of Peptides HFRWPGP (ACTH 6–9 PGP), KKRRPGP, and PyrRP in Cultured Cortical Neurons at Glutamate Excitotoxicity. Dokl. Biochem. Biophys. 2020, 491, 62–66. [Google Scholar] [CrossRef] [PubMed]
- Akimov, M.G.; Fomina-Ageeva, E.V.; Dudina, P.V.; Andreeva, L.A.; Myasoyedov, N.F.; Bezuglov, V.V. ACTH(6–9)PGP Peptide Protects SH-SY5Y Cells from H2O2, tert-Butyl Hydroperoxide, and Cyanide Cytotoxicity via Stimulation of Proliferation and Induction of Prosurvival-Related Genes. Molecules 2021, 26, 1878. [Google Scholar] [CrossRef]
- Filippenkov, I.B.; Remizova, J.A.; Denisova, A.E.; Stavchansky, V.V.; Golovina, K.D.; Gubsky, L.V.; Limborska, S.A.; Dergunova, L.V. Comparative Use of Contralateral and Sham-Operated Controls Reveals Traces of a Bilateral Genetic Response in the Rat Brain after Focal Stroke. Int. J. Mol. Sci. 2022, 23, 7308. [Google Scholar] [CrossRef] [PubMed]
- Koizumi, J.; Yoshida, Y.; Nakazawa, T.; Ooneda, G. Experimental studies of ischemic brain edema. Nosotchu 1986, 8, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Ashmarin, I.; Nezavibatko, V.; Levitskaya, N.; Koshelev, V.; Kamensky, A. Design and Investigation of an ACTH(4–10) Analog Lacking D-Amino Acids and Hydrophobic Radicals. Neurosci. Res. Commun. 1995, 16, 105–112. [Google Scholar]
- Miasoedova, N.F.; Skvortsova, V.I.; Nasonov, E.L.; Zhuravleva, E.I.; Grivennikov, I.A.; Arsen’eva, E.L.; Sukhanov, I.I. Investigation of mechanisms of neuro-protective effect of semax in acute period of ischemic stroke. Zhurnal Nevrol. Psihiatr. Im. S.S. Korsakova 1999, 99, 15–19. [Google Scholar]
- Medvedeva, E.V.; Dmitrieva, V.G.; Povarova, O.V.; Limborska, S.A.; Skvortsova, V.I.; Myasoedov, N.F.; Dergunova, L.V. Tripeptide Pro-Gly-Pro affects rat-brain transcriptome during focal ischemia. Mol. Biol. 2014, 48, 238–247. [Google Scholar] [CrossRef]
- Shubina, T.A.; Grigor’eva, M.E.; Lyapina, L.A.; Obergan, T.Y.; Myasoedov, N.F.; Andreeva, L.A. Hypoglycemic and anticoagulant effects of tetrapeptide Pro-Gly-Pro-Leu in hypercholesterolemia. Bull. Exp. Biol. Med. 2014, 158, 30–33. [Google Scholar] [CrossRef]
- Pfaffl, M.W.; Tichopad, A.; Prgomet, C.; Neuvians, T.P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol. Lett. 2004, 26, 509–515. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef] [PubMed]
- Babicki, S.; Arndt, D.; Marcu, A.; Liang, Y.; Grant, J.R.; Maciejewski, A.; Wishart, D.S. Heatmapper: Web-enabled heat mapping for all. Nucleic Acids Res. 2016, 44, W147–W153. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software Environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef]
- Filippenkov, I.B.; Remizova, J.A.; Denisova, A.E.; Stavchansky, V.V.; Golovina, K.D.; Gubsky, L.V.; Limborska, S.A.; Dergunova, L.V. Differential gene expression in the contralateral hemisphere of the rat brain after focal ischemia. Sci. Rep. 2023, 13, 573. [Google Scholar] [CrossRef] [PubMed]
- National Center for Biotechnology Information. Available online: http://www.ncbi.nlm.nih.gov/bioproject/916856 (accessed on 5 January 2023).
- Giulian, D.; Corpuz, M.; Chapman, S.; Mansouri, M.; Robertson, C. Reactive mononuclear phagocytes release neurotoxins after ischemic and traumatic injury to the central nervous system. J. Neurosci. Res. 1993, 36, 681–693. [Google Scholar] [CrossRef]
- Lehnardt, S.; Massillon, L.; Follett, P.; Jensen, F.E.; Ratan, R.; Rosenberg, P.A.; Volpe, J.J.; Vartanian, T. Activation of innate immunity in the CNS triggers neurodegeneration through a Toll-like receptor 4-dependent pathway. Proc. Natl. Acad. Sci. USA 2003, 100, 8514–8519. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Chopp, M.; Powers, C. Temporal profile of microglial response following transient (2 h) middle cerebral artery occlusion. Brain Res. 1997, 744, 189–198. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liu, H.; Zhang, L.; Wang, G.; Zhang, M.; Yu, Y. Neuroprotection of Dexmedetomidine against Cerebral Ischemia-Reperfusion Injury in Rats: Involved in Inhibition of NF-κB and Inflammation Response. Biomol. Ther. 2017, 25, 383–389. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marcheselli, V.L.; Hong, S.; Lukiw, W.J.; Tian, X.H.; Gronert, K.; Musto, A.; Hardy, M.; Gimenez, J.M.; Chiang, N.; Serhan, C.N.; et al. Novel Docosanoids Inhibit Brain Ischemia-Reperfusion-mediated Leukocyte Infiltration and Pro-inflammatory Gene Expression. J. Biol. Chem. 2003, 278, 43807–43817. [Google Scholar] [CrossRef] [Green Version]
- Nogawa, S.; Zhang, F.; Elizabeth Ross, M.; Iadecola, C. Cyclo-oxygenase-2 gene expression in neurons contributes to ischemic brain damage. J. Neurosci. 1997, 17, 2746–2755. [Google Scholar] [CrossRef] [Green Version]
- Iadecola, C.; Zhang, F.; Xu, S.; Casey, R.; Ross, M.E. Inducible Nitric Oxide Synthase Gene Expression in Brain following Cerebral Ischemia. J. Cereb. Blood Flow Metab. 1995, 15, 378–384. [Google Scholar] [CrossRef] [Green Version]
- Fury, W.; Park, K.W.; Wu, Z.; Kim, E.; Woo, M.S.; Bai, Y.; MacDonald, L.E.; Croll, S.D.; Cho, S. Sustained Increases in Immune Transcripts and Immune Cell Trafficking during the Recovery of Experimental Brain Ischemia. Stroke 2020, 51, 2514–2525. [Google Scholar] [CrossRef]
- Behrouzifar, S.; Vakili, A.; Barati, M. The Effects of Mouse Recombinant Resistin on mRNA Expression of Proinflammatory and Anti-Inflammatory Cytokines and Heat Shock Protein-70 in Experimental Stroke Model. J. Stroke Cerebrovasc. Dis. 2018, 27, 3272–3279. [Google Scholar] [CrossRef]
- Zhang, Y.Y.; Wang, K.; Liu, Y.U.N.E.; Wang, W.; Liu, A.F.; Zhou, J.; Li, C.; Zhang, Y.Q.; Zhang, A.P.; Lv, J.; et al. Identification of key transcription factors associated with cerebral ischemia-reperfusion injury based on gene-set enrichment analysis. Int. J. Mol. Med. 2019, 43, 2429–2439. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Johnsen, S.P.; Hundborg, H.H.; Sørensen, H.T.; Ørskov, H.; Tjønneland, A.; Overvad, K.; Jørgensen, J.O.L. Insulin-like growth factor (IGF) I, -II, and IGF binding protein-3 and risk of ischemic stroke. J. Clin. Endocrinol. Metab. 2005, 90, 5937–5941. [Google Scholar] [CrossRef] [PubMed]
- Giovannini, S.; Cesari, M.; Marzetti, E.; Leeuwenburgh, C.; Maggio, M.; Pahor, M. Effects of ACE-inhibition on IGF-1 and IGFBP-3 concentrations in older adults with high cardiovascular risk profile. J. Nutr. Health Aging 2010, 14, 457–460. [Google Scholar] [CrossRef] [Green Version]
- Dergunova, L.V.; Filippenkov, I.B.; Stavchansky, V.V.; Denisova, A.E.; Yuzhakov, V.V.; Mozerov, S.A.; Gubsky, L.V.; Limborska, S.A. Genome-wide transcriptome analysis using RNA-Seq reveals a large number of differentially expressed genes in a transient MCAO rat model. BMC Genom. 2018, 19, 655. [Google Scholar] [CrossRef] [Green Version]
- Dong, Y.; Benveniste, E.N. Immune function of astrocytes. Glia 2001, 36, 180–190. [Google Scholar] [CrossRef]
- Gusev, E.I.; Skvortsova, V.I.; Miasoedov, N.F.; Nezavibat’ko, V.N.; Zhuravleva, E.I.; Vanichkin, A.V. Effectiveness of semax in acute period of hemispheric ischemic stroke (a clinical and electrophysiological study). Zhurnal Nevrol. Psihiatr. Im. S.S. Korsakova 1997, 97, 26–34. [Google Scholar]
- Gusev, E.I.; Skvortsova, V.I.; Chukanova, E.I. Semax in prevention of disease progress and development of exacerbations in patients with cerebrovascular insufficiency. Zhurnal Nevrol. Psihiatr. Im. S.S. Korsakova 2005, 105, 35–40. [Google Scholar]
- Dergunova, L.V.; Dmitrieva, V.G.; Filippenkov, I.B.; Stavchansky, V.V.; Denisova, A.E.; Yuzhakov, V.V.; Sevan’kaeva, L.E.; Valieva, L.V.; Sudarkina, O.Y.; Gubsky, L.V.; et al. The Peptide Drug ACTH(4–7)PGP (Semax) Suppresses mRNA Transcripts Encoding Proinflammatory Mediators Induced by Reversible Ischemia of the Rat Brain. Mol. Biol. 2021, 55, 402–411. [Google Scholar] [CrossRef]
- Jayaraj, R.L.; Azimullah, S.; Beiram, R.; Jalal, F.Y.; Rosenberg, G.A.; Anrather, J.; Iadecola, C.; Tasaki, K.; Ruetzler, C.A.; Ohtsuki, T.; et al. Neuroinflammation in post-ischemic neurodegeneration of the brain: Friend, foe, or both? Int. J. Mol. Sci. 2021, 16, 267–270. [Google Scholar] [CrossRef]
- Anrather, J.; Iadecola, C. Inflammation and Stroke: An Overview. Neurotherapeutics 2016, 13, 661–670. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jayaraj, R.L.; Azimullah, S.; Beiram, R.; Jalal, F.Y.; Rosenberg, G.A. Neuroinflammation: Friend and foe for ischemic stroke. J. Neuroinflamm. 2019, 16, 142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shichita, T.; Ooboshi, H.; Yoshimura, A. Neuroimmune mechanisms and therapies mediating post-ischaemic brain injury and repair. Nat. Rev. Neurosci. 2023, 24, 36973481. [Google Scholar] [CrossRef] [PubMed]
- Chopp, M.; Zhang, R.L.; Chen, H.; Li, Y.; Jiang, N.; Rusche, J.R. Postischemic administration of an anti-Mac-1 antibody reduces ischemic cell damage after transient middle cerebral artery occlusion in rats. Stroke 1994, 25, 869–875. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garcia, J.H.; Liu, K.F.; Bree, M.P. Effects of CD11b/18 monoclonal antibody on rats with permanent middle cerebral artery occlusion. Am. J. Pathol. 1996, 148, 241. [Google Scholar]
- Lees, K.R.; Diener, H.C.; Asplund, K.; Krams, M. UK-279,276, a neutrophil inhibitory glycoprotein, in acute stroke: Tolerability and pharmacokinetics. Stroke 2003, 34, 1704–1709. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Zhang, Z.G.; Zhang, R.L.; Lu, M.; Krams, M.; Chopp, M. Effects of a selective CD11b/CD18 antagonist and recombinant human tissue plasminogen activator treatment alone and in combination in a rat embolic model of stroke. Stroke 2003, 34, 1790–1795. [Google Scholar] [CrossRef] [Green Version]
- Ohtsuki, T.; Ruetzler, C.A.; Tasaki, K.; Hallenbeck, J.M. Interleukin-1 mediates induction of tolerance to global ischemia in gerbil hippocampal CA1 neurons. J. Cereb. Blood Flow Metab. 1996, 16, 1137–1142. [Google Scholar] [CrossRef] [Green Version]
- Vyunova, T.V.; Andreeva, L.A.; Shevchenko, K.V.; Myasoedov, N.F. Synacton and individual activity of synthetic and natural corticotropins. J. Mol. Recognit. 2017, 30, e2597. [Google Scholar] [CrossRef]
- Vyunova, T.V.; Andreeva, L.A.; Shevchenko, K.V.; Shevchenko, V.P.; Bobrov, M.Y.; Bezuglov, V.V.; Myasoedov, N.F. Characteristic features of specific binding of pentapeptide HFPGP labeled at the C-terminal proline residue to rat forebrain plasma membranes. Dokl. Biochem. Biophys. 2014, 456, 101–103. [Google Scholar] [CrossRef]
- V’unova, T.V.; Andreeva, L.A.; Shevchenko, K.V.; Shevchenko, V.P.; Bobrov, M.Y.; Bezuglov, V.V.; Myasoedov, N.F. Binding of tripeptide Pro-Gly-Pro labeled at the C-terminal proline residue to plasma membranes of the rat forebrain. Dokl. Biol. Sci. 2008, 419, 95–96. [Google Scholar] [CrossRef]
- Dergunova, L.V.; Filippenkov, I.B.; Limborska, S.A.; Myasoedov, N.F. Pharmacotranscriptomics of peptide drugs with neuroprotective properties. Med. Res. Rev. 2021, 41, 754–769. [Google Scholar] [CrossRef]
- Shevchenko, K.V.; Nagaev, I.Y.; Andreeva, L.A.; Shevchenko, V.P.; Myasoedov, N.F. Stability of Proline-Containing Peptides in Biological Media. Biochem. Suppl. Ser. B Biomed. Chem. 2019, 13, 179–201. [Google Scholar] [CrossRef]
- Zolotarev, Y.A.; Dolotov, O.V.; Inozemtseva, L.S.; Dadayan, A.K.; Dorokhova, E.M.; Andreeva, L.A.; Alfeeva, L.Y.; Grivennikov, I.A.; Myasoedov, N.F. Degradation of the ACTH(4–10) analog Semax in the presence of rat basal forebrain cell cultures and plasma membranes. Amino Acids 2006, 30, 403–408. [Google Scholar] [CrossRef] [PubMed]
- Zolotarev, I.A.; Dadaian, A.K.; Dolotov, O.V.; Kozik, V.S.; Kost, N.V.; Sokolov, O.I.; Dorokhova, E.M.; Meshavkin, V.K.; Inozemtseva, L.S.; Gabaeva, M.V.; et al. Evenly tritium-labeled peptides and their in vivo and in vitro biodegradation. Bioorg. Khimiia 2006, 32, 183–191. [Google Scholar]
- Franks, A.; Airoldi, E.; Slavov, N. Post-transcriptional regulation across human tissues. PLoS Comput. Biol. 2017, 13, 28481885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buccitelli, C.; Selbach, M. mRNAs, proteins and the emerging principles of gene expression control. Nat. Rev. Genet. 2020, 21, 630–644. [Google Scholar] [CrossRef] [PubMed]
- Gry, M.; Rimini, R.; Strömberg, S.; Asplund, A.; Pontén, F.; Uhlén, M.; Nilsson, P. Correlations between RNA and protein expression profiles in 23 human cell lines. BMC Genom. 2009, 10, 19660143. [Google Scholar] [CrossRef] [Green Version]
- Eraslan, B.; Wang, D.; Gusic, M.; Prokisch, H.; Hallström, B.M.; Uhlén, M.; Asplund, A.; Pontén, F.; Wieland, T.; Hopf, T.; et al. Quantification and discovery of sequence determinants of protein-per-mRNA amount in 29 human tissues. Mol. Syst. Biol. 2019, 15, 30777893. [Google Scholar] [CrossRef]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Filippenkov, I.B.; Remizova, J.A.; Stavchansky, V.V.; Denisova, A.E.; Gubsky, L.V.; Myasoedov, N.F.; Limborska, S.A.; Dergunova, L.V. Synthetic Adrenocorticotropic Peptides Modulate the Expression Pattern of Immune Genes in Rat Brain following the Early Post-Stroke Period. Genes 2023, 14, 1382. https://doi.org/10.3390/genes14071382
Filippenkov IB, Remizova JA, Stavchansky VV, Denisova AE, Gubsky LV, Myasoedov NF, Limborska SA, Dergunova LV. Synthetic Adrenocorticotropic Peptides Modulate the Expression Pattern of Immune Genes in Rat Brain following the Early Post-Stroke Period. Genes. 2023; 14(7):1382. https://doi.org/10.3390/genes14071382
Chicago/Turabian StyleFilippenkov, Ivan B., Julia A. Remizova, Vasily V. Stavchansky, Alina E. Denisova, Leonid V. Gubsky, Nikolay F. Myasoedov, Svetlana A. Limborska, and Lyudmila V. Dergunova. 2023. "Synthetic Adrenocorticotropic Peptides Modulate the Expression Pattern of Immune Genes in Rat Brain following the Early Post-Stroke Period" Genes 14, no. 7: 1382. https://doi.org/10.3390/genes14071382





