Screening and Preliminary Identification of Asparagus officinalis Varieties under Low-Temperature Stress
Abstract
:1. Introduction
2. Plant Materials and Methods
2.1. Plant Materials and Low-Temperature Stress Treatment
2.2. Physiological Index Detection
2.3. Transcriptome Sequencing
2.4. Analysis of Transcriptomic Results
2.5. DEGs Identified Using qRT-PCR in A. officinalis
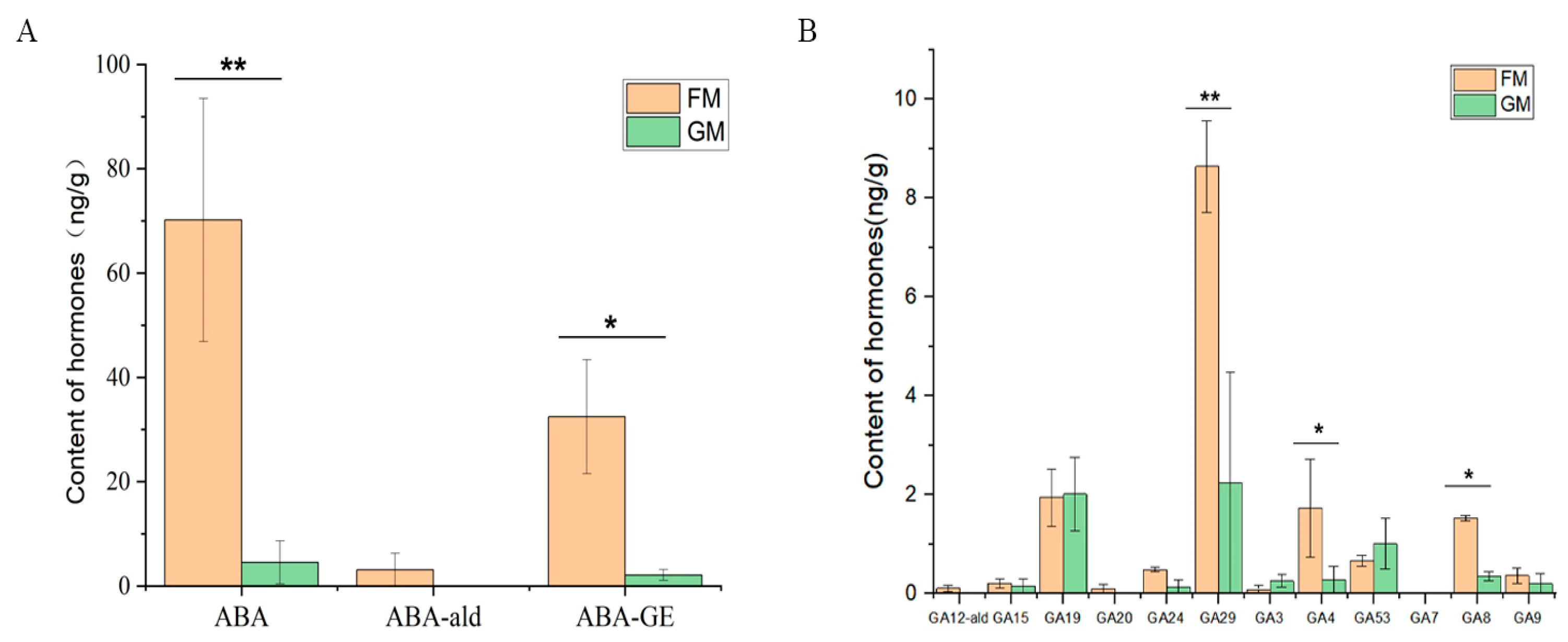
2.6. Determination of Endogenous Hormone Levels in A. officinalis
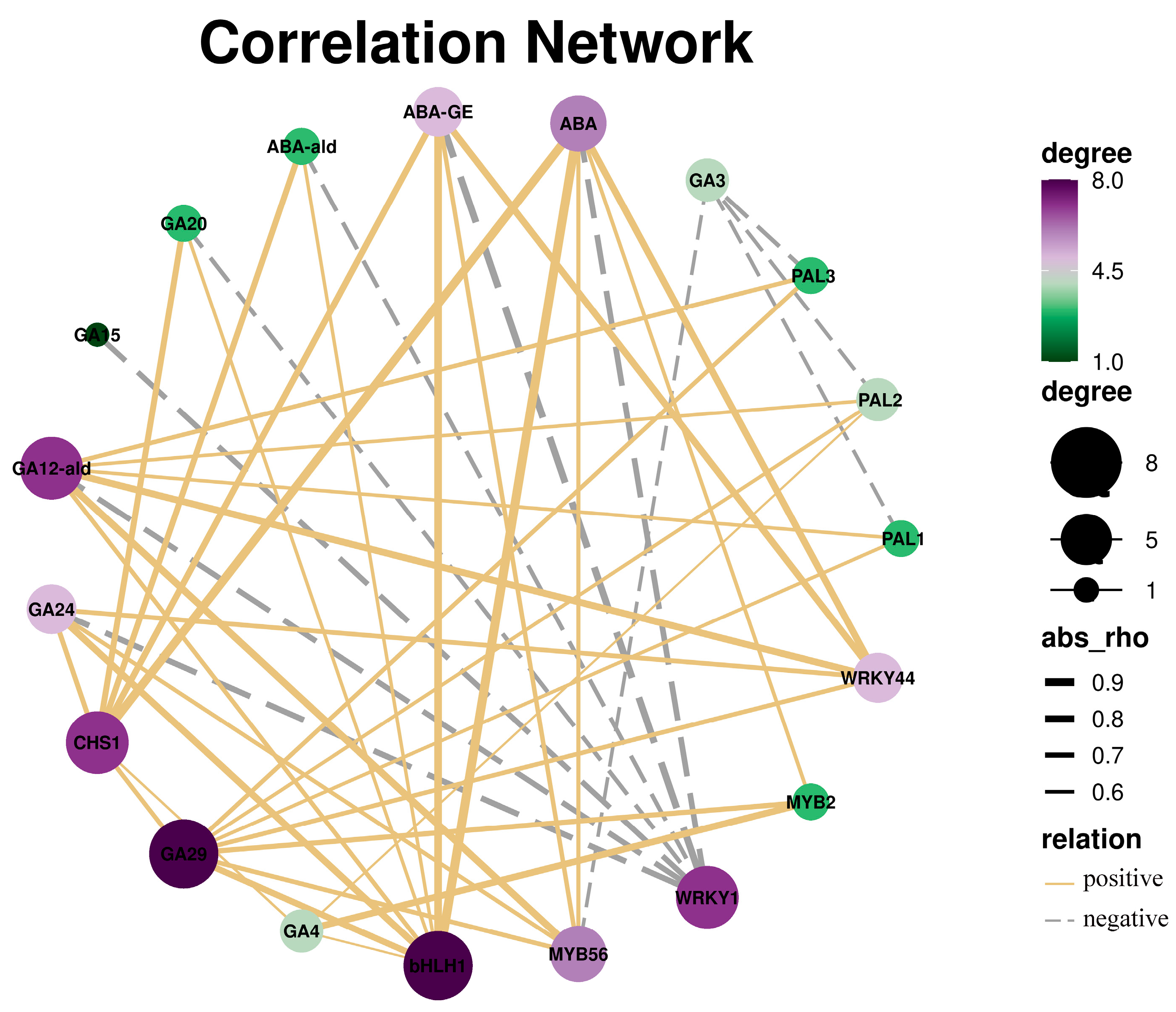
2.7. Combined Analysis of DEGs and Endogenous Hormones
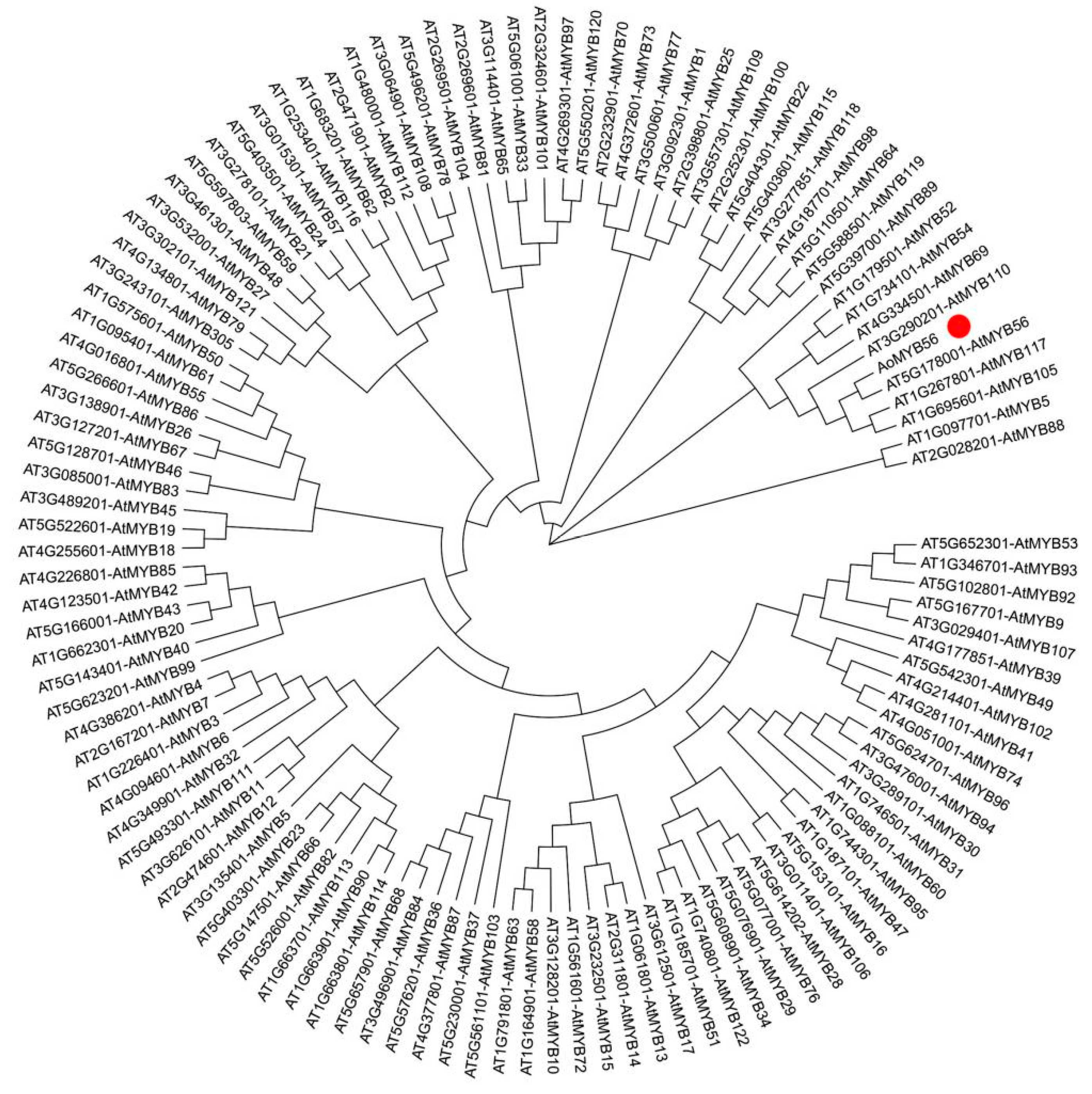
2.8. Phylogenetic Analysis between the AoMYB56 and MYB Genes in A. thaliana
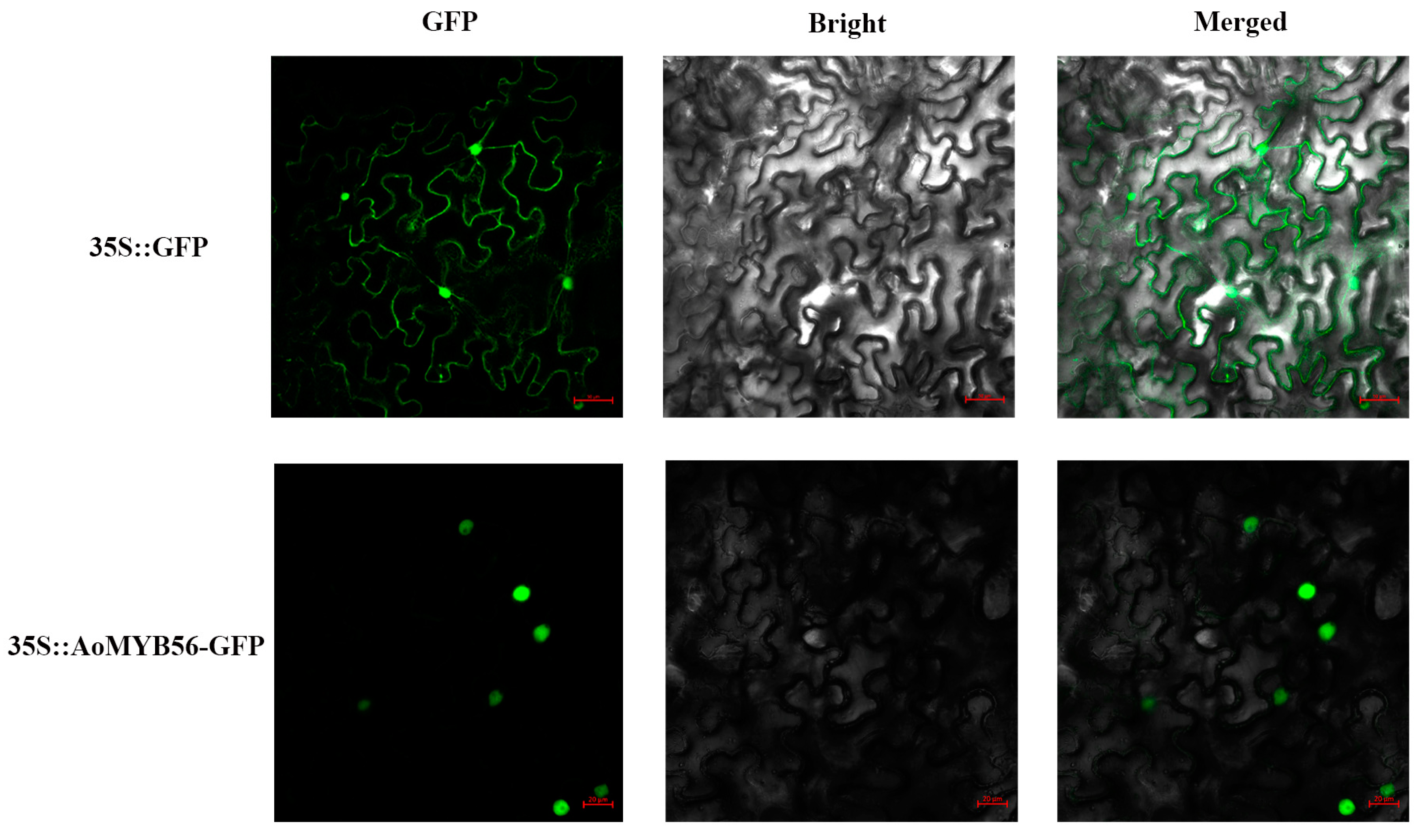
2.9. Subcellular Localisation of AoMYB56
3. Results
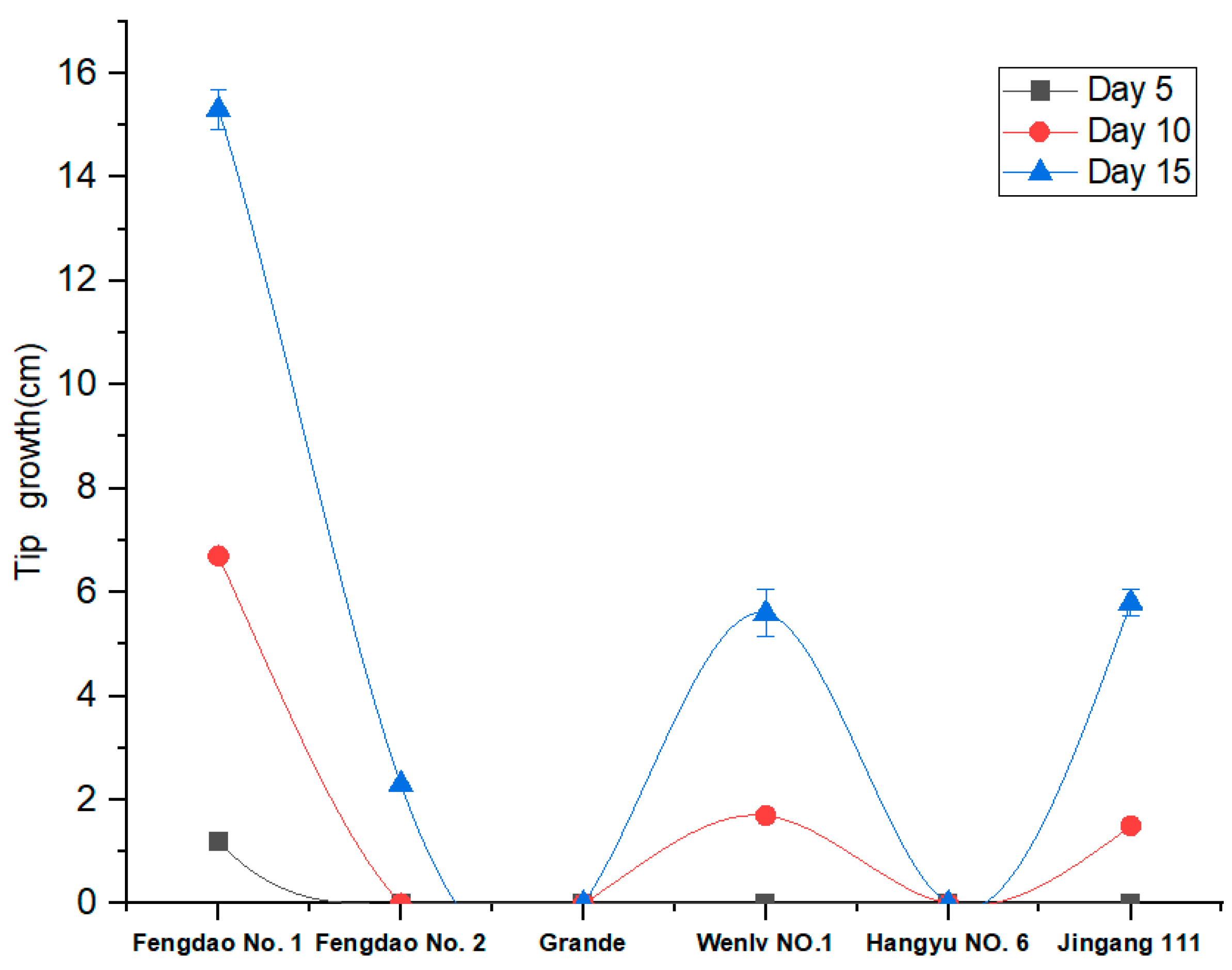
3.1. Growth of Six Asparagus Varieties under Low-Temperature Conditions
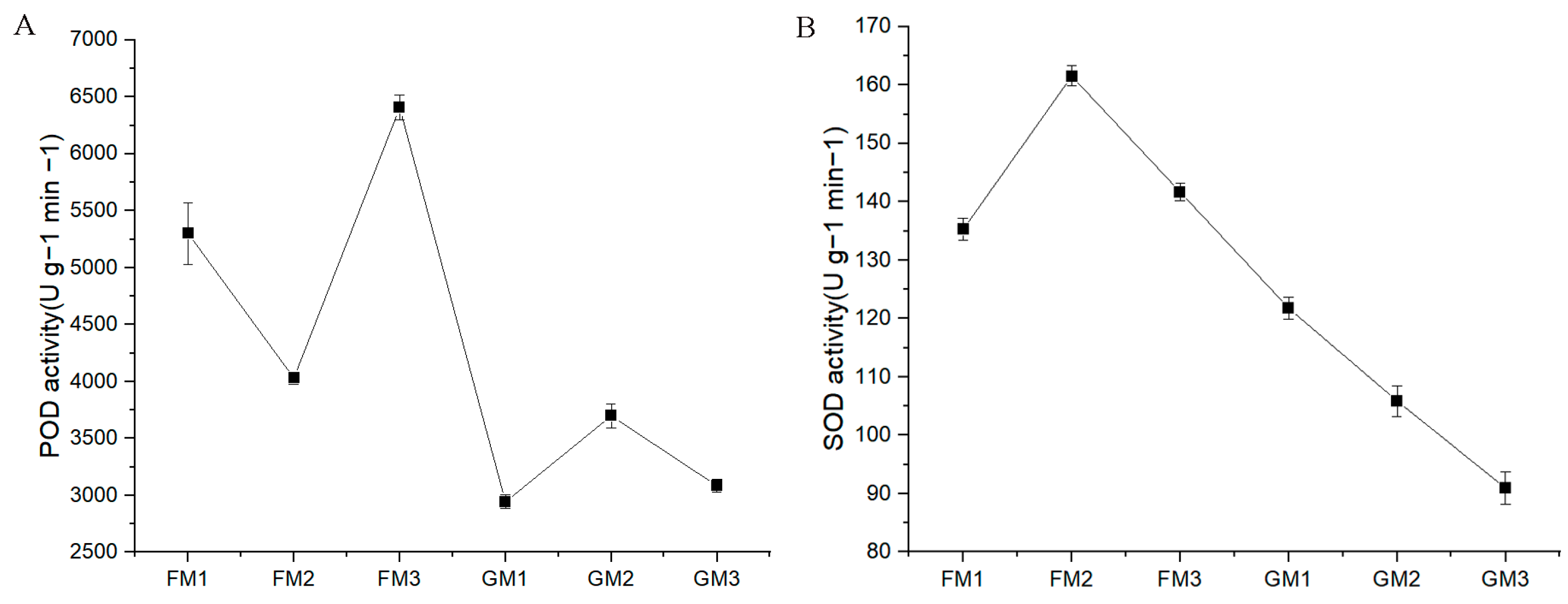
3.2. POD and SOD Activity Determination in “Fengdao No. 1” and “Grande”
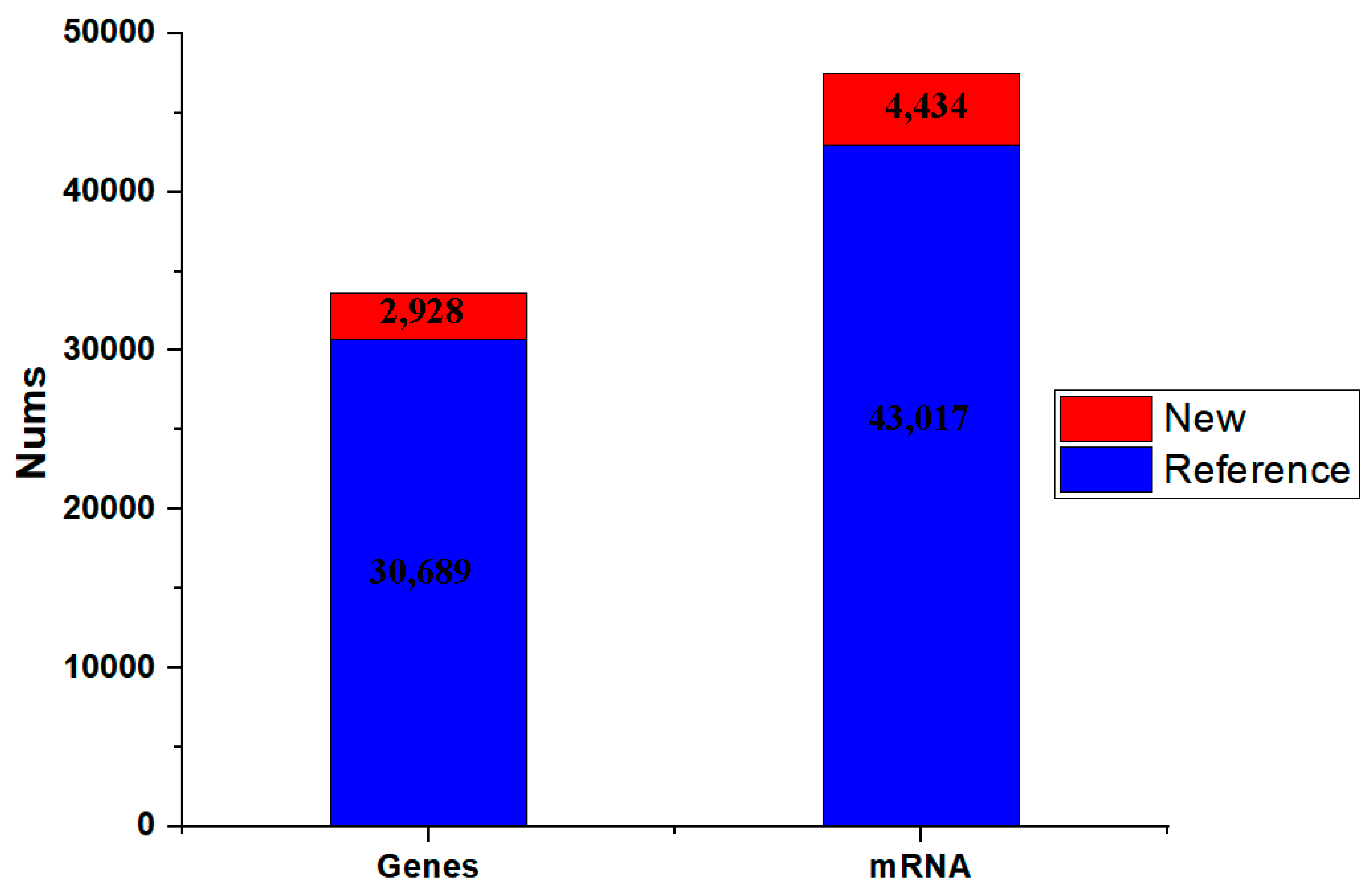
3.3. Transcriptome Sequencing Data
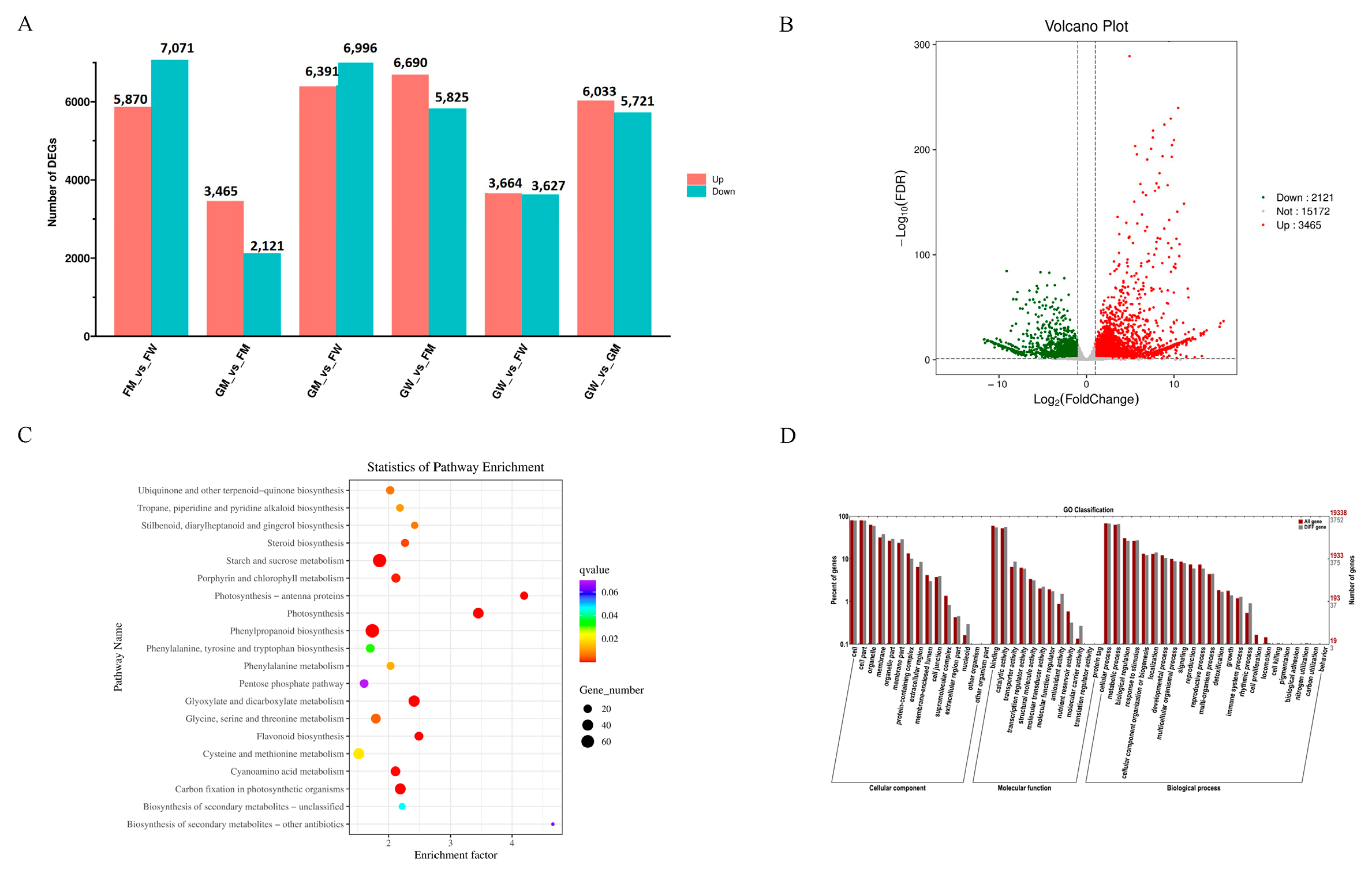
3.4. DEG Analysis between “Fengdao No. 1” and “Grande”
3.5. Candidate Key DEG Analysis and qRT-PCR Verification
3.6. Endogenous Hormone Analysis
3.7. Correlation Analysis of DEGs and Hormones
3.8. Preliminary Identification of AoMYB56
3.9. Subcellular Localization of AoMYB56
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yu, T.; Nassuth, A.; Peterson, R. Characterization of the interaction between the dark septate fungus Phialocephala fortinii and Asparagus officinalis roots. Can. J. Microbiol. 2001, 47, 741. [Google Scholar] [CrossRef] [PubMed]
- He, C.; Hsiang, T.; Wolyn, D. Induction of systemic disease resistance and pathogen defence responses in Asparagus officinalis inoculated with nonpathogenic strains of Fusarium oxysporum. Plant Pathol. 2010, 5, 225–230. [Google Scholar] [CrossRef]
- Gilmour, S.; Zarka, D.; Stockinger, E. Low temperature regulation of the Arabidopsis CBF family of AP2 transcriptional activators as an early step in cold-induced COR gene expression. Plant J. 2010, 16, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Song, Z.; Lai, X.; Chen, H.; Wang, L.; Pang, X.; Hao, Y.; Lu, W.; Chen, W.; Zhu, X.; Li, X. Role of MaABI5-like in abscisic acid-induced cold tolerance of ‘Fenjiao’ banana fruit. Hortic. Res. 2022, 9, uhac130. [Google Scholar] [CrossRef]
- Kreps, J.; Wu, Y.; Chang, H. Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold Stress. Plant Physiol. 2002, 130, 2129–2141. [Google Scholar] [CrossRef]
- Jiang, G.; Wang, S.; Xie, J.; Tan, P.; Han, L. Discontinuous low temperature stress and plant growth regulators during the germination period promote roots growth in alfalfa (Medicago sativa L.). Plant Physiol. Biochem. 2023, 11, 197–208. [Google Scholar] [CrossRef]
- Yang, C.; Dong, A.; Deng, L. Deciphering the change pattern of lipid metabolism in Saccharomyces cerevisiae responding to low temperature. Biochem. Eng. J. 2023, 12, 190–201. [Google Scholar] [CrossRef]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Molecular response to dehydration and low temperature: Differences and cross-talk between two stress signaling pathways. Curr. Opin. Plant Biol. 2000, 3, 217–223. [Google Scholar] [CrossRef]
- Renaut, J.; Hausman, J.; Wisniewski, M. Proteomics and low-temperature studies: Bridging the gap between gene expression and metabolism. Physiol. Plant. 2010, 126, 97–109. [Google Scholar] [CrossRef]
- Guan, Y.; Hwarari, D.; Korboe, H.; Ahmad, B.; Cao, Y.; Movahedi, A.; Yang, L. Low temperature stress-induced perception and molecular signaling pathways in plants. Environ. Exp. Bot. 2023, 207, 105–119. [Google Scholar] [CrossRef]
- Cao, F.; Yoshioka, K.; Desveaux, D. The roles of ABA in plant-pathogen interactions. J. Plant Res. 2011, 124, 489–499. [Google Scholar] [CrossRef]
- Teng, Z.; Lyu, J.; Chen, Y.; Zhang, J.; Ye, N. Effects of stress-induced ABA on root architecture development: Positive and negative actions. Crop J. 2023, 11, 1072–1079. [Google Scholar] [CrossRef]
- Alwutayd, K.; Rawat, A.; Sheikh, A.; Almeida-Trapp, M.; Veluchamy, A.; Jalal, R.; Karampelias, M.; Froehlich, K.; Alzaed, W.; Tabassum, N.; et al. Microbe-induced drought tolerance by ABA-mediated root architecture and epigenetic reprogramming. EMBO Rep. 2023, 24, e56754. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Fujita, Y.; Maruyama, K. Four Arabidopsis AREB/ABF transcription factors function predominantly in gene expression downstream of SnRK2 kinases in abscisic acid signalling in response to osmotic stress. Plant Cell Environ. 2015, 38, 35–49. [Google Scholar] [CrossRef] [PubMed]
- Gupta, K.; Wani, S.; Razzaq, A. Abscisic acid:role in fruit development and ripening. Front. Plant Sci. 2022, 13, 817500. [Google Scholar] [CrossRef] [PubMed]
- Zhan, W.; Wang, Y.; Duan, W.; Li, A.; Miao, Y.; Wang, H.; Meng, J.; Liu, H.; Niu, L.; Pan, L.; et al. Preliminary analysis, combined with omics of chilling injury mechanism of peach fruits with different cold sensitivities during postharvest cold storage. Horticulturae 2024, 10, 46. [Google Scholar] [CrossRef]
- Narusaka, Y.; Nakashima, K.; Shinwari, Z. Interaction between two cis-acting elements, ABRE and DRE, in ABA-dependent expression of Arabidopsis rd29A gene in response to dehydration and high-salinity stresses. Plant J. 2003, 34, 137–148. [Google Scholar] [CrossRef]
- Akihiro, M.; Junko, I.; Taeko, M. Arabidopsis transcriptome analysis under drought, cold, high-Salinity and ABA treatment conditions using a tiling array. Plant Cell Physiol. 2008, 8, 12–20. [Google Scholar]
- Sybilska, E.; Daszkowska-Golec, A. A complex signaling trio in seed germination: Auxin-JA-ABA. Trends Plant Sci. 2023, 8. [Google Scholar] [CrossRef]
- Oh, J.; Hong, S.; Lee, Y. Modulation of gene expressions and enzyme activities of methionine sulfoxide reductases by cold, ABA or high salt treatments in Arabidopsis. Plant Sci. 2005, 169, 1030–1036. [Google Scholar] [CrossRef]
- Jing, Y.; Lin, Z.; Jing, C. Effects of exogenous ABA on cold resistance and tender seedlings growth of winter wheat dongnongdongmai 1 in cold area. J. Triticeae Crops 2008, 28, 883–887. [Google Scholar]
- Yubero, M.; Enriqueta, M.; Medina, N. Identification of a strawberry gene encoding a non-specific lipid transfer protein that responds to ABA, wounding and cold stress. J. Exp. Bot. 2003, 389, 1865–1877. [Google Scholar] [CrossRef] [PubMed]
- Liang, J.; Li, J.; Liu, C.; Pan, W.Q.; Wu, W.J.; Shi, W.J.; Wang, L.J.; Yi, M.F.; Wu, J. GhbZIP30-GhCCCH17 module accelerates corm dormancy release by reducing endogenous ABA under cold storage in Gladiolus. Plant Cell Environ. 2023, 11, 14–30. [Google Scholar] [CrossRef] [PubMed]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef] [PubMed]
- Katiyar, A.; Smita, S.; Lenka, K.; Rajwanshi, R.; Chinnusamy, V.; Bansal, K.C. Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis. BMC Genom. 2012, 13, 544. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Chen, D.; Dai, Q.; Zhang, H.; Liu, Y.; Shen, W.; Zhu, B.; Cui, C.; Tan, C. Genome-wide identification of R2R3-MYB gene family and association with anthocyanin biosynthesis in Brassica species. BMC Genom. 2022, 23, 441. [Google Scholar] [CrossRef] [PubMed]
- An, P.; Wang, F.; Zhang, W.; Xu, H.F.; Bi, S.Q.; You, C.X.; Hao, Y.J. An apple MYB transcription factor regulates cold tolerance and anthocyanin accumulation and undergoes MIEL1-mediated degradation. Plant Biotechnol. J. 2020, 18, 337–353. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.; Yu, K.J.; Shi, Y.; Wang, J.; Duan, C.Q. Transcription factor VviMYB86 oppositely regulates proantho cyanidin and anthocyanin biosynthesis in grape berries. Front. Plant Sci. 2020, 11, 613677. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Li, J.; Hu, H.; Guo, J.; Liu, A.; Zhang, B. Acotton MYB transcription factor, GbMYB5, is positively involved in plant adaptive response to drought stress. Plant Cell Physiol. 2015, 56, 917–929. [Google Scholar] [CrossRef]
- Hong, Y.; Ahmad, N.; Zhang, J.; Lv, Y.; Zhang, X.; Ma, X.; Xiuming, L.; Na, Y. Genome-wide analysis and transcriptional reprogrammings of MYB superfamily revealed positive insights into abiotic stress responses and anthocyanin accumulation in Carthamus tinctorius L. Mol. Genet. Genom. 2022, 297, 125–145. [Google Scholar] [CrossRef]
- Masrur, R.; Allan, F.; Daiqing, H. Multiple roles of the transcription factor AtMYBR1/AtMYB44 in ABA signaling, stress responses, and leaf senescence. BMC Plant Biol. 2013, 13, 192. [Google Scholar]
- Medina, L.; Cumplido, G.; Amil, F. MYB10 plays a major role in the regulation of flavonoid/phenylpropanoid metabolism during ripening of Fragaria × ananassa fruits. J. Exp. Bot. 2014, 65, 401–417. [Google Scholar] [CrossRef]
- Ye, Y.; Wang, J.; Ni, Z.; Meng, X.; Xu, L. Small RNA and degradome sequencing reveal roles of miRNAs in strobilus development in masson pine (Pinus massoniana). Ind. Crop. Prod. 2020, 154, 112724. [Google Scholar] [CrossRef]
- Yang, K.; Liang, Z.; Wu, C. Analysis on differentially expressed genes in watermelon rind color based on RNA–Seq. J. Cent. South Univ. 2016, 23, 2818–2826. [Google Scholar] [CrossRef]
- Ye, Y.; Han, X.; Rong, H.; Qian, R.; Zheng, J.; Ni, Z.; Xu, L.A. Effect of the PmARF6 Gene from Masson Pine (Pinus massoniana) on the Development of Arabidopsis. Genes 2022, 13, 469. [Google Scholar] [CrossRef] [PubMed]
- Xu, Q.; Jian, H.; Zhao, W.; Li, J.; Zou, X.; Dong, X. Early weaning stress induces intestinal microbiota disturbance, mucosal barrier dysfunction and inflammation response activation in pigeon squabs. Front. Microbiol. 2022, 13, 866–877. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Zhu, J.; Zhu, J. Cold stress regulation of gene expression in plants. Trends Plant Sci. 2007, 12, 444–451. [Google Scholar] [CrossRef] [PubMed]
- Choudhury, F.; Rivero, R.; Blumwald, E.; Mittler, R. Reactive oxygen species, abiotic stress and stress combination. Plant. J. 2017, 90, 856–867. [Google Scholar] [CrossRef] [PubMed]
- Rabbani, M.; Maruyama, K.; Abe, H.; Khan, M.A.; Katsura, K.; Ito, Y.; Yoshiwara, K.; Seki, M.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Monitoring expression profiles of rice genes under cold, drought, and high-salinity stresses and abscisic acid application using cDNA microarray and RNA gel-blot analyses. Plant Physiol. 2003, 133, 175–185. [Google Scholar] [CrossRef]
- Gao, J.; Zhang, Y.; Xu, C.; Wang, X.; Wang, P.; Huang, S. Abscisic acid collaborates with lignin and flavonoid to improve pre-silking drought tolerance by tuning stem elongation and ear development in maize (Zea mays L.). Plant J. 2023, 12, 9–18. [Google Scholar] [CrossRef]
- Song, J.; Sajad, S.; Xia, D.; Jiang, S. Identification of F-box gene family in Brassica oleracea and expression analysis in response to low-temperature stress. Plant Physiol. Biochem. 2023, 199, 107717. [Google Scholar] [CrossRef]
- Ren, X.; Chen, Z.; Liu, Y.; Zhang, H.; Zhang, M.; Liu, Q.; Hong, X.; Zhu, J.K.; Gong, Z. ABO3, a WRKY transcription factor, mediates plant responses to abscisic acid and drought tolerance in Arabidopsis. Plant J. 2010, 63, 417–429. [Google Scholar] [CrossRef]
- Ruth, F. Abscisic Acid synthesis and response. Am. Soc. Plant Biol. 2013, 11, e0166. [Google Scholar]
- Iuchi, S.; Kobayashi, M.; Taji, T.; Naramoto, M.; Seki, M.; Kato, T.; Tabata, S.; Kakubari, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 2010, 27, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Cai, Z.; Li, R.; Zhao, Z.; Chen, Y.; Lu, C. Association between dormancy release and phenolic metabolic alteration induced by low temperature in Cardiocrinumgiganteum bulbs. J. Trop. Subtrop. Bot. 2024, 32, 209–218. [Google Scholar]
- Zhang, Y.; Liang, W.; Shi, J.; Xu, J.; Zhang, D. MYB56 encoding a R2R3 MYB transcription factor regulates seed size in Arabidopsis thaliana. J. Integr. Plant Biol. 2013, 55, 1166–1178. [Google Scholar] [CrossRef] [PubMed]
- Abe, H. Arabidopsis AtMYC2 (bHLH) and AtMYB2 (MYB) Function as transcriptional activators in abscisic acid signaling. Plant Cell 2003, 15, 63–78. [Google Scholar] [CrossRef] [PubMed]
- Xie, X.; Li, S.; Zhang, R.; Zhao, J.; Chen, Y.C.; Zhao, Q.; Yao, Y.X.; You, C.X.; Zhang, X.S.; Hao, Y.J. The bHLH transcription factor MdbHLH3 promotes anthocyanin accumulation and fruit colouration in response to low temperature in apples. Plant Cell Environ. 2012, 35, 1884–1897. [Google Scholar] [CrossRef]
- Yang, Y.; Jiang, M.; Feng, J.; Wu, C.; Shan, W.; Kuang, J.; Chen, J.; Hu, Z.; Lu, W. Transcriptome analysis of low-temperature-affected ripening revealed MYB transcription factors-mediated regulatory network in banana fruit. Food Res. Int. 2021, 1, 148. [Google Scholar] [CrossRef]
- Fujita, Y.; Fujita, M.; Shinozaki, K. ABA-mediated transcriptional regulation in response to osmotic stress in plants. J. Plant Res. 2011, 124, 509–525. [Google Scholar] [CrossRef]
- Seo, P.J.; Xiang, F.; Qiao, M.; Park, J.Y.; Lee, Y.N.; Kim, S.G.; Lee, Y.H.; Park, W.J.; Park, C.M. The MYB96 transcription factor mediates abscisic acid signaling during drought stress response in Arabidopsis. Plant Physiol. 2009, 151, 275–289. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Zhou, H.; Xiong, C.; Peng, Z.; Du, W.; Li, H.; Wang, L.; Ruan, C. Genome-wide analysis R2R3-MYB transcription factors in Xanthoceras sorbifolium Bunge and functional analysis of XsMYB30 in drought and salt stresses tolerance. Ind. Crops Prod. 2022, 17, 178–185. [Google Scholar] [CrossRef]

| Sample | ReadSum | BaseSum | GC (%) | N (%) | Q20 (%) | Q30 (%) |
|---|---|---|---|---|---|---|
| FM1 | 23,817,457 | 7,145,237,100 | 45.35 | 0 | 98.25 | 94.73 |
| FM2 | 19,199,653 | 5,759,895,900 | 46.01 | 0 | 98.24 | 94.74 |
| FM3 | 20,242,141 | 6,072,642,300 | 45.31 | 0 | 98.1 | 94.4 |
| GM1 | 22,487,796 | 6,746,338,800 | 45.89 | 0 | 98.26 | 94.66 |
| GM2 | 22,890,147 | 6,867,044,100 | 46.61 | 0 | 98.4 | 95.16 |
| GM3 | 24,023,866 | 7,207,159,800 | 45.76 | 0 | 98.28 | 94.8 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ye, Y.; Wen, S.; Ying, J.; Cai, Y.; Qian, R. Screening and Preliminary Identification of Asparagus officinalis Varieties under Low-Temperature Stress. Genes 2024, 15, 486. https://doi.org/10.3390/genes15040486
Ye Y, Wen S, Ying J, Cai Y, Qian R. Screening and Preliminary Identification of Asparagus officinalis Varieties under Low-Temperature Stress. Genes. 2024; 15(4):486. https://doi.org/10.3390/genes15040486
Chicago/Turabian StyleYe, Youju, Shuangshuang Wen, Jiali Ying, Yunfei Cai, and Renjuan Qian. 2024. "Screening and Preliminary Identification of Asparagus officinalis Varieties under Low-Temperature Stress" Genes 15, no. 4: 486. https://doi.org/10.3390/genes15040486






