Mutant CAG Repeats Effectively Targeted by RNA Interference in SCA7 Cells
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Oligonucleotides and Transfection
2.3. Reverse Transcription Polymerase Chain Reaction and Quantitative Reverse Transcription Polymerase Chain Reaction
2.4. Western Blot
2.5. Immunofluorescence
2.6. Statistical Analysis
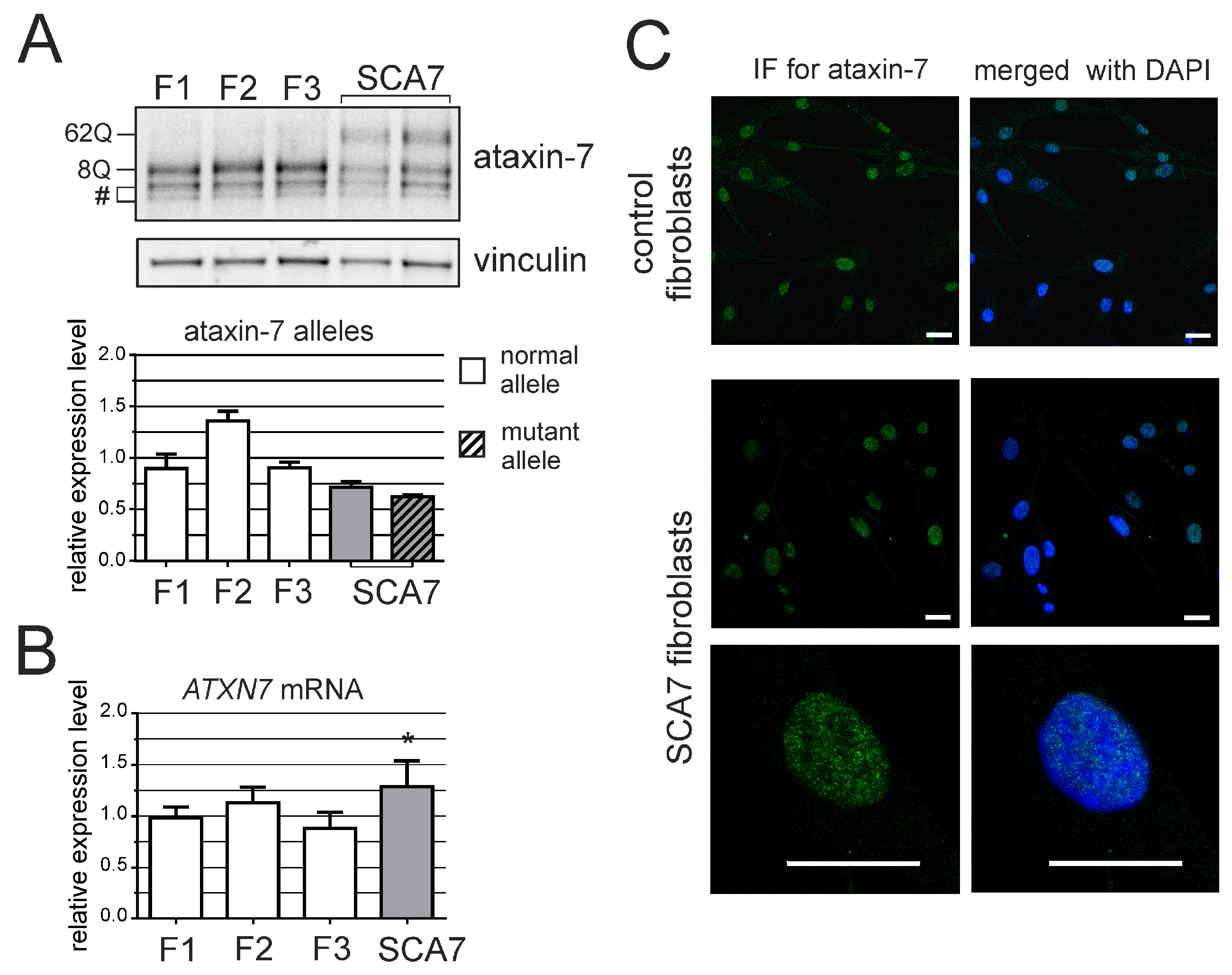
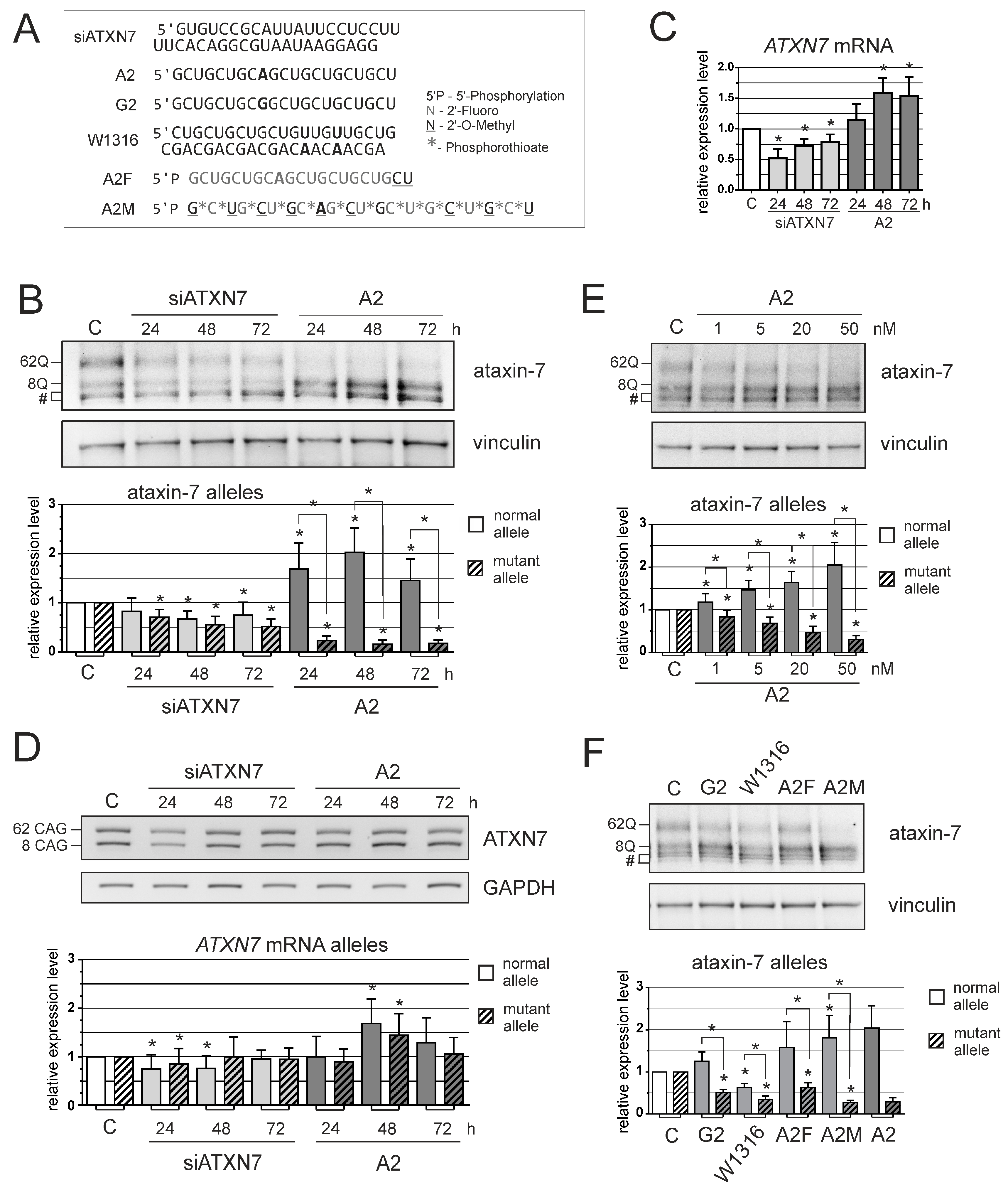
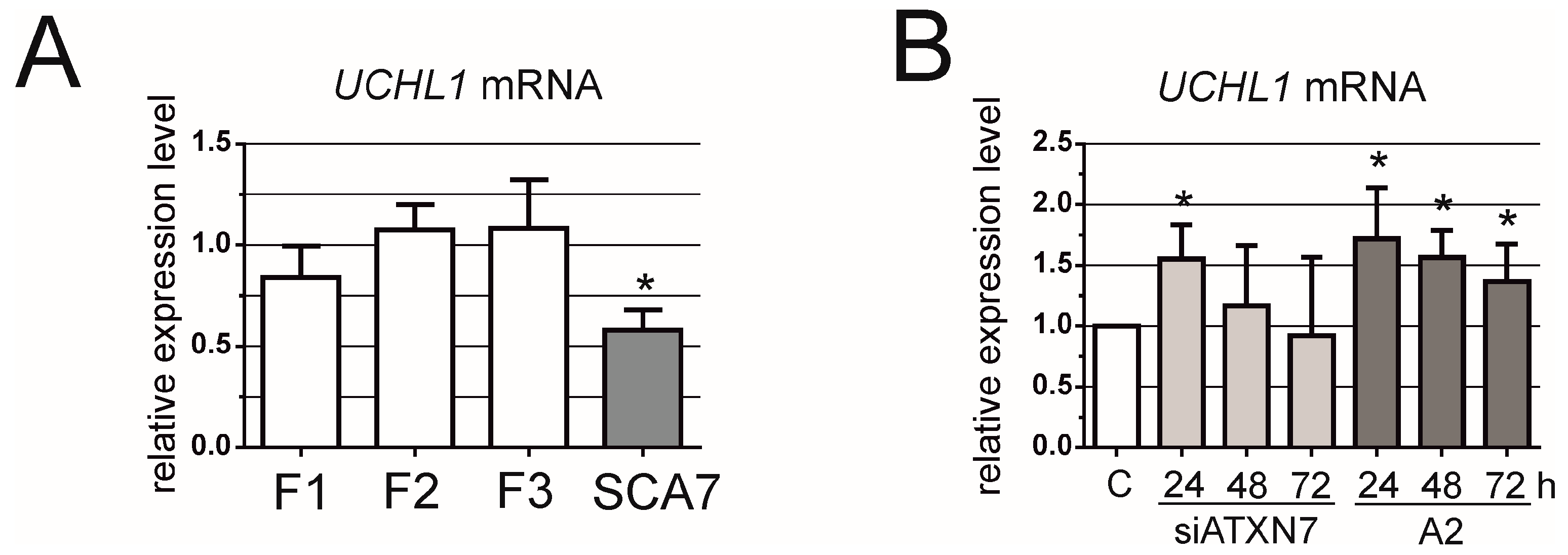
3. Results
4. Discussion
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Fan, H.-C.; Ho, L.-I.; Chi, C.-S.; Chen, S.-J.; Peng, G.-S.; Chan, T.-M.; Lin, S.-Z.; Harn, H.-J. Polyglutamine (PolyQ) Diseases: Genetics to Treatments. Cell Transplant. 2014, 23, 441–458. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, Y.; Tagawa, K.; Oka, T.; Sasabe, T.; Ito, H.; Shiwaku, H.; La Spada, A.R.; Okazawa, H. Ataxin-7 associates with microtubules and stabilizes the cytoskeletal network. Hum. Mol. Genet. 2012, 21, 1099–1110. [Google Scholar] [CrossRef] [PubMed]
- Helmlinger, D.; Hardy, S.; Sasorith, S.; Klein, F.; Robert, F.; Weber, C.; Miguet, L.; Potier, N.; Van-Dorsselaer, A.; Wurtz, J.-M.; et al. Ataxin-7 is a subunit of GCN5 histone acetyltransferase-containing complexes. Hum. Mol. Genet. 2004, 13, 1257–1265. [Google Scholar] [CrossRef] [PubMed]
- Tan, J.Y.; Vance, K.W.; Varela, M.A.; Sirey, T.; Watson, L.M.; Curtis, H.J.; Marinello, M.; Alves, S.; Steinkraus, B.R.; Cooper, S.; et al. Cross-talking noncoding RNAs contribute to cell-specific neurodegeneration in SCA7. Nat. Struct. Mol. Biol. 2014, 21, 955–961. [Google Scholar] [CrossRef] [PubMed]
- Young, J.E.; Gouw, L.; Propp, S.; Sopher, B.L.; Taylor, J.; Lin, A.; Hermel, E.; Logvinova, A.; Chen, S.F.; Chen, S.; et al. Proteolytic cleavage of ataxin-7 by caspase-7 modulates cellular toxicity and transcriptional dysregulation. J. Biol. Chem. 2007, 282, 30150–30160. [Google Scholar] [CrossRef] [PubMed]
- Alves, S.; Cormier-Dequaire, F.; Marinello, M.; Marais, T.; Muriel, M.-P.; Beaumatin, F.; Charbonnier-Beaupel, F.; Tahiri, K.; Seilhean, D.; El Hachimi, K.; et al. The autophagy/lysosome pathway is impaired in SCA7 patients and SCA7 knock-in mice. Acta Neuropathol. 2014, 128, 705–722. [Google Scholar] [CrossRef] [PubMed]
- Alves, S.; Marais, T.; Biferi, M.-G.; Furling, D.; Marinello, M.; El Hachimi, K.; Cartier, N.; Ruberg, M.; Stevanin, G.; Brice, A.; et al. Lentiviral vector-mediated overexpression of mutant ataxin-7 recapitulates SCA7 pathology and promotes accumulation of the FUS/TLS and MBNL1 RNA-binding proteins. Mol. Neurodegener. 2016, 11, 58. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Garden, G.A.; La Spada, A.R. Molecular pathogenesis and cellular pathology of spinocerebellar ataxia type 7 neurodegeneration. Cerebellum 2008, 7, 138–149. [Google Scholar] [CrossRef] [PubMed]
- Rüb, U.; Schöls, L.; Paulson, H.; Auburger, G.; Kermer, P.; Jen, J.C.; Seidel, K.; Korf, H.W.; Deller, T. Clinical features, neurogenetics and neuropathology of the polyglutamine spinocerebellar ataxias type 1, 2, 3, 6 and 7. Prog. Neurobiol. 2013, 104, 38–66. [Google Scholar] [CrossRef] [PubMed]
- Matsui, M.; Corey, D.R. Allele-selective inhibition of trinucleotide repeat genes. Drug Discov. Today 2012, 17, 443–450. [Google Scholar] [CrossRef] [PubMed]
- Watson, L.M.; Wood, M.J. RNA therapy for polyglutamine neurodegenerative diseases. Expert Rev. Mol. Med. 2012, 14, e3. [Google Scholar] [CrossRef] [PubMed]
- Fiszer, A.; Krzyzosiak, W.J. Oligonucleotide-based strategies to combat polyglutamine diseases. Nucleic Acids Res. 2014, 42, 6787–6810. [Google Scholar] [CrossRef] [PubMed]
- Scholefield, J.; Greenberg, L.J.; Weinberg, M.S.; Arbuthnot, P.B.; Abdelgany, A.; Wood, M.J. Design of RNAi hairpins for mutation-specific silencing of ataxin-7 and correction of a SCA7 phenotype. PLoS ONE 2009, 4, e7232. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scholefield, J.; Watson, L.; Smith, D.; Greenberg, J.; Wood, M.J. Allele-specific silencing of mutant Ataxin-7 in SCA7 patient-derived fibroblasts. Eur. J. Hum. Genet. 2014, 22, 1369–1375. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, P.S.; Bhattarai, S.; Singh, P.; Boudreau, R.L.; Thompson, S.; LaSpada, A.R.; Drack, A.V.; Davidson, B.L. RNA Interference-Based Therapy for Spinocerebellar Ataxia Type 7 Retinal Degeneration. PLoS ONE 2014, 9, e95362. [Google Scholar] [CrossRef] [PubMed]
- Ramachandran, P.S.; Boudreau, R.L.; Schaefer, K.A.; La Spada, A.R.; Davidson, B.L. Nonallele specific silencing of ataxin-7 improves disease phenotypes in a mouse model of SCA7. Mol. Ther. 2014, 22, 1635–1642. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Liu, J.; Corey, D.R. Allele-Selective Inhibition of Huntingtin Expression by Switching to an miRNA-like RNAi Mechanism. Chem. Biol. 2010, 17, 1183–1188. [Google Scholar] [CrossRef] [PubMed]
- Fiszer, A.; Mykowska, A.; Krzyzosiak, W.J. Inhibition of mutant huntingtin expression by RNA duplex targeting expanded CAG repeats. Nucleic Acids Res. 2011, 39, 5578–5585. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Gagnon, K.T.; Liu, J.; Watts, J.K.; Syeda-Nawaz, J.; Bennett, C.F.; Swayze, E.E.; Randolph, J.; Chattopadhyaya, J.; Corey, D.R. Allele-selective inhibition of ataxin-3 (ATX3) expression by antisense oligomers and duplex RNAs. Biol. Chem. 2011, 392, 315–325. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Pendergraff, H.; Narayanannair, K.J.; Lackey, J.G.; Kuchimanchi, S.; Rajeev, K.G.; Manoharan, M.; Hu, J.; Corey, D.R. RNA duplexes with abasic substitutions are potent and allele-selective inhibitors of huntingtin and ataxin-3 expression. Nucleic Acids Res. 2013, 41, 8788–8801. [Google Scholar] [CrossRef] [PubMed]
- Fiszer, A.; Olejniczak, M.; Galka-Marciniak, P.; Mykowska, A.; Krzyzosiak, W.J. Self-duplexing CUG repeats selectively inhibit mutant huntingtin expression. Nucleic Acids Res. 2013, 41, 10426–10437. [Google Scholar] [CrossRef] [PubMed]
- Aiba, Y.; Hu, J.; Liu, J.; Xiang, Q.; Martinez, C.; Corey, D.R. Allele-Selective Inhibition of Huntingtin and Ataxin-3 Expression by RNA Duplexes Containing Unlocked Nucleic Acid (UNA) Substitutions. Biochemistry 2013, 52, 9329–9338. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Yu, D.; Aiba, Y.; Pendergraff, H.; Swayze, E.E.; Lima, W.F.; Hu, J.; Prakash, T.P.; Corey, D.R. ss-siRNAs allele selectively inhibit ataxin-3 expression: Multiple mechanisms for an alternative gene silencing strategy. Nucleic Acids Res. 2013, 41, 9570–9583. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Liu, J.; Narayanannair, K.J.; Lackey, J.G.; Kuchimanchi, S.; Rajeev, K.G.; Manoharan, M.; Swayze, E.E.; Lima, W.F.; Prakash, T.P.; et al. Allele-selective inhibition of mutant Atrophin-1 expression by duplex and single-stranded RNAs. Biochemistry 2014, 53, 4510–4518. [Google Scholar] [CrossRef] [PubMed]
- Hu, J.; Liu, J.; Yu, D.; Chu, Y.; Corey, D.R. Mechanism of allele-selective inhibition of huntingtin expression by duplex RNAs that target CAG repeats: Function through the RNAi pathway. Nucleic Acids Res. 2012, 40, 11270–11280. [Google Scholar] [CrossRef] [PubMed]
- Fiszer, A.; Ellison-Klimontowicz, M.E.; Krzyzosiak, W.J. Silencing of genes responsible for polyQ diseases using chemically modified single-stranded siRNAs. Acta Biochim. Pol. 2016, in press. [Google Scholar] [CrossRef] [PubMed]
- McClorey, G.; Wood, M.J. An overview of the clinical application of antisense oligonucleotides for RNA-targeting therapies. Curr. Opin. Pharmacol. 2015, 24, 52–58. [Google Scholar] [CrossRef] [PubMed]
- Jinek, M.; Chylinski, K.; Fonfara, I.; Hauer, M.; Doudna, J.A.; Charpentier, E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012, 337, 816–821. [Google Scholar] [CrossRef] [PubMed]
- Maeder, M.L.; Gersbach, C.A. Genome-editing Technologies for Gene and Cell Therapy. Mol. Ther. 2016, 24, 430–446. [Google Scholar] [CrossRef] [PubMed]
- Furrer, S.A.; Waldherr, S.M.; Mohanachandran, M.S.; Baughn, T.D.; Nguyen, K.-T.; Sopher, B.L.; Damian, V.A.; Garden, G.A.; La Spada, A.R. Reduction of mutant ataxin-7 expression restores motor function and prevents cerebellar synaptic reorganization in a conditional mouse model of SCA7. Hum. Mol. Genet. 2013, 22, 890–903. [Google Scholar] [CrossRef] [PubMed]
| Name | Sequence 5′-3′ |
|---|---|
| GAPDH F | GAAGGTGAAGGTCGGAGTC |
| GAPDH R | GAAGATGGTGATGGGATTTC |
| ATXN7 F | AGGTGTTCTTAGCGCATCCT |
| ATXN7 R | AGTGTGCCATCCATTTTCGG |
| ATXN7* F | ACCCTCCAAAGAAAAGGAGCG |
| ATXN7* R | AGCATCACTTCAGGACTGGG |
| UCHL1 F | GGAAGGCCAATGTCGGGTAG |
| UCHL1 R | GCAGGGTGTCCTCTGAACTG |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fiszer, A.; Wroblewska, J.P.; Nowak, B.M.; Krzyzosiak, W.J. Mutant CAG Repeats Effectively Targeted by RNA Interference in SCA7 Cells. Genes 2016, 7, 132. https://doi.org/10.3390/genes7120132
Fiszer A, Wroblewska JP, Nowak BM, Krzyzosiak WJ. Mutant CAG Repeats Effectively Targeted by RNA Interference in SCA7 Cells. Genes. 2016; 7(12):132. https://doi.org/10.3390/genes7120132
Chicago/Turabian StyleFiszer, Agnieszka, Joanna P. Wroblewska, Bartosz M. Nowak, and Wlodzimierz J. Krzyzosiak. 2016. "Mutant CAG Repeats Effectively Targeted by RNA Interference in SCA7 Cells" Genes 7, no. 12: 132. https://doi.org/10.3390/genes7120132





