Contradiction in Star-Allele Nomenclature of Pharmacogenes between Common Haplotypes and Rare Variants
Abstract
:1. Introduction
2. Materials and Methods
2.1. The 1000 Genomes Project
2.2. Functional Variant Determination
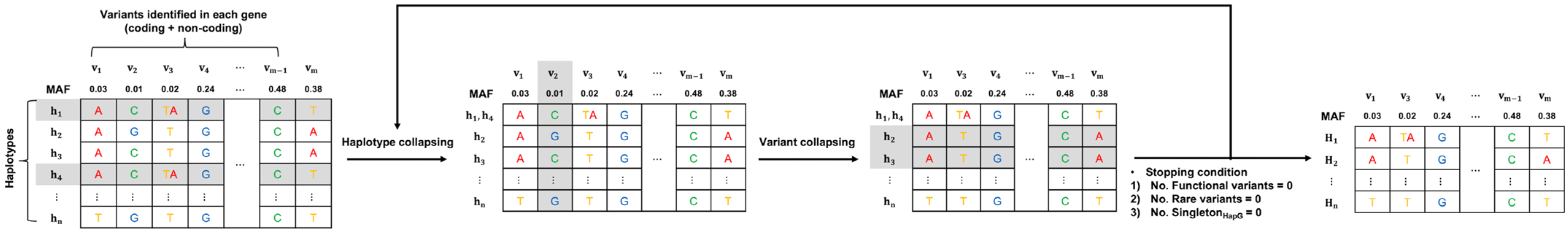
2.3. Constructing Haplogroups
2.4. Assignment of Star Alleles
2.5. Evaluation
2.6. Enrichment Analysis
2.7. Genomic Features of Star Alleles
3. Results
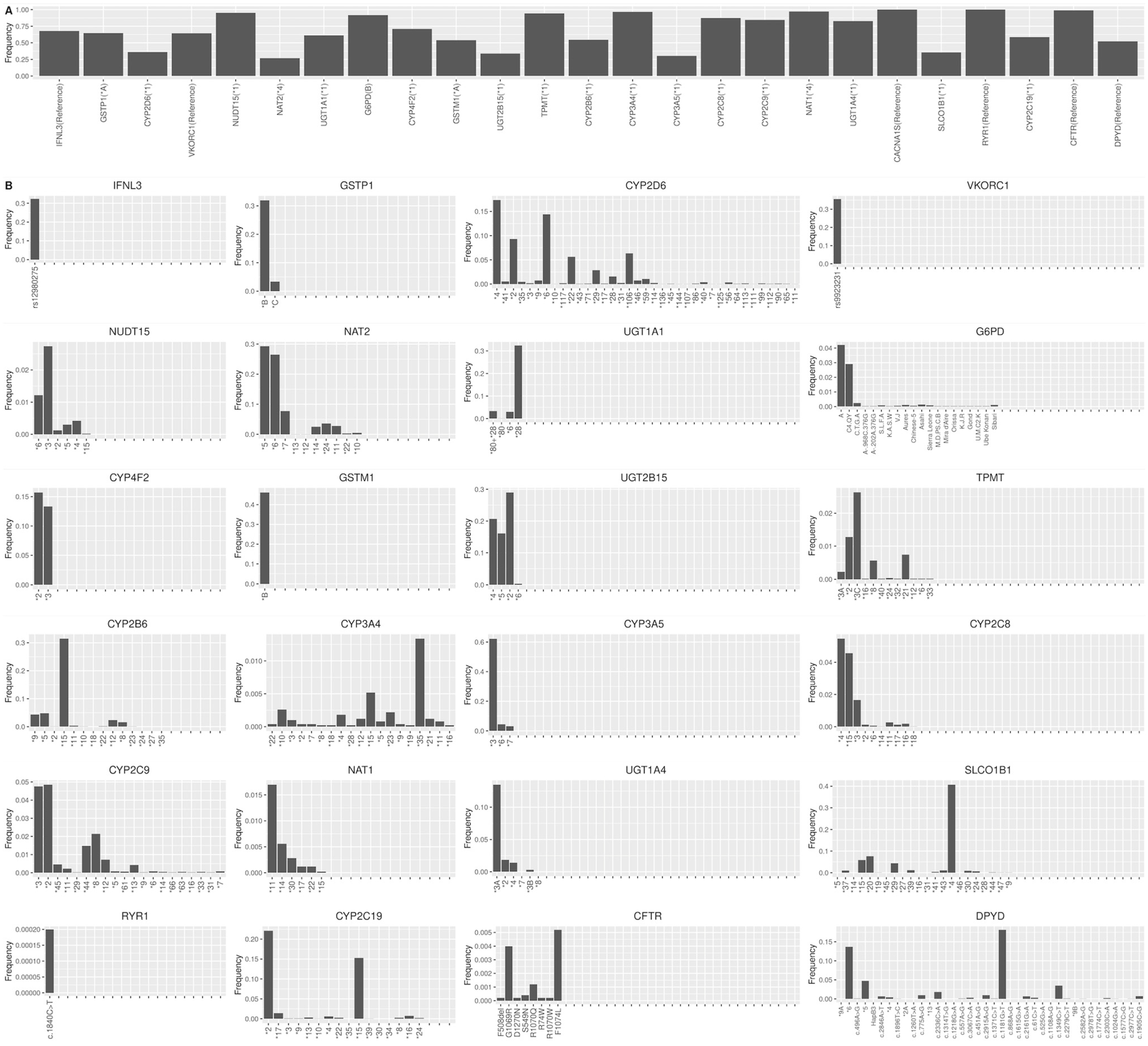
3.1. Haplogroup Construction
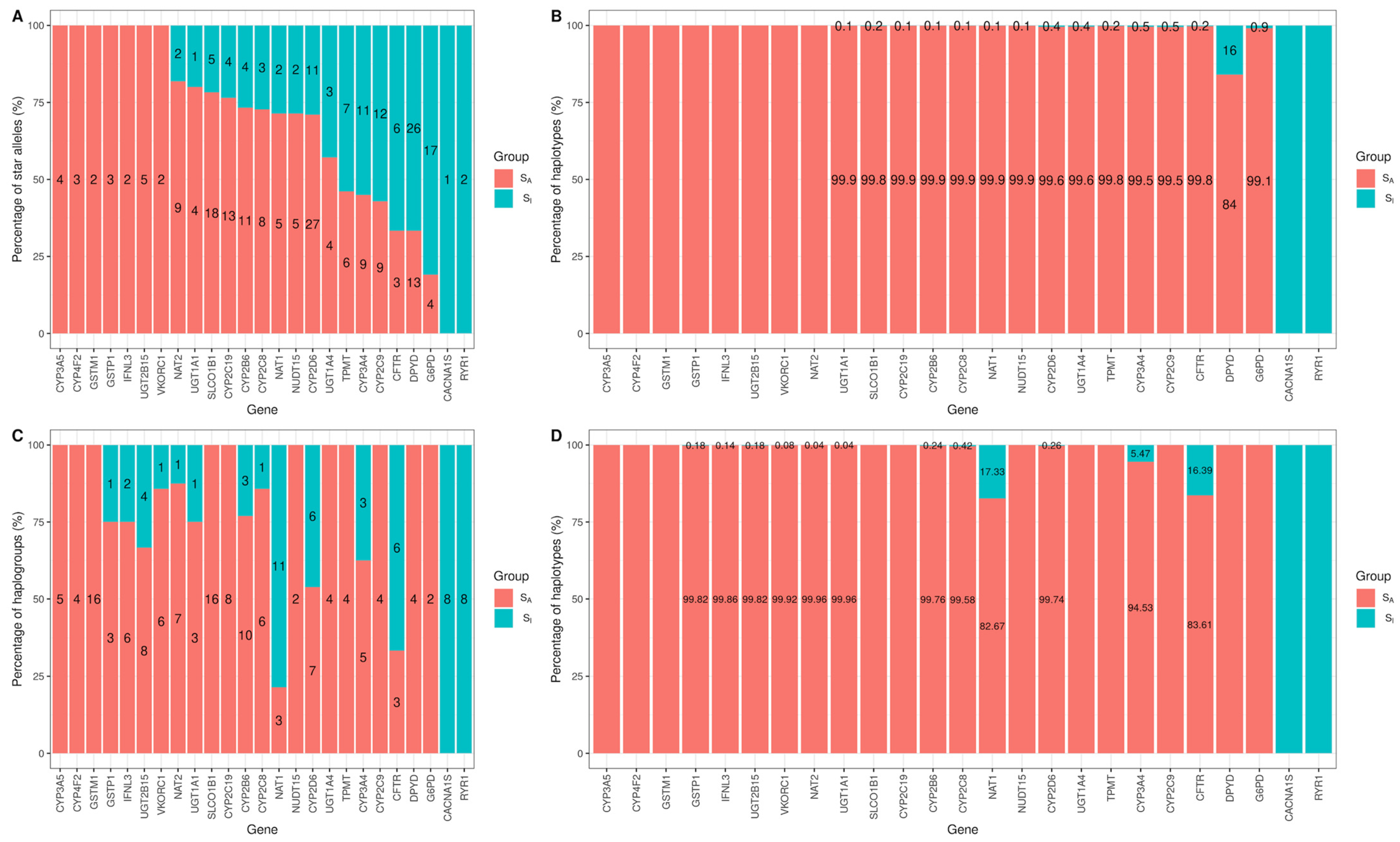
3.2. Evaluate Haplogroup Construction
3.3. Genomic Characterization of Star Alleles by Haplogroups
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Evans, W.E.; Johnson, J.A. Pharmacogenomics: The inherited basis for interindividual differences in drug response. Annu. Rev. Genom. Hum. Genet. 2001, 2, 9–39. [Google Scholar] [CrossRef] [PubMed]
- Schwab, M.; Schaeffeler, E. Pharmacogenomics: A key component of personalized therapy. Genome Med. 2012, 4, 93. [Google Scholar] [CrossRef] [PubMed]
- Cacabelos, R.; Cacabelos, N.; Carril, J.C. The role of pharmacogenomics in adverse drug reactions. Expert Rev. Clin. Pharmacol. 2019, 12, 407–442. [Google Scholar] [CrossRef] [PubMed]
- Schwarz, U.I.; Gulilat, M.; Kim, R.B. The Role of Next-Generation Sequencing in Pharmacogenetics and Pharmacogenomics. Cold Spring Harb. Perspect. Med. 2019, 9, a033027. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Tremmel, R.; Schaeffeler, E.; Schwab, M.; Lauschke, V.M. Challenges and opportunities associated with rare-variant pharmacogenomics. Trends Pharmacol. Sci. 2022, 43, 852–865. [Google Scholar] [CrossRef] [PubMed]
- Robarge, J.D.; Li, L.; Desta, Z.; Nguyen, A.; Flockhart, D.A. The star-allele nomenclature: Retooling for translational genomics. Clin. Pharmacol. Ther. 2007, 82, 244–248. [Google Scholar] [CrossRef] [PubMed]
- Nebert, D.W. Suggestions for the nomenclature of human alleles: Relevance to ecogenetics, pharmacogenetics and molecular epidemiology. Pharmacogenetics 2000, 10, 279–290. [Google Scholar] [CrossRef] [PubMed]
- Twesigomwe, D.; Wright, G.E.B.; Drogemoller, B.I.; da Rocha, J.; Lombard, Z.; Hazelhurst, S. A systematic comparison of pharmacogene star allele calling bioinformatics algorithms: A focus on CYP2D6 genotyping. NPJ Genom. Med. 2020, 5, 30. [Google Scholar] [CrossRef]
- Relling, M.V.; Klein, T.E. CPIC: Clinical Pharmacogenetics Implementation Consortium of the Pharmacogenomics Research Network. Clin. Pharmacol. Ther. 2011, 89, 464–467. [Google Scholar] [CrossRef]
- Swen, J.J.; Wilting, I.; de Goede, A.L.; Grandia, L.; Mulder, H.; Touw, D.J.; de Boer, A.; Conemans, J.M.; Egberts, T.C.; Klungel, O.H.; et al. Pharmacogenetics: From bench to byte. Clin. Pharmacol. Ther. 2008, 83, 781–787. [Google Scholar] [CrossRef]
- Lyon, E.; Gastier Foster, J.; Palomaki, G.E.; Pratt, V.M.; Reynolds, K.; Sabato, M.F.; Scott, S.A.; Vitazka, P.; working group of the Molecular Genetics Subcommittee on behalf of the American College of Medical Genetics and Genomics ACMG) Laboratory Quality Assurance Committee. Laboratory testing of CYP2D6 alleles in relation to tamoxifen therapy. Genet. Med. 2012, 14, 990–1000. [Google Scholar] [CrossRef] [PubMed]
- Abdullah-Koolmees, H.; van Keulen, A.M.; Nijenhuis, M.; Deneer, V.H.M. Pharmacogenetics Guidelines: Overview and Comparison of the DPWG, CPIC, CPNDS, and RNPGx Guidelines. Front. Pharmacol. 2020, 11, 595219. [Google Scholar] [CrossRef] [PubMed]
- Pratt, V.M.; Cavallari, L.H.; Del Tredici, A.L.; Gaedigk, A.; Hachad, H.; Ji, Y.; Kalman, L.V.; Ly, R.C.; Moyer, A.M.; Scott, S.A.; et al. Recommendations for Clinical CYP2D6 Genotyping Allele Selection: A Joint Consensus Recommendation of the Association for Molecular Pathology, College of American Pathologists, Dutch Pharmacogenetics Working Group of the Royal Dutch Pharmacists Association, and the European Society for Pharmacogenomics and Personalized Therapy. J. Mol. Diagn. 2021, 23, 1047–1064. [Google Scholar] [CrossRef] [PubMed]
- Lasorsa, V.A.; Montella, A.; Cantalupo, S.; Tirelli, M.; de Torres, C.; Aveic, S.; Tonini, G.P.; Iolascon, A.; Capasso, M. Somatic Mutations Enriched in Cis-Regulatory Elements Affect Genes Involved in Embryonic Development and Immune System Response in Neuroblastoma. Cancer Res. 2022, 82, 1193–1207. [Google Scholar] [CrossRef] [PubMed]
- Balasubramanian, S.; Fu, Y.; Pawashe, M.; McGillivray, P.; Jin, M.; Liu, J.; Karczewski, K.J.; MacArthur, D.G.; Gerstein, M. Using ALoFT to determine the impact of putative loss-of-function variants in protein-coding genes. Nat. Commun. 2017, 8, 382. [Google Scholar] [CrossRef] [PubMed]
- Genomes Project, C.; Auton, A.; Brooks, L.D.; Durbin, R.M.; Garrison, E.P.; Kang, H.M.; Korbel, J.O.; Marchini, J.L.; McCarthy, S.; McVean, G.A.; et al. A global reference for human genetic variation. Nature 2015, 526, 68–74. [Google Scholar] [CrossRef]
- Fairley, S.; Lowy-Gallego, E.; Perry, E.; Flicek, P. The International Genome Sample Resource (IGSR) collection of open human genomic variation resources. Nucleic Acids Res. 2020, 48, D941–D947. [Google Scholar] [CrossRef] [PubMed]
- Mauleekoonphairoj, J.; Chamnanphon, M.; Khongphatthanayothin, A.; Sutjaporn, B.; Wandee, P.; Poovorawan, Y.; Nademanee, K.; Pongpanich, M.; Chariyavilaskul, P. Phenotype prediction and characterization of 25 pharmacogenes in Thais from whole genome sequencing for clinical implementation. Sci. Rep. 2020, 10, 18969. [Google Scholar] [CrossRef]
- McLaren, W.; Gil, L.; Hunt, S.E.; Riat, H.S.; Ritchie, G.R.; Thormann, A.; Flicek, P.; Cunningham, F. The Ensembl Variant Effect Predictor. Genome Biol. 2016, 17, 122. [Google Scholar] [CrossRef]
- Kircher, M.; Witten, D.M.; Jain, P.; O’Roak, B.J.; Cooper, G.M.; Shendure, J. A general framework for estimating the relative pathogenicity of human genetic variants. Nat. Genet. 2014, 46, 310–315. [Google Scholar] [CrossRef]
- Lee, S.B.; Shin, J.Y.; Kwon, N.J.; Kim, C.; Seo, J.S. ClinPharmSeq: A targeted sequencing panel for clinical pharmacogenetics implementation. PLoS ONE 2022, 17, e0272129. [Google Scholar] [CrossRef]
- Meilă, M. Comparing Clusterings by the Variation of Information. In Learning Theory and Kernel Machines. Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2003; pp. 173–187. [Google Scholar]
- Scrucca, L.; Fop, M.; Murphy, T.B.; Raftery, A.E. mclust 5: Clustering, Classification and Density Estimation Using Gaussian Finite Mixture Models. R J. 2016, 8, 289–317. [Google Scholar] [CrossRef]
- Takezaki, N.; Nei, M. Genetic distances and reconstruction of phylogenetic trees from microsatellite DNA. Genetics 1996, 144, 389–399. [Google Scholar] [CrossRef]
- Davydov, E.V.; Goode, D.L.; Sirota, M.; Cooper, G.M.; Sidow, A.; Batzoglou, S. Identifying a high fraction of the human genome to be under selective constraint using GERP++. PLoS Comput. Biol. 2010, 6, e1001025. [Google Scholar] [CrossRef]
- Ng, P.C.; Henikoff, S. SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 2003, 31, 3812–3814. [Google Scholar] [CrossRef]
- Adzhubei, I.A.; Schmidt, S.; Peshkin, L.; Ramensky, V.E.; Gerasimova, A.; Bork, P.; Kondrashov, A.S.; Sunyaev, S.R. A method and server for predicting damaging missense mutations. Nat. Methods 2010, 7, 248–249. [Google Scholar] [CrossRef]
- Lopes, I.; Altab, G.; Raina, P.; de Magalhaes, J.P. Gene Size Matters: An Analysis of Gene Length in the Human Genome. Front. Genet. 2021, 12, 559998. [Google Scholar] [CrossRef]
- Shin, S.-H.; Choi, S.S. Lengths of coding and noncoding regions of a gene correlate with gene essentiality and rates of evolution. Genes Genom. 2015, 37, 365–374. [Google Scholar] [CrossRef]
- Maranville, J.C.; Cox, N.J. Pharmacogenomic variants have larger effect sizes than genetic variants associated with other dichotomous complex traits. Pharmacogenom. J. 2016, 16, 388–392. [Google Scholar] [CrossRef]
- Whirl-Carrillo, M.; Huddart, R.; Gong, L.; Sangkuhl, K.; Thorn, C.F.; Whaley, R.; Klein, T.E. An Evidence-Based Framework for Evaluating Pharmacogenomics Knowledge for Personalized Medicine. Clin. Pharmacol. Ther. 2021, 110, 563–572. [Google Scholar] [CrossRef]
- Gaedigk, A.; Ingelman-Sundberg, M.; Miller, N.A.; Leeder, J.S.; Whirl-Carrillo, M.; Klein, T.E.; PharmVar Steering, C. The Pharmacogene Variation (PharmVar) Consortium: Incorporation of the Human Cytochrome P450 (CYP) Allele Nomenclature Database. Clin. Pharmacol. Ther. 2018, 103, 399–401. [Google Scholar] [CrossRef]
- Rogers, M.F.; Shihab, H.A.; Mort, M.; Cooper, D.N.; Gaunt, T.R.; Campbell, C. FATHMM-XF: Accurate prediction of pathogenic point mutations via extended features. Bioinformatics 2018, 34, 511–513. [Google Scholar] [CrossRef]



| Gene | Gene Length a | Haplogroups (Variant) | Observed Star Alleles b (Variant) | TotalStar Alleles c (Variant) | VI |
|---|---|---|---|---|---|
| IFNL3 | 1.40 | 8 (4) | 2 (1) | 4 (3) | 0.98 |
| GSTP1 | 3.06 | 4 (2) | 3 (2) | 4 (2) | 1.37 |
| CYP2D6 | 4.42 | 13 (4) | 38 (37) | 131 (125) | 1.76 |
| VKORC1 | 5.14 | 7 (5) | 2 (1) | 2 (1) | 1.04 |
| NUDT15 | 9.66 | 2 (1) | 7 (5) | 20 (18) | 1.23 |
| NAT2 | 9.97 | 8 (3) | 11 (10) | 18 (17) | 0.96 |
| UGT1A1 | 13.05 | 4 (2) | 5 (3) | 9 (6) | 1.36 |
| G6PD | 16.18 | 2 (1) | 21 (19) | 186 (182) | 1.38 |
| CYP4F2 | 20.10 | 4 (2) | 3 (2) | 4 (2) | 2.00 |
| GSTM1 | 21.23 | 16 (4) | 2 (1) | 3 (1) | 3.24 |
| UGT2B15 | 24.00 | 12 (4) | 5 (3) | 11 (4) | 1.35 |
| TPMT | 26.76 | 4 (2) | 13 (12) | 46 (45) | 1.47 |
| CYP2B6 | 27.10 | 13 (4) | 15 (7) | 37 (35) | 2.10 |
| CYP3A4 | 27.29 | 8 (3) | 20 (19) | 33 (32) | 2.28 |
| CYP3A5 | 31.81 | 5 (3) | 4 (3) | 9 (8) | 0.88 |
| CYP2C8 | 32.73 | 7 (3) | 11 (11) | 18 (18) | 2.40 |
| CYP2C9 | 50.73 | 4 (2) | 21 (20) | 71 (69) | 1.57 |
| NAT1 | 53.21 | 14 (5) | 7 (6) | 11 (10) | 2.27 |
| UGT1A4 | 54.52 | 4 (2) | 7 (10) | 12 (12) | 1.98 |
| CACNA1S | 73.05 | 8 (3) | 1 (0) | 3 (2) | 2.37 |
| SLCO1B1 | 108.05 | 16 (4) | 23 (16) | 44 (31) | 3.06 |
| RYR1 | 153.87 | 8 (3) | 2 (1) | 49 (48) | 1.27 |
| CYP2C19 | 165.11 | 8 (3) | 17 (17) | 36 (33) | 2.15 |
| CFTR | 250.19 | 9 (4) | 9 (8) | 41 (40) | 2.05 |
| DPYD | 843.31 | 4 (2) | 39 (39) | 83 (83) | 4.08 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ahn, S.H.; Park, Y.; Kim, J.H. Contradiction in Star-Allele Nomenclature of Pharmacogenes between Common Haplotypes and Rare Variants. Genes 2024, 15, 521. https://doi.org/10.3390/genes15040521
Ahn SH, Park Y, Kim JH. Contradiction in Star-Allele Nomenclature of Pharmacogenes between Common Haplotypes and Rare Variants. Genes. 2024; 15(4):521. https://doi.org/10.3390/genes15040521
Chicago/Turabian StyleAhn, Se Hwan, Yoomi Park, and Ju Han Kim. 2024. "Contradiction in Star-Allele Nomenclature of Pharmacogenes between Common Haplotypes and Rare Variants" Genes 15, no. 4: 521. https://doi.org/10.3390/genes15040521




