Host-Derived Artificial MicroRNA as an Alternative Method to Improve Soybean Resistance to Soybean Cyst Nematode
Abstract
:1. Introduction
2. Materials and Methods
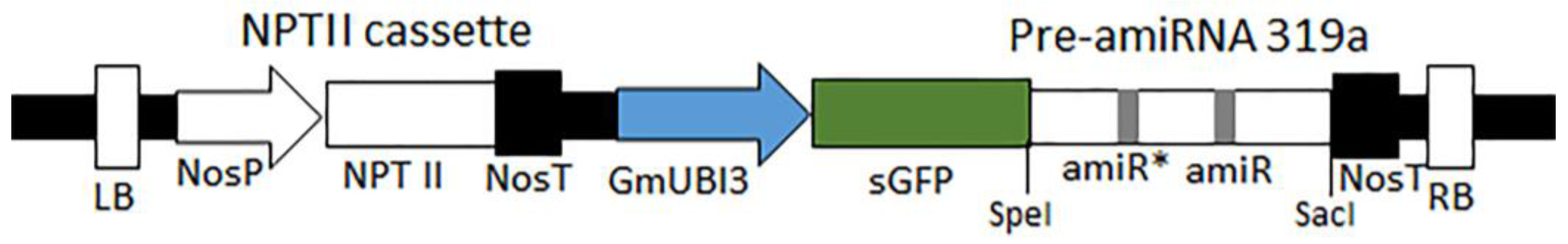
2.1. Plasmids, Vector Design, and Construction
2.2. Hairy Root Transformation of Soybean
2.3. RNA Isolation
2.4. RT-qPCR Analysis
2.5. SCN Bioassay
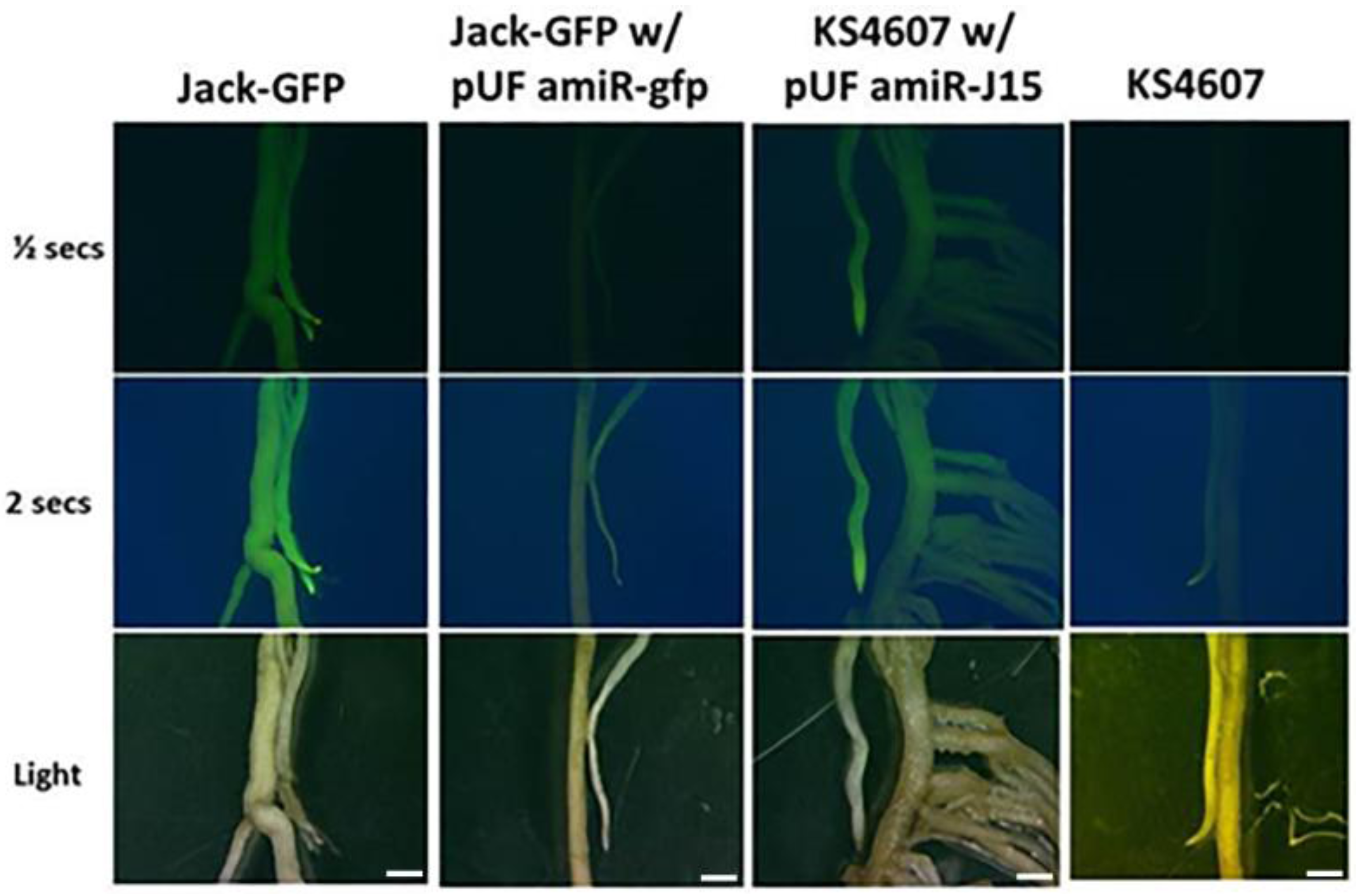
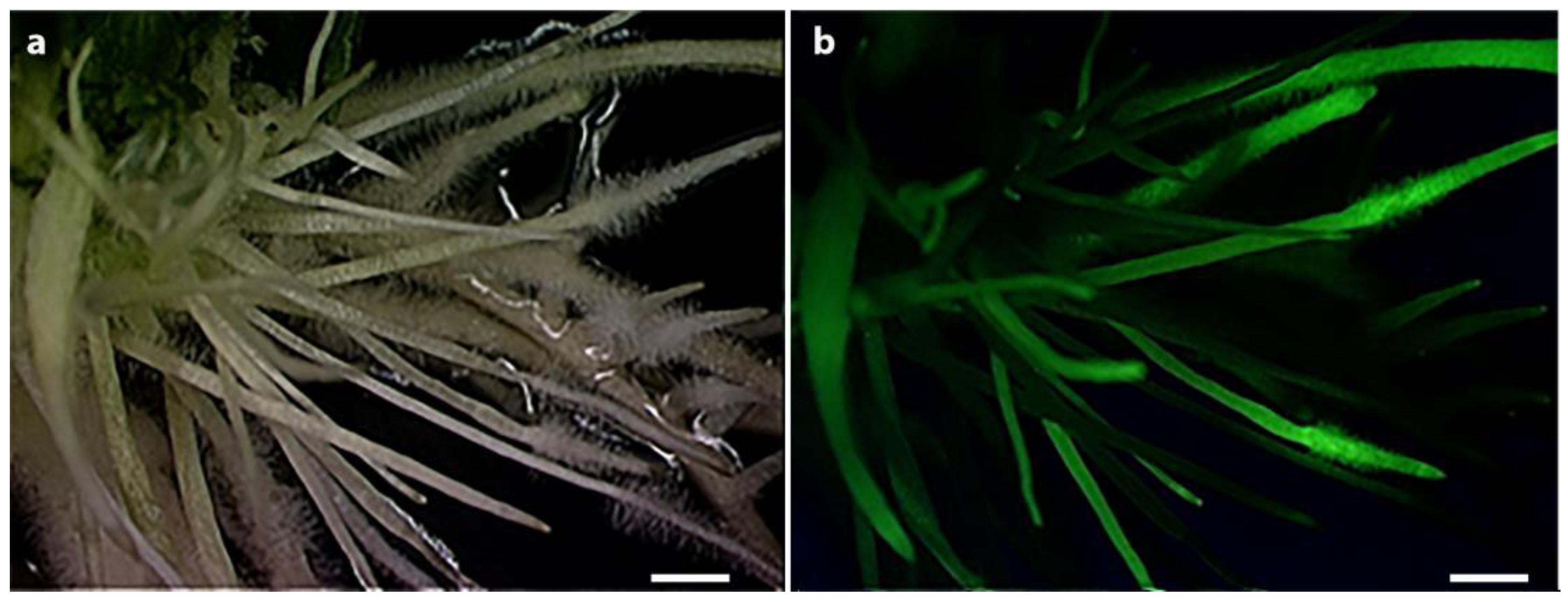
2.6. GFP Imaging
3. Results and Discussion
3.1. Identification of miRNA Targets
3.2. Construction of an amiRNA Gene Silencing Vector
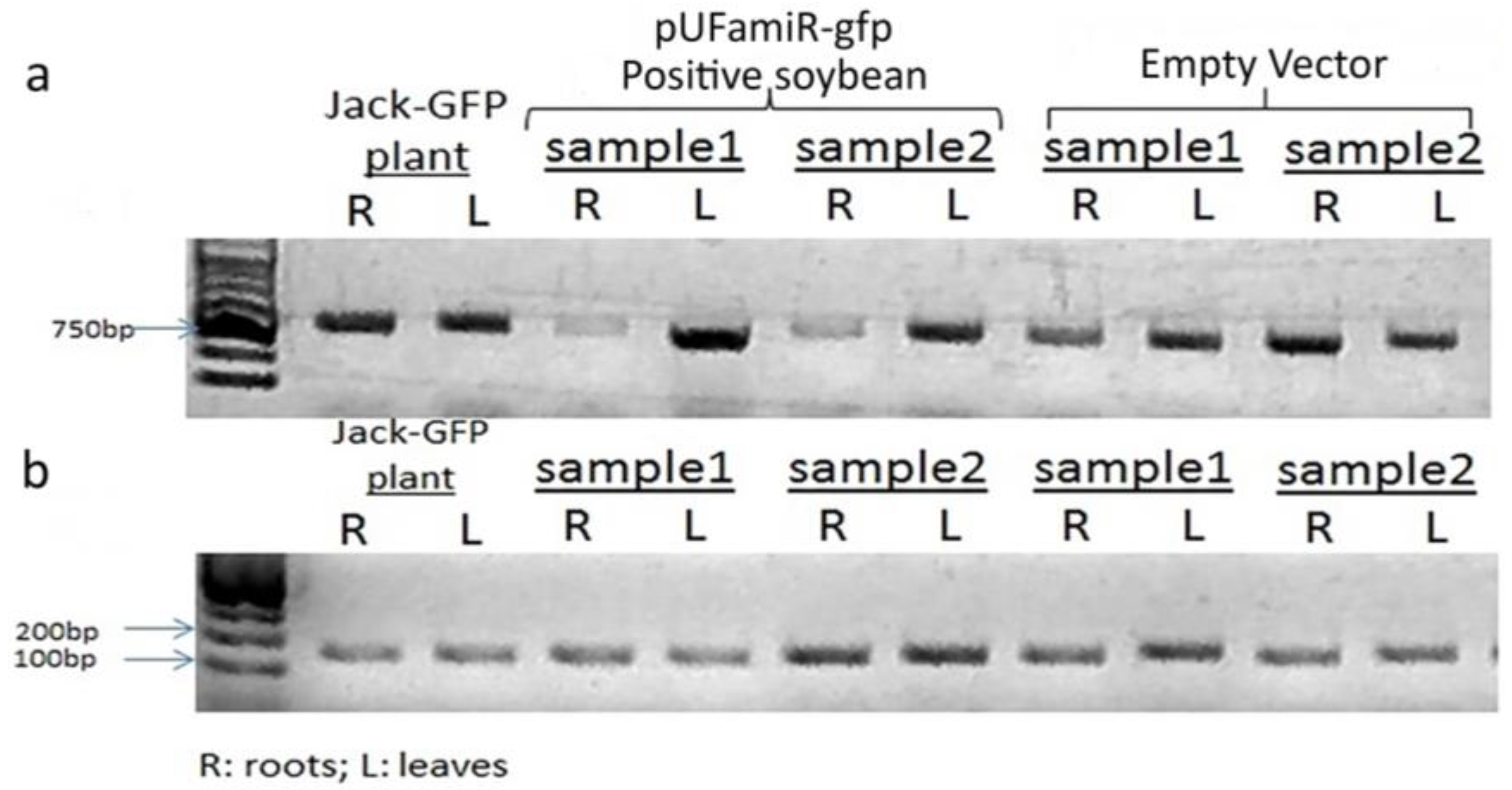
3.3. Confirmation of the Functioinality of the pUFamiR Vector
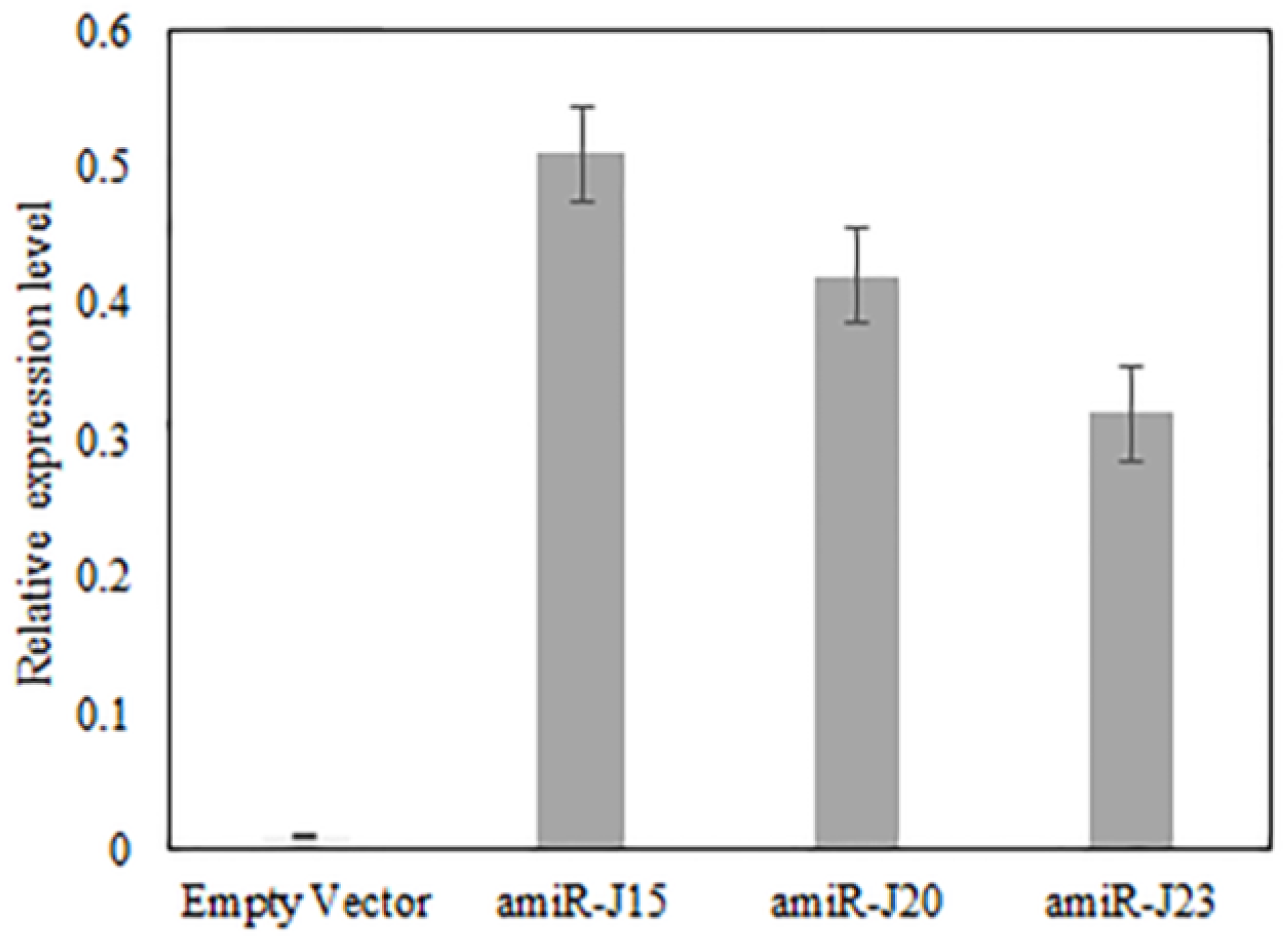
3.4. Expression of amiR Constructs Targeting Nematode Genes in Soybean Hairy Roots
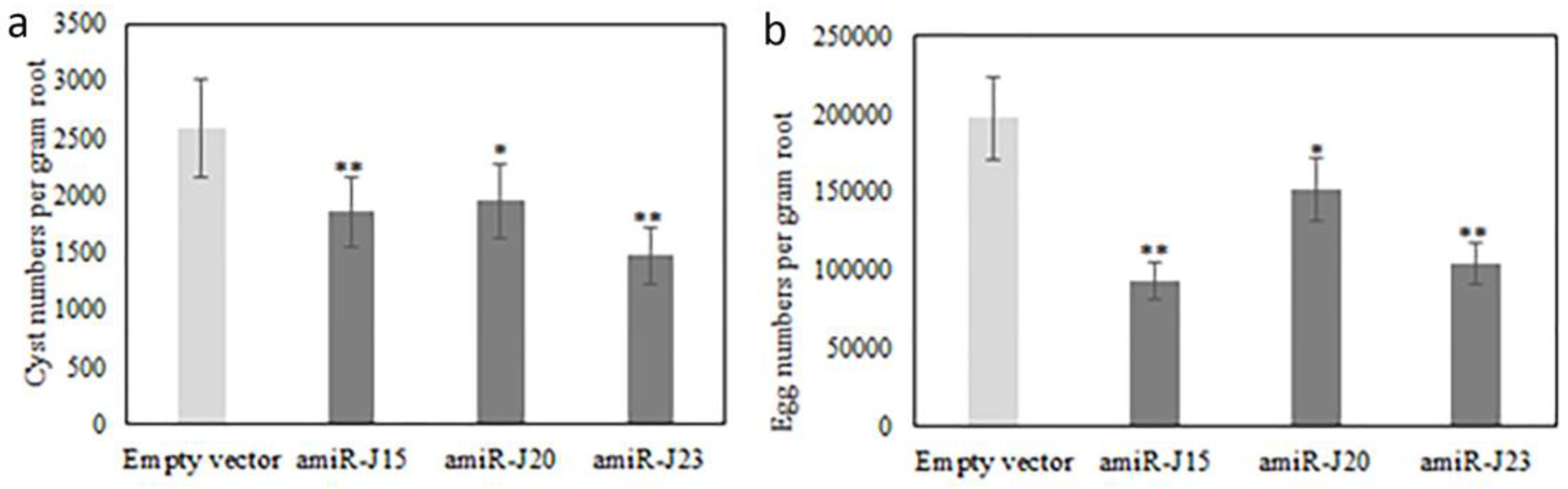
3.5. Bioassay of SCN Production on Transgenic Hairy Roots Expressing pUFamiR Constructs Targeting Nematodes Genes
3.6. Downregulation of Candidate Genes in Nematodes Feeding on Composite Hairy Roots Expressing the pUFamiR Constructs
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Koenning, S.R.; Wrather, J.A. Suppression of soybean yield potential in the continental united states from plant diseases estimated from 2006 to 2009. Plant Health Prog. 2010. [Google Scholar] [CrossRef]
- Davis, E.L.; Tylka, G.L. Soybean cyst nematode disease. Plant Health Instr. 2000. [Google Scholar] [CrossRef]
- Dong, K.; Barker, K.R.; Opperman, C.H. Genetics of soybean-Heterodera glycines interactions. J. Nematol. 1997, 29, 509–522. [Google Scholar] [PubMed]
- Klink, V.P.; Matthews, B.F. Emerging approaches to broaden resistance of soybean to soybean cyst nematode as supported by gene expression studies. Plant Physiol. 2009, 151, 1017–1022. [Google Scholar] [CrossRef] [PubMed]
- Agrawal, N.; Dasaradhi, P.V.N.; Mohmmed, A.; Malhotra, P.; Bhatnagar, R.K.; Mukherjee, S.K. RNA interference: Biology, mechanism, and applications. Microbiol. Mol. Biol. Rev. 2003, 67, 657–685. [Google Scholar] [CrossRef] [PubMed]
- Moazed, D. Small RNAs in transcriptional gene silencing and genome defence. Nature 2009, 457, 413–420. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, M.; Sharma, D.; Trivedi, P.K. Artificial microRNA mediated gene silencing in plants: Progress and perspectives. Plant Mol. Biol. 2014, 86, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Yogindran, S.; Rajam, M.V. RNAi for Crop Improvement. In Plant Biology and Biotechnology: Volume II: Plant Genomics and Biotechnology; Bahadur, B., Venkat Rajam, M., Sahijram, L., Krishnamurthy, K.V., Eds.; Springer India: New Delhi, India, 2015; pp. 623–637. [Google Scholar]
- Eamens, A.; Curtin, S.J.; Waterhouse, P.M. RNA Silencing in Plants. In Plant Developmental Biology—Biotechnological Perspectives: Volume 2; Pua, E.C., Davey, M.R., Eds.; Springer: Berlin/Heidelberg, Germany, 2010; pp. 277–294. [Google Scholar]
- Zhang, J.; Khan, S.A.; Hasse, C.; Ruf, S.; Heckel, D.G.; Bock, R. Full crop protection from an insect pest by expression of long double-stranded RNAs in plastids. Science 2015, 347, 991–994. [Google Scholar] [CrossRef] [PubMed]
- Govindarajulu, M.; Epstein, L.; Wroblewski, T.; Michelmore, R.W. Host-induced gene silencing inhibits the biotrophic pathogen causing downy mildew of lettuce. Plant Biotechnol. J. 2015, 13, 875–883. [Google Scholar] [CrossRef] [PubMed]
- Cheng, W.; Song, X.-S.; Li, H.-P.; Cao, L.-H.; Sun, K.; Qiu, X.-L.; Xu, Y.-B.; Yang, P.; Huang, T.; Zhang, J.-B.; et al. Host-induced gene silencing of an essential chitin synthase gene confers durable resistance to Fusarium head blight and seedling blight in wheat. Plant Biotechnol. J. 2015, 13, 1335–1345. [Google Scholar] [CrossRef] [PubMed]
- Nandety, R.S.; Kuo, Y.-W.; Nouri, S.; Falk, B.W. Emerging strategies for rna interference (rnai) applications in insects. Bioengineered 2015, 6, 8–19. [Google Scholar] [CrossRef] [PubMed]
- Voinnet, O. Origin, biogenesis, and activity of plant microRNAs. Cell 2009, 136, 669–687. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Todd, T.C.; Lee, J.; Trick, H.N. Biotechnological application of functional genomics towards plant-parasitic nematode control. Plant Biotechnol. J. 2011, 9, 936–944. [Google Scholar] [CrossRef] [PubMed]
- Devers, E.A.; Teply, J.; Reinert, A.; Gaude, N.; Krajinski, F. An endogenous artificial microRNA system for unraveling the function of root endosymbioses related genes in medicago truncatula. BMC Plant Biol. 2013, 13, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Warthmann, N.; Chen, H.; Ossowski, S.; Weigel, D.; Hervé, P. Highly specific gene silencing by artificial miRNAs in rice. PLoS ONE 2008, 3, e1829. [Google Scholar] [CrossRef] [PubMed]
- Molesini, B.; Pii, Y.; Pandolfini, T. Fruit improvement using intragenesis and artificial microRNA. Trends Biotechnol. 2012, 30, 80–88. [Google Scholar] [CrossRef] [PubMed]
- Niu, Q.-W.; Lin, S.-S.; Reyes, J.L.; Chen, K.-C.; Wu, H.-W.; Yeh, S.-D.; Chua, N.-H. Expression of artificial microRNAs in transgenic Arabidopsis thaliana confers virus resistance. Nat. Biotechnol. 2006, 24, 1420–1428. [Google Scholar] [CrossRef] [PubMed]
- Ai, T.; Zhang, L.; Gao, Z.; Zhu, C.X.; Guo, X. Highly efficient virus resistance mediated by artificial microRNAs that target the suppressor of PVX and PVY in plants. Plant Biol. 2011, 13, 304–316. [Google Scholar] [CrossRef] [PubMed]
- Duan, C.-G.; Wang, C.-H.; Fang, R.-X.; Guo, H.-S. Artificial microRNAs highly accessible to targets confer efficient virus resistance in plants. J. Virol. 2008, 82, 11084–11095. [Google Scholar] [CrossRef] [PubMed]
- Qu, J.; Ye, J.; Fang, R. Artificial microRNA-mediated virus resistance in plants. J. Virol. 2007, 81, 6690–6699. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.-Z.; Han, Q.-J.; Jiang, F.; Sun, R.-Z.; Fan, Z.-H.; Zhu, C.-X.; Wen, F.-J. Effects of the sequence characteristics of miRNAs on multi-viral resistance mediated by single amiRNAs in transgenic tobacco. Plant Physiol. Biochem. 2014, 77, 90–98. [Google Scholar] [CrossRef] [PubMed]
- Mitter, N.; Zhai, Y.; Bai, A.X.; Chua, K.; Eid, S.; Constantin, M.; Mitchell, R.; Pappu, H.R. Evaluation and identification of candidate genes for artificial microRNA-mediated resistance to tomato spotted wilt virus. Virus Res. 2016, 211, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Song, X.; Wang, G.; Yang, K.; Wang, Y.; Niu, L.; Chen, X.; Fang, R. Plant-generated artificial small RNAs mediated aphid resistance. PLoS ONE 2014, 9, e97410. [Google Scholar] [CrossRef] [PubMed]
- Chiera, J.M.; Bouchard, R.A.; Dorsey, S.L.; Park, E.; Buenrostro-Nava, M.T.; Ling, P.P.; Finer, J.J. Isolation of two highly active soybean (Glycine max (L.) merr.) promoters and their characterization using a new automated image collection and analysis system. Plant Cell Rep. 2007, 26, 1501–1509. [Google Scholar] [CrossRef] [PubMed]
- Schwab, R.; Ossowski, S.; Riester, M.; Wart hmann, N.; Weigel, D. Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 2006, 18, 1121–1133. [Google Scholar] [CrossRef] [PubMed]
- pRS300 plasmid. Available online: https://www.Addgene.Org/22846/ (accessed on 5 December 2016).
- Li, J.; Todd, T.C.; Trick, H.N. Rapid in planta evaluation of root expressed transgenes in chimeric soybean plants. Plant Cell Rep. 2009, 29, 113–123. [Google Scholar] [CrossRef] [PubMed]
- Paolacci, A.R.; Tanzarella, O.A.; Porceddu, E.; Ciaffi, M. Identification and validation of reference genes for quantitative RT-PCR normalization in wheat. BMC Mol. Biol. 2009. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, C.; Ridzon, D.A.; Broomer, A.J.; Zhou, Z.; Lee, D.H.; Nguyen, J.T.; Barbisin, M.; Xu, N.L.; Mahuvakar, V.R.; Andersen, M.R.; et al. Real-time quantification of micrornas by stem-loop RT-PCR. Nucleic Acids Res. 2005, 33, e179. [Google Scholar] [CrossRef] [PubMed]
- Varkonyi-Gasic, E.; Wu, R.; Wood, M.; Walton, E.F.; Hellens, R.P. Protocol: A highly sensitive RT-PCR method for detection and quantification of micrornas. Plant Methods 2007. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆Ct method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Sawa, M.; Suetsugu, S.; Sugimoto, A.; Miki, H.; Yamamoto, M.; Takenawa, T. Essential role of the C. elegans arp2/3 complex in cell migration during ventral enclosure. J. Cell Sci. 2003, 116, 1505–1518. [Google Scholar] [CrossRef] [PubMed]
- Berridge, M.J. Inositol trisphosphate and calcium signalling. Nature 1993, 361, 315–325. [Google Scholar] [CrossRef] [PubMed]
- Tiganis, T.; Bennett Anton, M. Protein tyrosine phosphatase function: The substrate perspective. Biochem. J. 2007, 402, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Polet, D.; Lambrechts, A.; Ono, K.; Mah, A.; Peelman, F.; Vandekerckhove, J.; Baillie, D.L.; Ampe, C.; Ono, S. Caenorhabditis elegans expresses three functional profilins in a tissue-specific manner. Cell Motil. Cytoskelet. 2006, 63, 14–28. [Google Scholar] [CrossRef] [PubMed]
- Hernandez-Garcia, C.M.; Martinelli, A.P.; Bouchard, R.A.; Finer, J.J. A soybean (Glycine max) polyubiquitin promoter gives strong constitutive expression in transgenic soybean. Plant Cell Rep. 2009, 28, 837–849. [Google Scholar] [CrossRef] [PubMed]
- Fellmann, C.; Hoffmann, T.; Sridhar, V.; Hopfgartner, B.; Muhar, M.; Roth, M.; Lai Dan, Y.; Barbosa Inês, A.M.; Kwon Jung, S.; Guan, Y.; et al. An optimized microrna backbone for effective single-copy RNAi. Cell Rep. 2013, 5, 1704–1713. [Google Scholar] [CrossRef] [PubMed]
- Fowler, D.K.; Williams, C.; Gerritsen, A.T.; Washbourne, P. Improved knockdown from artificial microRNAs in an enhanced miR-155 backbone: A designer’s guide to potent multi-target RNAi. Nucleic Acids Res. 2016, 44, e48. [Google Scholar] [CrossRef] [PubMed]
- Kereszt, A.; Li, D.; Indrasumunar, A.; Nguyen, C.D.T.; Nontachaiyapoom, S.; Kinkema, M.; Gresshoff, P.M. Agrobacterium rhizogenes-mediated transformation of soybean to study root biology. Nat. Protoc. 2007, 2, 948–952. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Todd, T.C.; Oakley, T.R.; Lee, J.; Trick, H.N. Host-derived suppression of nematode reproductive and fitness genes decreases fecundity of Heterodera glycines ichinohe. Planta 2010, 232, 775–785. [Google Scholar] [CrossRef] [PubMed]
- Bhagwat, B.; Chi, M.; Su, L.; Tang, H.; Tang, G.; Xiang, Y. An in vivo transient expression system can be applied for rapid and effective selection of artificial microRNA constructs for plant stable genetic transformation. J. Genet. Genom. 2013, 40, 261–270. [Google Scholar] [CrossRef] [PubMed]
- Makhzoum, A.B.; Sharma, P.; Bernards, M.A.; Trémouillaux-Guiller, J. Hairy Roots: An Ideal Platform for Transgenic Plant Production and Other Promising Applications. In Phytochemicals, Plant Growth, and the Environment; Gang, D.R., Ed.; Springer: New York, NY, USA, 2013; pp. 95–142. [Google Scholar]
- Liu, S.; Kandoth, P.K.; Warren, S.D.; Yeckel, G.; Heinz, R.; Alden, J.; Yang, C.; Jamai, A.; El-Mellouki, T.; Juvale, P.S.; et al. A soybean cyst nematode resistance gene points to a new mechanism of plant resistance to pathogens. Nature 2012, 492, 256–260. [Google Scholar] [CrossRef] [PubMed]
- Ossowski, S.; Schwab, R.; Weigel, D. Gene silencing in plants using artificial microRNAs and other small rnas. Plant J. 2008, 53, 674–690. [Google Scholar] [CrossRef] [PubMed]
- Ithal, N.; Recknor, J.; Nettleton, D.; Maier, T.; Baum, T.J.; Mitchum, M.G. Developmental transcript profiling of cyst nematode feeding cells in soybean roots. Mol. Plant Microbe Interact. 2007, 20, 510–525. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Pilot, G. Testing the efficiency of plant artificial microRNAs By transient expression in nicotiana benthamiana reveals additional action at the translational level. Front. Plant Sci. 2014. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Chronis, D.; de la Torre, C.M.; Smeda, J.; Wang, X.; Mitchum, M.G. Enhanced resistance to soybean cyst nematode Heterodera glycines in transgenic soybean by silencing putative CLE receptors. Plant Biotechnol. J. 2015, 13, 801–810. [Google Scholar] [CrossRef] [PubMed]
- Sindhu, A.S.; Maier, T.R.; Mitchum, M.G.; Hussey, R.S.; Davis, E.L.; Baum, T.J. Effective and specific in planta RNAi in cyst nematodes: Expression interference of four parasitism genes reduces parasitic success. J. Exp. Bot. 2009, 60, 315–324. [Google Scholar] [CrossRef] [PubMed]
- Dutta, T.K.; Banakar, P.; Rao, U. The status of RNAi-based transgenic research in plant nematology. Front. Microbiol. 2015. [Google Scholar] [CrossRef] [PubMed]
- Steeves, R.M.; Todd, T.C.; Essig, J.S.; Trick, H.N. Transgenic soybeans expressing siRNAs specific to a major sperm protein gene suppress Heterodera glycines reproduction. Funct. Plant Biol. 2006, 33, 991–999. [Google Scholar] [CrossRef]
- Klink, V.P.; Kim, K.-H.; Martins, V.; MacDonald, M.H.; Beard, H.S.; Alkharouf, N.W.; Lee, S.-K.; Park, S.-C.; Matthews, B.F. A correlation between host-mediated expression of parasite genes as tandem inverted repeats and abrogation of development of female Heterodera glycines cyst formation during infection of glycine max. Planta 2009, 230, 53–71. [Google Scholar] [CrossRef] [PubMed]
- Fairbairn, D.J.; Cavallaro, A.S.; Bernard, M.; Mahalinga-Iyer, J.; Graham, M.W.; Botella, J.R. Host-delivered RNAi: An effective strategy to silence genes in plant parasitic nematodes. Planta 2007, 226, 1525–1533. [Google Scholar] [CrossRef] [PubMed]
- Patel, N.; Hamamouch, N.; Li, C.; Hewezi, T.; Hussey, R.S.; Baum, T.J.; Mitchum, M.G.; Davis, E.L. A nematode effector protein similar to annexins in host plants. J. Exp. Bot. 2010, 61, 235–248. [Google Scholar] [CrossRef] [PubMed]
- Williamson, V.M.; Gleason, C.A. Plant–nematode interactions. Curr. Opin. Plant Biol. 2003, 6, 327–333. [Google Scholar] [CrossRef]
- Kyndt, T.; Ji, H.; Vanholme, B.; Gheysen, G. Transcriptional silencing of RNAi constructs against nematode genes in Arabidopsis. Nematology 2013, 15, 519–528. [Google Scholar] [CrossRef]


© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, B.; Li, J.; Oakley, T.R.; Todd, T.C.; Trick, H.N. Host-Derived Artificial MicroRNA as an Alternative Method to Improve Soybean Resistance to Soybean Cyst Nematode. Genes 2016, 7, 122. https://doi.org/10.3390/genes7120122
Tian B, Li J, Oakley TR, Todd TC, Trick HN. Host-Derived Artificial MicroRNA as an Alternative Method to Improve Soybean Resistance to Soybean Cyst Nematode. Genes. 2016; 7(12):122. https://doi.org/10.3390/genes7120122
Chicago/Turabian StyleTian, Bin, Jiarui Li, Thomas R. Oakley, Timothy C. Todd, and Harold N. Trick. 2016. "Host-Derived Artificial MicroRNA as an Alternative Method to Improve Soybean Resistance to Soybean Cyst Nematode" Genes 7, no. 12: 122. https://doi.org/10.3390/genes7120122





