New Insights into Phasmatodea Chromosomes
Abstract
:1. Introduction
2. Material and Methods
2.1. Stick Insects Studied
2.2. Chromosome Preparation
2.3. C-Banding
2.4. DNA Probe Preparation
2.5. Fluorescence In Situ Hybridization
2.6. Microscopy
3. Results
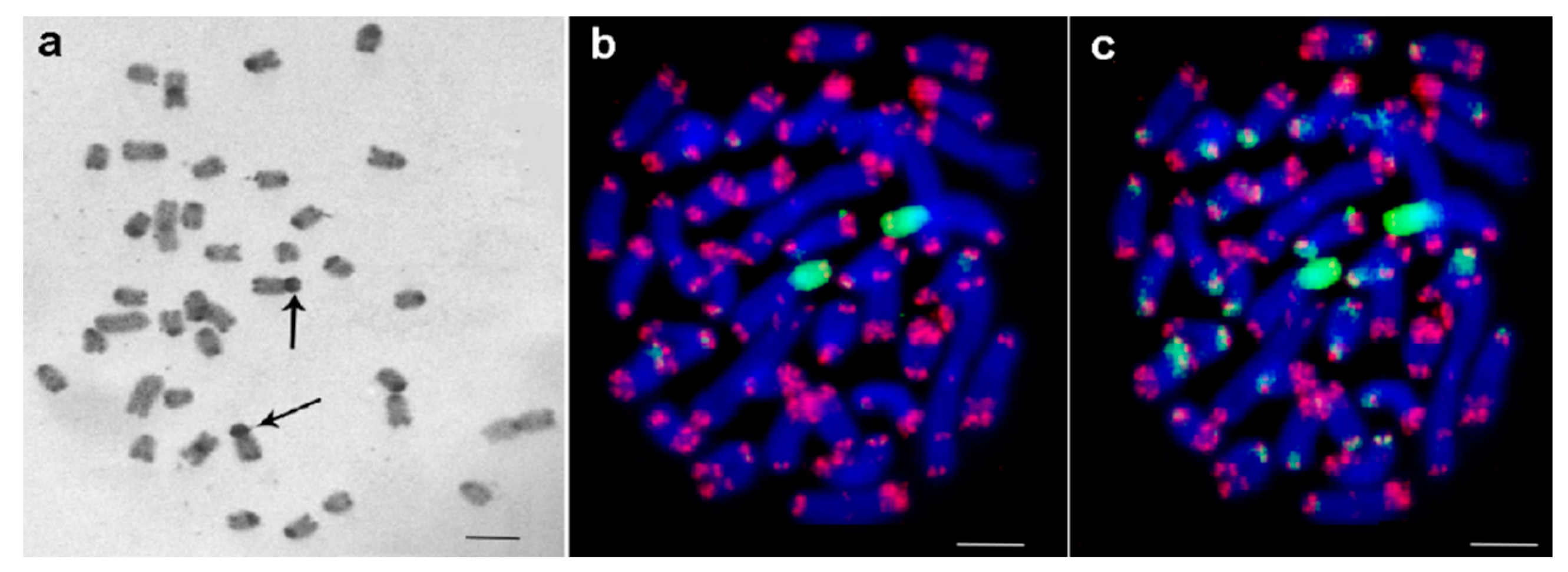
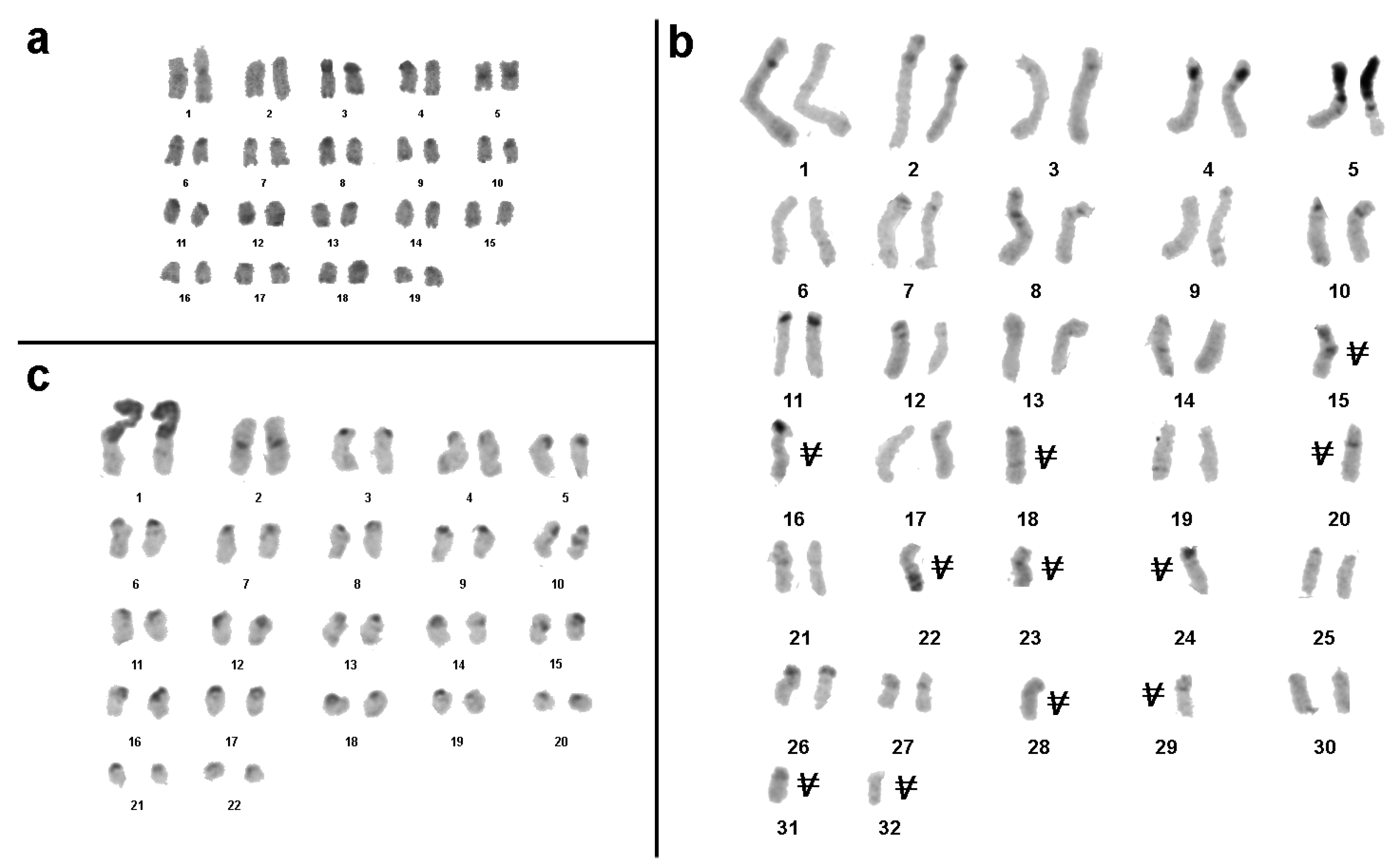
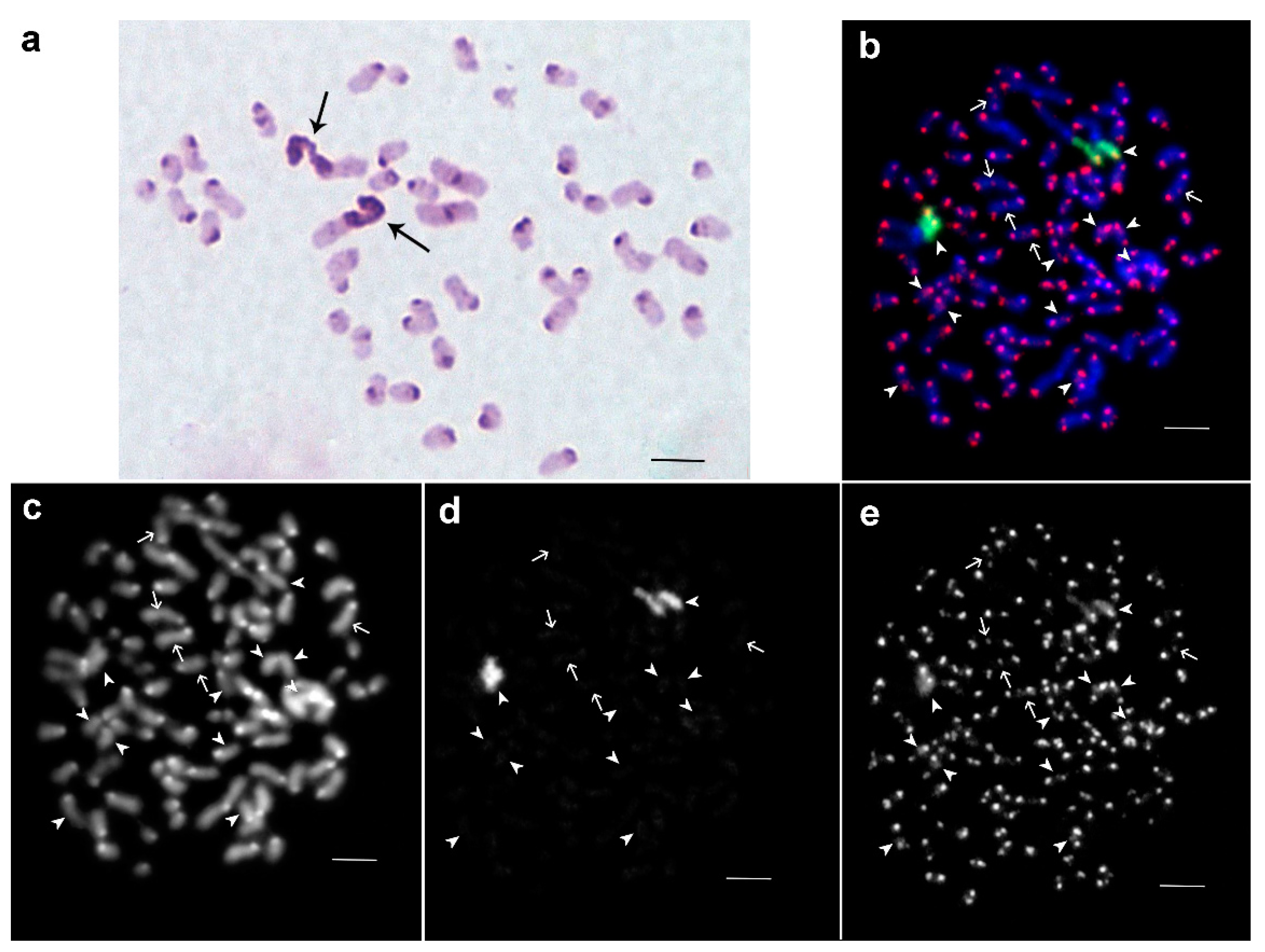
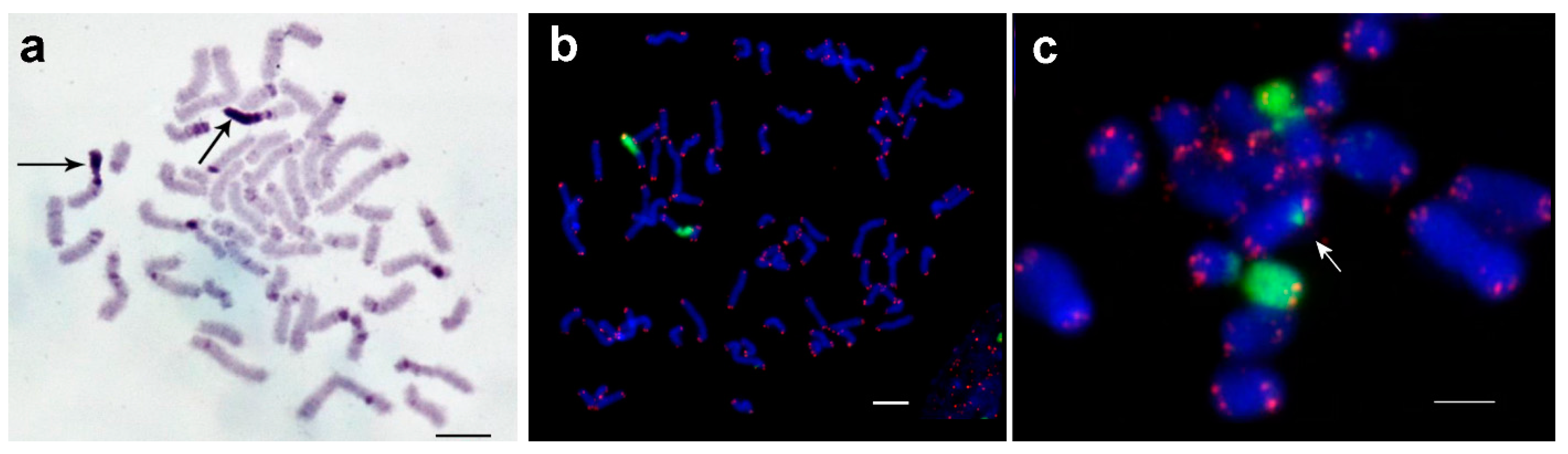
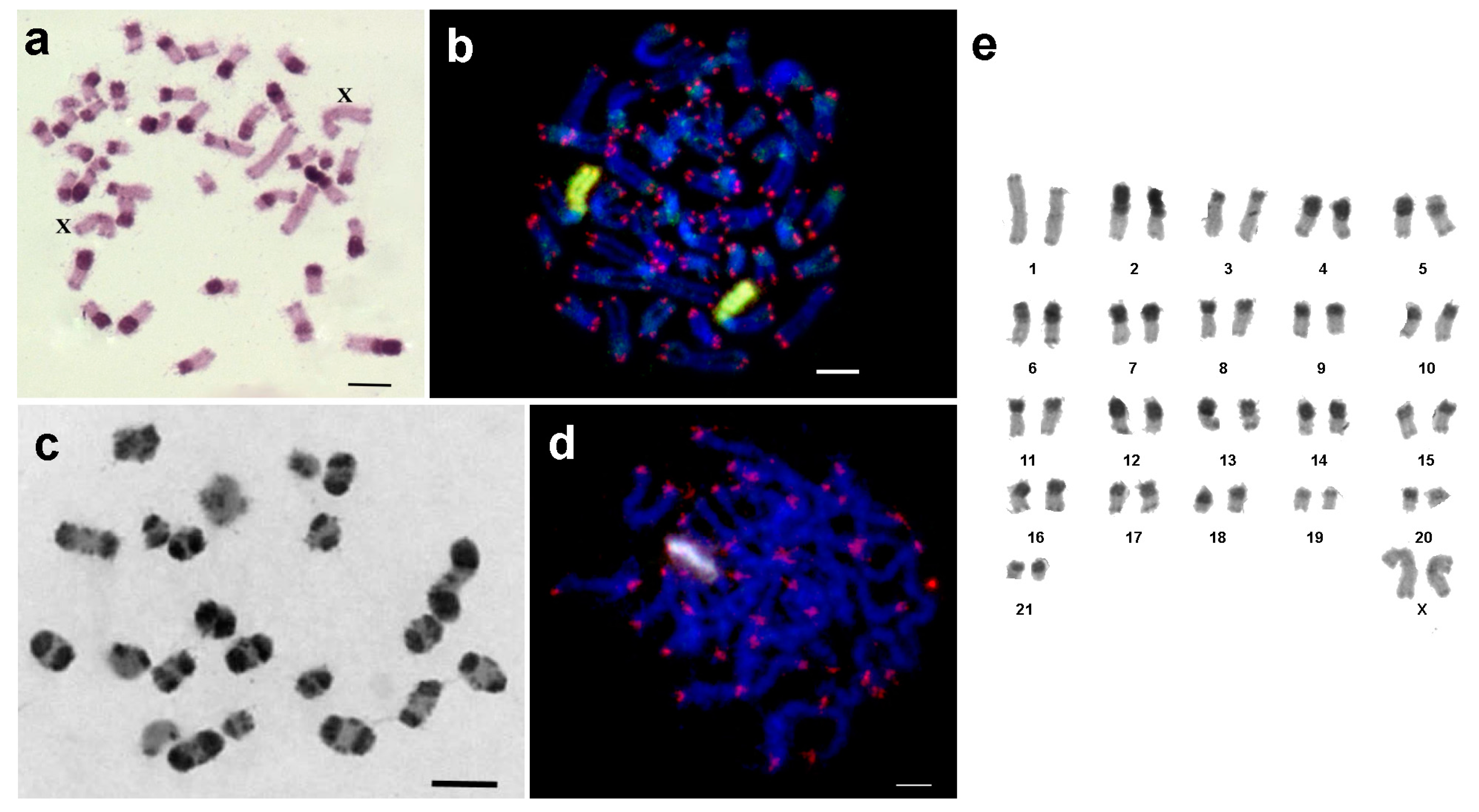
3.1. Medauroidea extradentata
3.2. Sungaya inexpectata
3.3. Sipyloidea sipylus
3.4. Phaenopharos khaoyaiensis
3.5. Peruphasma schultei
4. Discussion
4.1. Does Parthenogenetic Reproduction Lead to New Lineages Characterized by Chromosomal Evolution?
4.2. Does the Diversity of C-Banding Patterns in Different Taxa Indicate Intense Amplification and/or Transposition of Mobile Elements as Well as Different Types of Repeats during Chromosomal Evolution?
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Blackmon, H.; Ross, L.; Bachtrog, D. Sex determination, sex chromosomes, and karyotype evolution in insects. J. Hered. 2017, 108, 78–93. [Google Scholar] [CrossRef] [PubMed]
- Conle, O.; Hennemann, F. Studies on neotropical Phasmatodea 1: A remarkable new species of Peruphasma Conle & Hennemann, 2002 from Northern Peru (Phasmatodea: Pseudophasmatidae: Pseudophasmatinae). Zootaxa 2005, 1068, 59–68. [Google Scholar]
- Von Wattenwyl, K.B. Die Insektenfamilie der Phasmiden, II. Phasmidae Anareolatae; Wilhelm Engelmann: Leipzig, Germany, 1907. [Google Scholar]
- Wodarz, D. Effect of stem cell turnover rates on protection against cancer and aging. J. Theor. Biol. 2007, 245, 449–458. [Google Scholar] [CrossRef] [PubMed]
- Gregory, T.R. Animal Genome Size Database. Available online: http://www.genomesize.com (accessed on 20 August 2009).
- Husemann, M.; Namkung, S.; Habel, J.C.; Danyel, P.D.; Hochkirch, A.; Hochkirch, A. Phylogenetic analyses of band-winged grasshoppers (Orthoptera, Acrididae, Oedipodinae) reveal convergence of wing morphology. Zool. Scr. 2012, 41, 515–526. [Google Scholar] [CrossRef]
- Misof, B.; Liu, S.; Meusemann, K.; Peters, R.S.; Donath, A.; Mayer, C.; Frandsen, P.B.; Ware, J.; Flouri, T.; Beutel, R.G.; et al. Phylogenomics resolves the timing and pattern of insect evolution. Science 2014, 346, 763–767. [Google Scholar] [CrossRef] [PubMed]
- Milani, L.; Scali, V.; Passamonti, M. The Clonopsis gallica puzzle: Mendelian species, polyploid parthenogens with karyotype re-diploidization and clonal androgens in Moroccan stick insects (Phasmida). J. Zool. Syst. Evol. Res. 2009, 47, 132–140. [Google Scholar] [CrossRef]
- Milani, L.; Scali, V.; Passamonti, M. Speciation through androgenesis in the stick insect genus Clonopsis (Insecta Phasmatodea). J. Zool. Syst. Evol. Res. 2015, 53, 116–123. [Google Scholar] [CrossRef]
- Bergerard, J. Etude de la Parthénogenèse Facultative de Clitumnus Extradentatus Br. (Phasmidae); Bulletin Biologique de la France et de la Belgique: Paris, France, 1958; Volume 92, pp. 7–182. [Google Scholar]
- Samtschrecke. Available online: http://tierdoku.com/index.php?title=Samtschrecke (accessed on 20 August 2009).
- Scali, V.; Milani, L.; Passamonti, M. Clonopsis gallica, a stick insect parthenogen exploiting different egg maturation mechanisms over its range. Invertebr. Reprod. Dev. 2010, 54, 143–150. [Google Scholar] [CrossRef]
- John, B.; Rentz, D.C.F.; Contreras, N. Extensive chromosome variation in the stick insect genus Sipyloidea Brunner von Wattenwil (Phyllidae: Necrosciinae) within Australia, and descriptions of three new species. Invertebr. Taxon. 1987, 1, 603–630. [Google Scholar]
- Scali, V. Metasexual Stick Insects: Model Pathways to Losing Sex and Bringing It Back. In Lost Sex; Schoen, I., Martens, K., Dijk, P.V., Eds.; Springer: Dordrecht, The Netherlands, 2009; pp. 317–345. [Google Scholar]
- White, M.D.J. Insect 2. In Animal Cytogenetics; John, B., Ed.; Gebrüder Bornträger: Berlin, Germany, 1976; pp. 1–75. [Google Scholar]
- Tinti, F.; Scali, V. C-banding, Ag-NOR localization and chromosomal repatterning in Sardinian Bacillus atticus (Insecta Phasmatodea). Boll. Zool. 1991, 58, 235–243. [Google Scholar] [CrossRef]
- Manaresi, S.M.; Marescalchi, O.; Scali, V. The chromosome complement of the hybrid Bacillus whitei complex (Insecta Phasmatodea) I. The paleo- and neo-standard karyotypes. Cytologia 1992, 57, 101–109. [Google Scholar] [CrossRef]
- Manaresi, S.M.; Marescalchi, O.; Scali, V. The chromosome complement of the hybrid Bacillus whitei complex (Insecta Phasmatodea). II. The repatterned cytotypes. Cytologia 1992, 57, 111–119. [Google Scholar] [CrossRef]
- Ghiselli, F.; Milani, L.; Scali, V.; Passamonti, M. The Leptynia hispanica species complex (Insecta Phasmida): Polyploidy, parthenogenesis, hybridization and more. Mol. Ecol. 2007, 16, 4256–4268. [Google Scholar] [CrossRef] [PubMed]
- Scali, V.; Milani, L.; Passamonti, M. Revision of the stick insect genus Leptynia: Description of new taxa, speciation mechanism and phylogeography. Contrib. Zool. 2012, 81, 25–42. [Google Scholar]
- Scali, V. Descrizione di due specie incipienti di insetti stecco (Phasmatodea) del complesso Leptynia attenuata Pantel: L. montana n. sp. e L. caprai n. sp. Redia 1996, 79, 123–136. [Google Scholar]
- Scali, V.; Coluccia, E.; Deidda, F.; Lobina, C.; Deiana, A.M.; Salvadori, S. Co-localization of ribosomal and telomeric sequences in Leptynia (Insecta: Phasmatodea). Ital. J. Zool. 2016, 83, 285–290. [Google Scholar] [CrossRef]
- Manaresi, S.; Marescalchi, O.; Scali, V. Ag-detected NOR and C-banding patterns in Bacillus rossius (Insecta Phasmatodea) from Sicily. Caryologia 1991, 44, 265–286. [Google Scholar] [CrossRef]
- Marescalchi, O.; Scali, V. Karyotypes and Ag-NORs of five Heteronemiidae (Insecta Phasmatodea) from Somalia. Boll. Zool. 1993, 60, 53–61. [Google Scholar] [CrossRef]
- Makino, S. An Atlas of the Chromosome Number in Animals; Iowa State Collq. Press: Ames, IA, USA, 1951; Volume 1, pp. 113–219. [Google Scholar]
- Zompro, O. Zur Entdeckung von Sungaya inexpectata. Arthropoda 2008, 16, 41. [Google Scholar]
- Westwood, J.O. Catalogue of the Orthopterous Insects in the Collection of the British Museum. Part I. Phasmidae; Printer by order of the Trustees: London, UK, 1859; Volume 138. [Google Scholar]
- Pijnacker, L.P. Oogenesis in the parthenogenetic stick insect Sipyloidea sipylus Westwood (Orthoptera, Phasmidae). Genetica 1967, 38, 504–515. [Google Scholar]
- More, E. Parthenogenesis explained. Phasmid Stud. 1996, 5, 62–69. [Google Scholar]
- Zompro, O. Neue Stabschrecken aus Thailand (Insecta: Phasmatodea). TenDenZen Suppl. 2000, 1999, 49–60. [Google Scholar]
- Bugrov, A.G.; Warchalowska-Sliwa, E.; Ito, G.; Akimoto, S. C-banded karyotypes of some Podismini grasshoppers (Orthoptera, Acrididae) from Japan. Cytologia 2000, 65, 351–358. [Google Scholar] [CrossRef]
- Bugrov, A.G.; Karamysheva, T.V.; Rubtsov, D.N.; Andreenkova, O.V.; Rubtsov, N.B. Comparative FISH analysis of distribution of B chromosome repetitive DNA in A and B chromosomes in two subspecies of Podisma sapporensis (Orthoptera, Acrididae). Cytogenet. Genome Res. 2004, 106, 284–288. [Google Scholar] [CrossRef] [PubMed]
- Sumner, A.T. A simple technique for demonstrating centromeric heterochromatin. Exp. Cell Res. 1972, 75, 304–306. [Google Scholar] [CrossRef]
- Ijdo, J.W.; Wells, R.A.; Baldini, A.; Reeders, S.T. Improved telomere detection using a telomere repeat probe (TTAGGG)n generated by PCR. Nucleic Acids Res. 1991, 19, 4780. [Google Scholar] [CrossRef] [PubMed]
- Malygin, A.A.; Graifer, D.M.; Zenkova, M.A.; Mamaev, S.V.; Karpova, G.G. Affinity modification of 80S ribosomes from human placenta by derivatives of tri- and hexauridylates as mRNA analogs. Mol. Biol. 1992, 26, 142–149. [Google Scholar]
- Rubtsov, N.B.; Karamysheva, T.V.; Andreenkova, O.V.; Bochkaerev, M.N.; Kartavtseva, I.V.; Roslik, G.V.; Borissov, Y.M. Comparative analysis of micro and macro B chromosomes in the Korean field mouse Apodemus peninsulae (Rodentia, Murinae) performed by chromosome microdissection and FISH. Cytogenet. Genome Res. 2004, 106, 289–294. [Google Scholar] [CrossRef] [PubMed]
- Zadesenets, K.S.; Vizoso, D.B.; Schlatter, A.; Konopatskaia, I.D.; Berezikov, E.; Schaerer, L.; Rubtsov, N.B. Evidence for karyotype polymorphism in the free-living flatworm, Macrostomum lignano, a model organism for evolutionary and developmental biology. PLoS ONE 2016, 11, e0164915. [Google Scholar] [CrossRef] [PubMed]
- Zadesenets, K.S.; Ershov, N.I.; Berezikov, E.; Rubtsov, N.B. Chromosome evolution in the free-living flatworms: First evidence of intrachromosomal rearrangements in karyotype evolution of Macrostomum lignano (Platyhelminthes, Turbellaria). Genes 2017, 8, 298. [Google Scholar] [CrossRef] [PubMed]
- Zadesenets, K.S.; Schaerer, L.; Rubtsov, N.B. New insights into the karyotype evolution of the free-living flatworm Macrostomum lignano (Platyhelminthes, Turbellaria). Sci. Rep. 2017, 7, 6066. [Google Scholar] [CrossRef] [PubMed]
- Yano, C.F.; Bertollo, L.A.; Rebordinos, L.; Merlo, M.A.; Liehr, T.; Portela-Bens, S.; Cioffi, M.B. Evolutionary dynamics of rDNAs and U2 small nuclear DNAs in Triportheus (Characiformes, Triportheidae): High variability and particular syntenic organization. Zebrafish 2017, 14, 146–154. [Google Scholar] [CrossRef] [PubMed]
- Palacios-Gimenez, O.M.; Carvalho, C.R.; Ferrari Soares, F.A.; Cabral-de-Mello, D.C. Contrasting the chromosomal organization of repetitive DNAs in two Gryllidae Crickets with highly divergent karyotypes. PLoS ONE 2015, 10, e0143540. [Google Scholar] [CrossRef] [PubMed]
- Biltueva, L.S.; Prokopov, D.Y.; Makunin, A.I.; Komissarov, A.S.; Kudryavtseva, A.V.; Lemskaya, N.A.; Vorobieva, N.V.; Serdyukova, N.A.; Romanenko, S.A.; Gladkikh, O.L.; et al. Genomic organization and physical mapping of tandemly arranged repetitive DNAs in sterlet (Acipenser ruthenus). Cytogenet. Genome Res. 2017, 152, 148–157. [Google Scholar] [CrossRef] [PubMed]
- Palomeque, T.; Lorite, P. Satellite DNA in insects: A review. Heredity 2008, 100, 564–573. [Google Scholar] [CrossRef] [PubMed]
- Liehr, T. Benign & Pathological Chromosomal Imbalances, Microscopic and Submicroscopic Copy Number Variations (CNVs) in Genetics and Counseling, 1st ed.; Academic Press: New York, NY, USA, 2014. [Google Scholar]
- Bugrov, A.G.; Karamysheva, T.V.; Perepelov, E.A.; Elisaphenko, E.A.; Rubtsov, D.N.; Warchałowska-Sliwa, E.; Tatsuta, H.; Rubtsov, N.B. DNA content of the B chromosomes in grasshopper Podisma kanoi Storozh. (Orthoptera, Acrididae). Chromosome Res. 2007, 15, 315–325. [Google Scholar] [CrossRef] [PubMed]
- Zhdanova, N.S.; Karamisheva, T.V.; Minina, J.; Astakhova, N.M.; Lansdorp, P.; Kammori, M.; Rubtsov, N.B.; Searle, J.B. Unusual distribution pattern of telomeric repeats in the shrews Sorex araneus and Sorex granarius. Chromosome Res. 2005, 13, 617–625. [Google Scholar] [CrossRef] [PubMed]
- Zhdanova, N.S.; Minina, J.M.; Karamisheva, T.V.; Draskovic, I.; Rubtsov, N.B.; Londoño-Vallejo, J.A. The very long telomeres in Sorex granarius (Soricidae, Eulipothyphla) contain ribosomal DNA. Chromosome Res. 2007, 15, 881–890. [Google Scholar] [CrossRef] [PubMed]
- Zhdanova, N.S.; Draskovic, I.; Minina, J.M.; Karamysheva, T.V.; Novo, C.L.; Liu, W.-Y.; Porreca, R.M.; Gibaud, A.; Zvereva, M.E.; Skvortsov, D.A.; et al. Recombinogenic telomeres in diploid Sorex granarius (Soricidae, Eulipotyphla) fibroblast cells. Mol. Cell. Biol. 2014, 34, 2786–2799. [Google Scholar] [CrossRef] [PubMed]
- Zhdanova, N.S.; Minina, Y.; Karamisheva, T.V.; Rubtsov, N.B. Distribution of telomeric and ribosomal DNA on the chromosomes of two related species, Sorex araneus and Sorex granarius (Soricidae, Eulipotiphla). Russ. J. Teriol. 2007, 6, 7–13. [Google Scholar] [CrossRef]
- Shimizu, N. Extrachromosomal double minutes and chromosomal homogeneously staining regions as probes for chromosome research. Cytogenet. Genome Res. 2009, 124, 312–326. [Google Scholar] [CrossRef] [PubMed]
- Bolzán, A.D. Interstitial telomeric sequences in vertebrate chromosomes: Origin, function, instability and evolution. Mutat. Res. 2017, 773, 51–65. [Google Scholar] [CrossRef] [PubMed]

| Species | Embryos Prepared | Metaphases Obtained | Metaphases per Embryo |
|---|---|---|---|
| Medauroidea extradentata | 28 | 112 | 4.0 |
| Sungaya inexpectata | 68 | 107 | 1.7 |
| Sipyloidea sipylus | 15 | 28 | 1.9 |
| Phaenopharos khaoyaiensis | 31 | 59 | 1.9 |
| Peruphasma schultei | 10 | 22 | 2.2 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liehr, T.; Buleu, O.; Karamysheva, T.; Bugrov, A.; Rubtsov, N. New Insights into Phasmatodea Chromosomes. Genes 2017, 8, 327. https://doi.org/10.3390/genes8110327
Liehr T, Buleu O, Karamysheva T, Bugrov A, Rubtsov N. New Insights into Phasmatodea Chromosomes. Genes. 2017; 8(11):327. https://doi.org/10.3390/genes8110327
Chicago/Turabian StyleLiehr, Thomas, Olesya Buleu, Tatyana Karamysheva, Alexander Bugrov, and Nikolai Rubtsov. 2017. "New Insights into Phasmatodea Chromosomes" Genes 8, no. 11: 327. https://doi.org/10.3390/genes8110327
APA StyleLiehr, T., Buleu, O., Karamysheva, T., Bugrov, A., & Rubtsov, N. (2017). New Insights into Phasmatodea Chromosomes. Genes, 8(11), 327. https://doi.org/10.3390/genes8110327






